NEK6 Regulates Redox Balance and DNA Damage Response in DU-145 Prostate Cancer Cells
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture
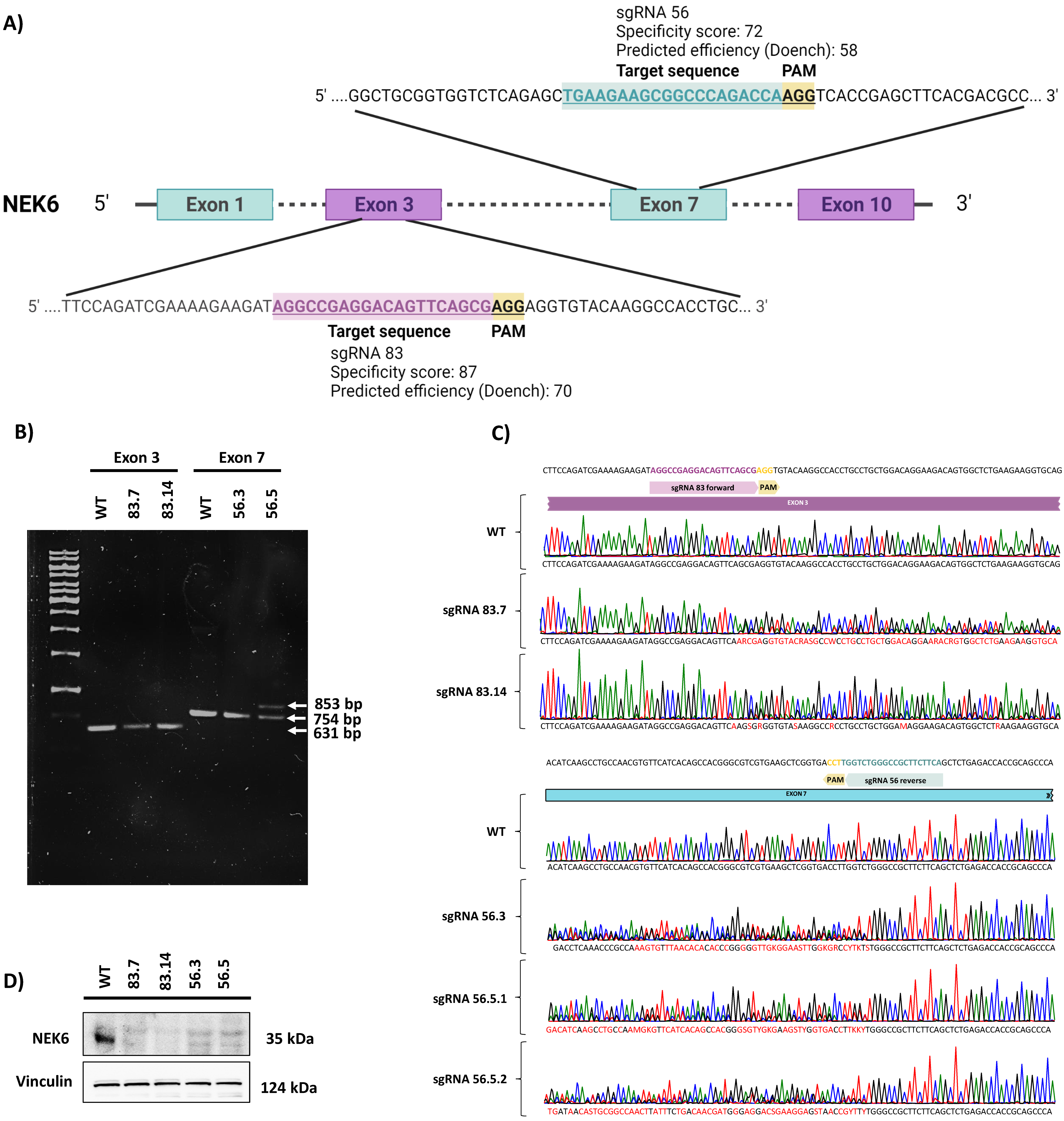
2.2. NEK6 Knockout (NEK6-KO) Generation Using CRISPR-Cas9 System
2.3. DNA Genomic Extraction, Conventional PCR and RT-qPCR
- NEK6 genomic 83fwdF 5′ CAGCAGAGTCCCTCCTTCACCTTAGAG 3′;
- NEK6 genomic 83fwdR 5′ GATGGGTGGACATGGTATGAACCTCAG 3′;
- NEK6 genomic 56revF 5′ ACGTAGGCTGCTTCATGGAC 3′;
- NEK6 genomic 56revR 5′ GCCACAGCTGATTCCCTTCT 3′.
- SOD1_Forward: 5′ GTTTCCGTTGCAGTCCTCG 3′;
- SOD1_Reverse: 5′ GGTCCATTACTTTCCTTCTGCTC 3′;
- SOD2_Forward: 5′AAGGAACGGGGACACTTACAAA 3′;
- SOD2_Reverse: 5′AGCAGTGGAATAAGGCCTGTTG 3′;
- PRDX3_Forward: 5′ GCCACATGAACATCGCACTCTTG 3′;
- PRDX3_Reverse: 5′ ACTGGGAGATCGTTGACGCTCA 3′;
- β-actin_Forward: 5′ GCCGCCAGCTCACCAT 3′;
- β-actin_Reverse: 5′ CCACGATGGAGGGGAAGAC 3′.
2.4. Cell Treatment
2.5. Colony Formation Assay
2.6. Measurement of Mitochondrial Membrane Potential (ΔΨm)
2.7. Proliferation and MTT Assay
2.8. ROS Detection
2.9. NEK6 Overexpression
2.10. Apoptosis Assay
2.11. Subcellular Fractionation
2.12. Western Blotting
2.13. Immunofluorescence Assay
2.14. Statistical and Biostatistical Analysis
3. Results
3.1. Generation of NEK6-KO in DU-145 Cells Using the CRISPR-Cas9 Gene-Editing System
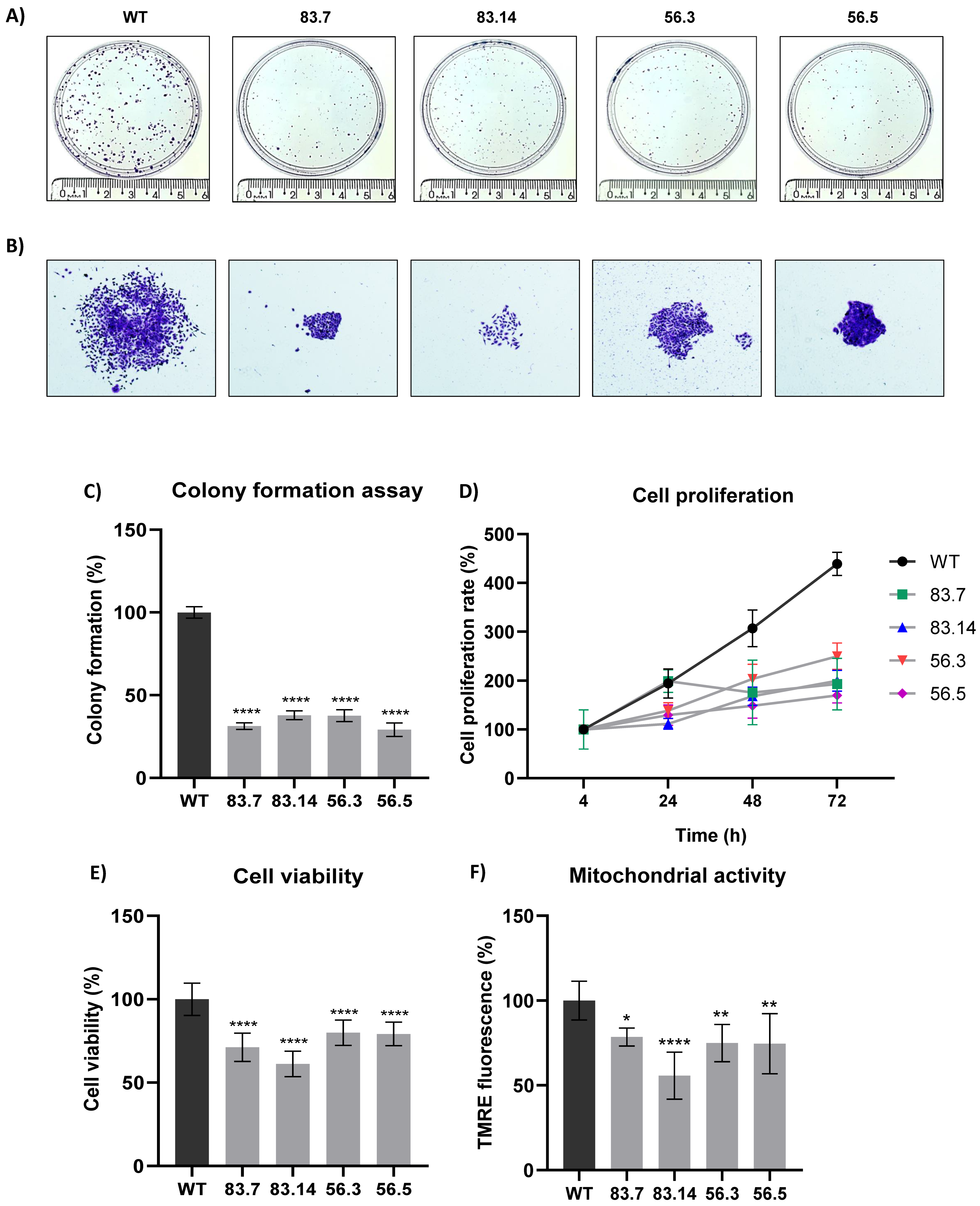
3.2. Targeted Deletion of NEK6 in DU-145 Cells Reduces Clonogenic Capacity, Cell Proliferation, and Mitochondrial Membrane Potential
3.3. Modulation in NEK6 Expression Alters ROS Levels and Antioxidant Defenses in DU-145 Cells
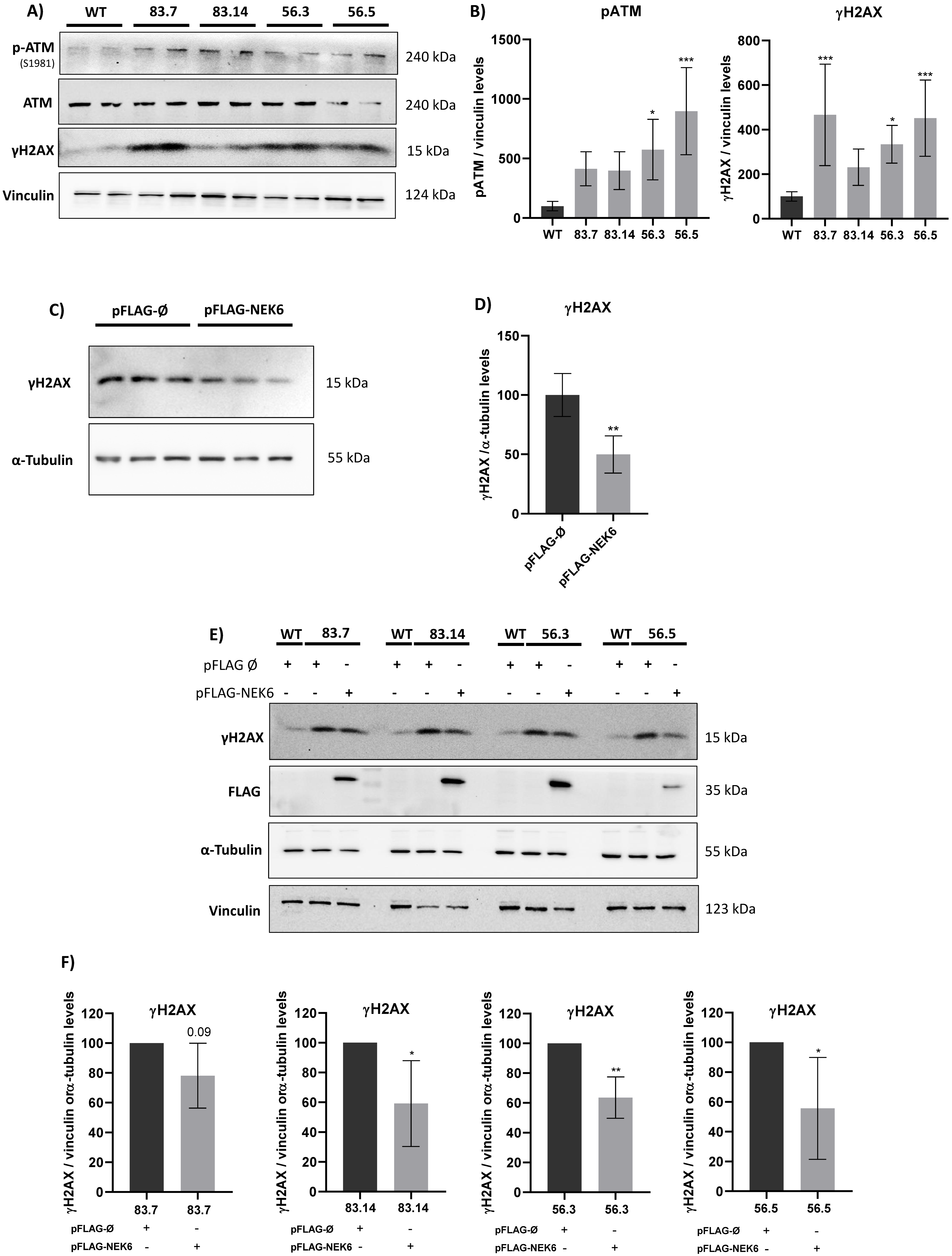
3.4. Targeted Deletion of NEK6 Increases DNA Damage Markers in DU-145 Cells
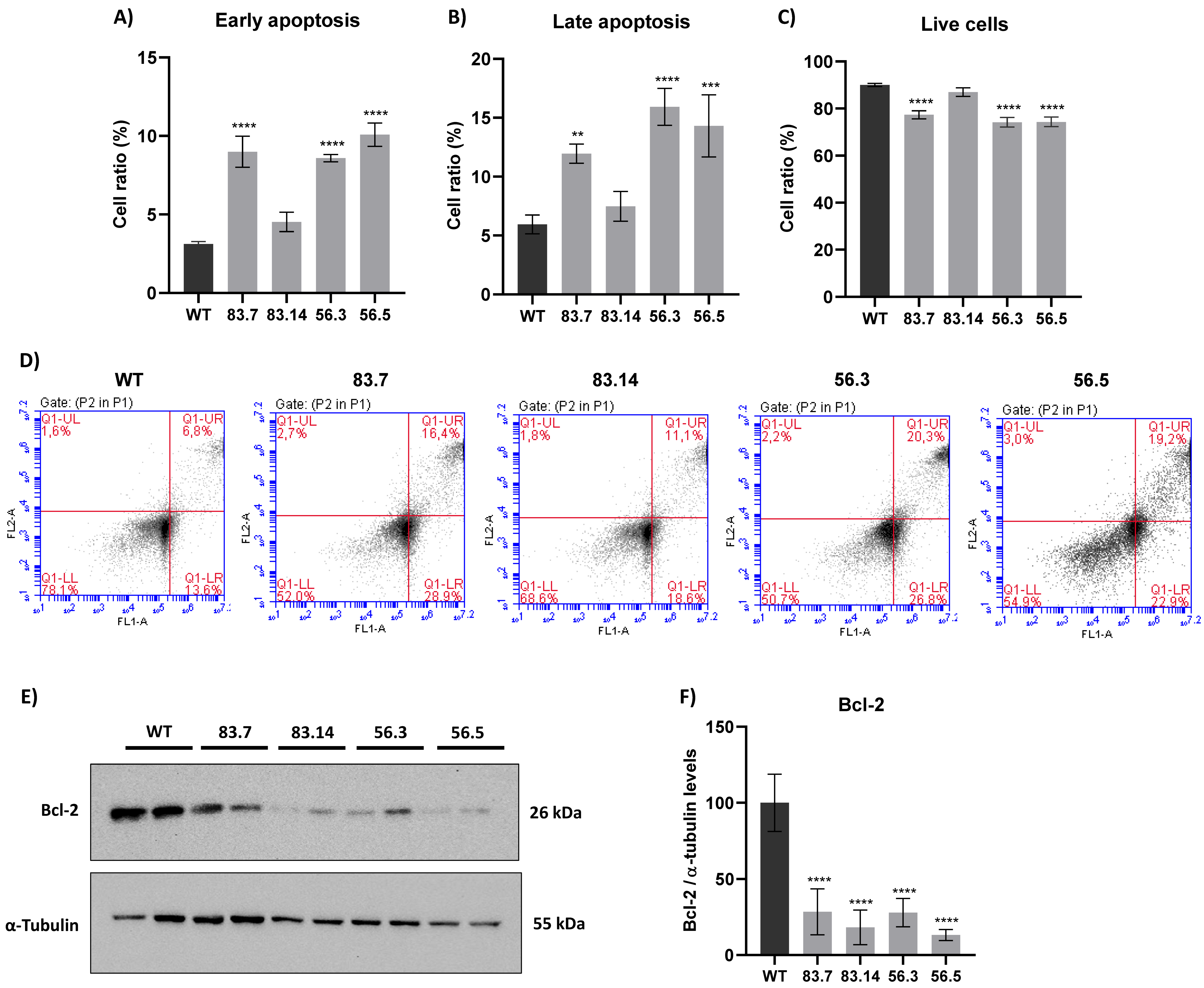
3.5. Targeted Deletion of NEK6 Induces Death in DU-145 Cells
3.6. Targeted Deletion of NEK6 Sensitizes DU-145 to Cisplatin through Impairment of Antioxidant Defenses and Increase of DNA Damage
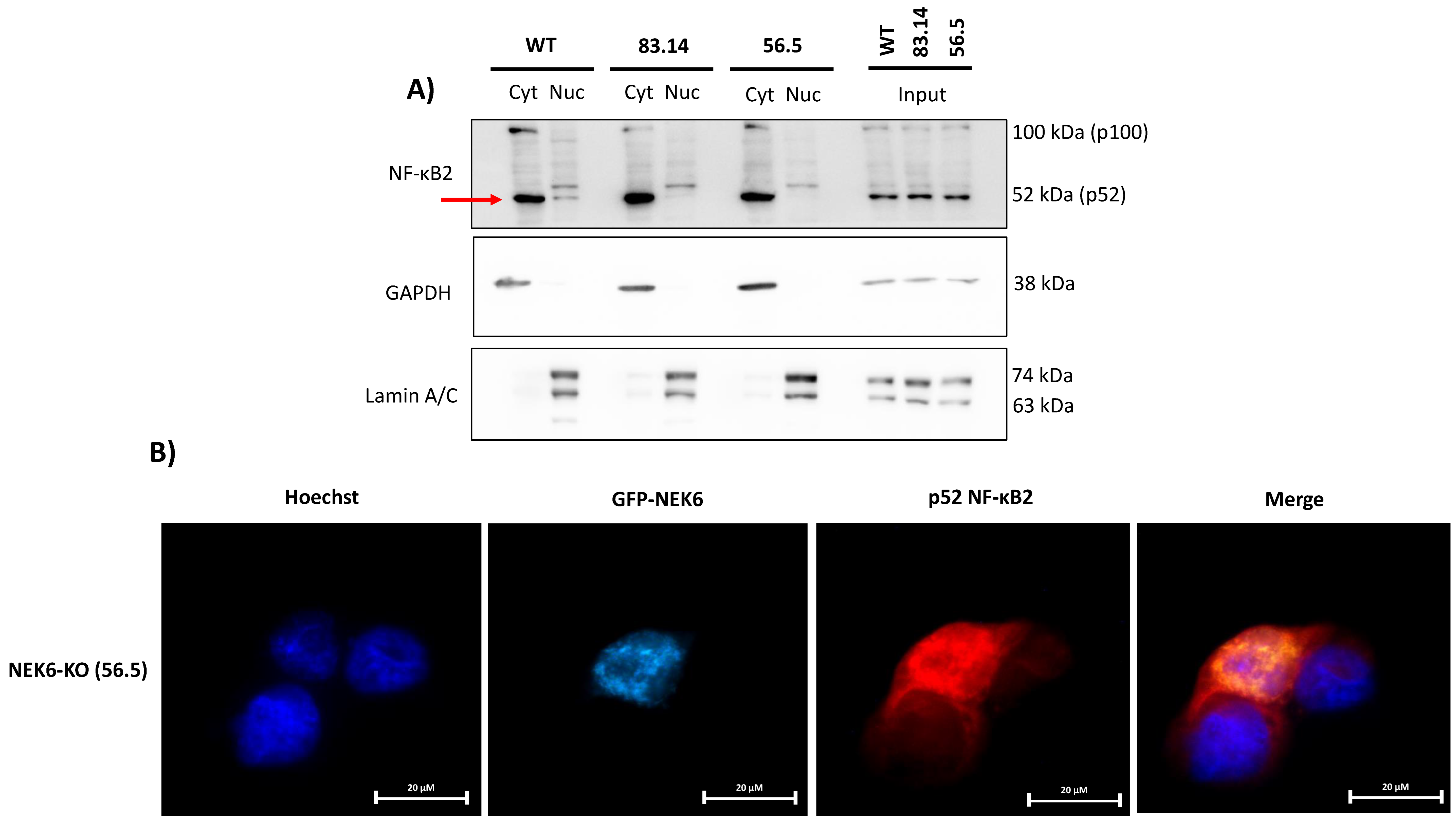
3.7. NEK6 May Be Involved in the NF-κB2 Translocation to the Nucleus
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rawla, P. Epidemiology of Prostate Cancer. World J. Oncol. 2019, 10, 63–89. [Google Scholar] [CrossRef]
- Chandrasekar, T.; Yang, J.C.; Gao, A.C.; Evans, C.P. Mechanisms of resistance in castration-resistant prostate cancer (CRPC). Transl. Androl. Urol. 2015, 4, 365–380. [Google Scholar] [CrossRef]
- O’Connell, M.J.; Krien, M.J.; Hunter, T. Never say never. The NIMA-related protein kinases in mitotic control. Trends Cell Biol. 2003, 13, 221–228. [Google Scholar] [CrossRef]
- Fry, A.M.; Bayliss, R.; Roig, J. Mitotic Regulation by NEK Kinase Networks. Front. Cell Dev. Biol. 2017, 5, 102. [Google Scholar] [CrossRef]
- Shalom, O.; Shalva, N.; Altschuler, Y.; Motro, B. The mammalian Nek1 kinase is involved in primary cilium formation. FEBS Lett. 2008, 582, 1465–1470. [Google Scholar] [CrossRef]
- Mahjoub, M.R.; Trapp, M.L.; Quarmby, L.M. NIMA-Related Kinases Defective in Murine Models of Polycystic Kidney Diseases Localize to Primary Cilia and Centrosomes. J. Am. Soc. Nephrol. 2005, 16, 3485–3489. [Google Scholar] [CrossRef] [PubMed]
- Coene, K.L.; Mans, D.A.; Boldt, K.; Gloeckner, C.J.; van Reeuwijk, J.; Bolat, E.; Roosing, S.; Letteboer, S.J.; Peters, T.A.; Cremers, F.P.; et al. The ciliopathy-associated protein homologs RPGRIP1 and RPGRIP1L are linked to cilium integrity through interaction with Nek4 serine/threonine kinase. Hum. Mol. Genet. 2011, 20, 3592–3605. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Pavan, I.; de Oliveira, A.P.; Dias, P.; Basei, F.; Issayama, L.; Ferezin, C.; Silva, F.; de Oliveira, A.R.; dos Reis Moura, A.; Martins, M.; et al. On Broken Ne(c)ks and Broken DNA: The Role of Human NEKs in the DNA Damage Response. Cells 2021, 10, 507. [Google Scholar] [CrossRef] [PubMed]
- de Oliveira, A.P.; Basei, F.L.; Slepicka, P.F.; Ferezin, C.D.C.; Melo-Hanchuk, T.D.; de Souza, E.E.; Lima, T.I.; dos Santos, V.T.; Mendes, D.; Silveira, L.R.; et al. NEK10 interactome and depletion reveal new roles in mitochondria. Proteome Sci. 2020, 18, 4. [Google Scholar] [CrossRef]
- Hanchuk, T.D.M.; Papa, P.F.; La Guardia, P.G.; Vercesi, A.E.; Kobarg, J. Nek5 interacts with mitochondrial proteins and interferes negatively in mitochondrial mediated cell death and respiration. Cell. Signal. 2015, 27, 1168–1177. [Google Scholar] [CrossRef] [PubMed]
- Panchal, N.K.; Prince, S.E. The NEK family of serine/threonine kinases as a biomarker for cancer. Clin. Exp. Med. 2022, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Li, L.; Zhang, Y.; Yang, H.; Wei, Y.; Zhang, L.; Liu, X.; Yu, L. Interaction of Pin1 with Nek6 and characterization of their expression correlation in Chinese hepatocellular carcinoma patients. Biochem. Biophys. Res. Commun. 2006, 341, 1059–1065. [Google Scholar] [CrossRef] [PubMed]
- Capra, M.; Nuciforo, P.G.; Confalonieri, S.; Quarto, M.; Bianchi, M.; Nebuloni, M.; Boldorini, R.; Pallotti, F.; Viale, G.; Gishizky, M.L.; et al. Frequent Alterations in the Expression of Serine/Threonine Kinases in Human Cancers. Cancer Res. 2006, 66, 8147–8154. [Google Scholar] [CrossRef] [PubMed]
- De Donato, M.; Fanelli, M.; Mariani, M.; Raspaglio, G.; Pandya, D.; He, S.; Fiedler, P.; Petrillo, M.; Scambia, G.; Ferlini, C. Nek6 and Hif-1α cooperate with the cytoskeletal gateway of drug resistance to drive outcome in serous ovarian cancer. Am. J. Cancer Res. 2015, 5, 1862–1877. [Google Scholar]
- He, Z.; Ni, X.; Xia, L.; Shao, Z. Overexpression of NIMA-related kinase 6 (NEK6) contributes to malignant growth and dismal prognosis in Human Breast Cancer. Pathol. Res. Pr. 2018, 214, 1648–1654. [Google Scholar] [CrossRef]
- Hong, Z.; Chen, Z.; Pan, J.; Shi, Z.; Wang, C.; Qiu, C. MicroRNA-323a-3p Negatively Regulates NEK6 in Colon Adenocarcinoma Cells. J. Oncol. 2022, 2022, 7007718. [Google Scholar] [CrossRef]
- Xu, J.; He, Q.; He, X.; Shao, Q.; Tao, H.; Ye, Z. Expression of NEK-6 in gastric cancer and its clinical significance. Zhonghua Wei Chang. Wai Ke Za Zhi = Chin. J. Gastrointest. Surg. 2015, 18, 1036–1040. Available online: http://www.ncbi.nlm.nih.gov/pubmed/26499152 (accessed on 8 November 2022).
- Wu, L.; Chen, Z.; Xing, Y. Retraction: MiR-506-3p inhibits cell proliferation, induces cell cycle arrest and apoptosis in retinoblastoma by directly targeting NEK6 by Lina Wu, Zhen Chen, Yiqiao Xing. Cell Biol. Int. 2018, 43, 1524. [Google Scholar] [CrossRef]
- Oliveira, A.P.D.P.D.; Issayama, L.K.K.; Pavan, I.; Silva, F.R.R.; Melo-Hanchuk, T.D.D.; Simabuco, F.M.M.; Kobarg, J. Checking NEKs: Overcoming a Bottleneck in Human Diseases. Molecules 2020, 25, 1778. [Google Scholar] [CrossRef]
- Nassirpour, R.; Shao, L.; Flanagan, P.; Abrams, T.; Jallal, B.; Smeal, T.; Yin, M.-J. Nek6 Mediates Human Cancer Cell Transformation and Is a Potential Cancer Therapeutic Target. Mol. Cancer Res. 2010, 8, 717–728. [Google Scholar] [CrossRef]
- Panchal, N.K.; Mohanty, S.; Prince, S.E. NIMA-related kinase-6 (NEK6) as an executable target in cancer. Clin. Transl. Oncol. 2022, 1–12. [Google Scholar] [CrossRef]
- Choudhury, A.D.; Schinzel, A.C.; Cotter, M.B.; Lis, R.T.; Labella, K.; Lock, Y.J.; Izzo, F.; Guney, I.; Bowden, M.; Li, Y.Y.; et al. Castration Resistance in Prostate Cancer Is Mediated by the Kinase NEK6. Cancer Res. 2016, 77, 753–765. [Google Scholar] [CrossRef] [PubMed]
- Sinha, K.; Das, J.; Pal, P.B.; Sil, P.C. Oxidative stress: The mitochondria-dependent and mitochondria-independent pathways of apoptosis. Arch. Toxicol. 2013, 87, 1157–1180. [Google Scholar] [CrossRef]
- Kumari, S.; Badana, A.K.; Murali, M.G.; Shailender, G.; Malla, R. Reactive Oxygen Species: A Key Constituent in Cancer Survival. Biomark. Insights 2018, 13, 1177271918755391. [Google Scholar] [CrossRef] [PubMed]
- Perillo, B.; Di Donato, M.; Pezone, A.; Di Zazzo, E.; Giovannelli, P.; Galasso, G.; Castoria, G.; Migliaccio, A. ROS in cancer therapy: The bright side of the moon. Exp. Mol. Med. 2020, 52, 192–203. [Google Scholar] [CrossRef] [PubMed]
- Reczek, C.R.; Chandel, N.S. The Two Faces of Reactive Oxygen Species in Cancer. Annu. Rev. Cancer Biol. 2017, 1, 79–98. [Google Scholar] [CrossRef]
- Zaidieh, T.; Smith, J.R.; Ball, K.E.; An, Q. ROS as a novel indicator to predict anticancer drug efficacy. BMC Cancer 2019, 19, 1224. [Google Scholar] [CrossRef]
- Sack, M.; Alili, L.; Karaman, E.; Das, S.; Gupta, A.; Seal, S.; Brenneisen, P. Combination of Conventional Chemotherapeutics with Redox-Active Cerium Oxide Nanoparticles—A Novel Aspect in Cancer Therapy. Mol. Cancer Ther. 2014, 13, 1740–1749. [Google Scholar] [CrossRef]
- Wang, H.; Bouzakoura, S.; de Mey, S.; Jiang, H.; Law, K.; Dufait, I.; Corbet, C.; Verovski, V.; Gevaert, T.; Feron, O.; et al. Auranofin radiosensitizes tumor cells through targeting thioredoxin reductase and resulting overproduction of reactive oxygen species. Oncotarget 2017, 8, 35728–35742. [Google Scholar] [CrossRef]
- Srinivas, U.S.; Tan, B.W.Q.; Vellayappan, B.A.; Jeyasekharan, A.D. ROS and the DNA damage response in cancer. Redox Biol. 2019, 25, 101084. [Google Scholar] [CrossRef]
- Yin, M.-J.; Shao, L.; Voehringer, D.; Smeal, T.; Jallal, B. The Serine/Threonine Kinase Nek6 Is Required for Cell Cycle Progression through Mitosis. J. Biol. Chem. 2003, 278, 52454–52460. [Google Scholar] [CrossRef]
- O’Regan, L.; Fry, A.M. The Nek6 and Nek7 Protein Kinases Are Required for Robust Mitotic Spindle Formation and Cytokinesis. Mol. Cell. Biol. 2009, 29, 3975–3990. [Google Scholar] [CrossRef] [PubMed]
- Concordet, J.-P.; Haeussler, M. CRISPOR: Intuitive guide selection for CRISPR/Cas9 genome editing experiments and screens. Nucleic Acids Res. 2018, 46, W242–W245. [Google Scholar] [CrossRef] [PubMed]
- Doench, J.G.; Fusi, N.; Sullender, M.; Hegde, M.; Vaimberg, E.W.; Donovan, K.F.; Smith, I.; Tothova, Z.; Wilen, C.; Orchard, R.; et al. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat. Biotechnol. 2016, 34, 184–191. [Google Scholar] [CrossRef] [PubMed]
- Hsu, P.D.; Scott, D.A.; Weinstein, J.A.; Ran, F.A.; Konermann, S.; Agarwala, V.; Li, Y.; Fine, E.J.; Wu, X.; Shalem, O.; et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol. 2013, 31, 827–832. [Google Scholar] [CrossRef]
- Ran, F.A.; Hsu, P.D.; Wright, J.; Agarwala, V.; Scott, D.A.; Zhang, F. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 2013, 8, 2281–2308. [Google Scholar] [CrossRef]
- Pavan, I.C.B.; Yokoo, S.; Granato, D.C.; Meneguello, L.; Carnielli, C.M.; Tavares, M.R.; Amaral, C.L.D.; de Freitas, L.B.; Leme, A.F.P.; Luchessi, A.D.; et al. Different interactomes for p70-S6K1 and p54-S6K2 revealed by proteomic analysis. PROTEOMICS 2016, 16, 2650–2666. [Google Scholar] [CrossRef] [PubMed]
- Schober, P.; Boer, C.; Schwarte, L.A. Correlation Coefficients: Appropriate Use and Interpretation. Anesth. Analg. 2018, 126, 1763–1768. [Google Scholar] [CrossRef]
- Belham, C.; Roig, J.; Caldwell, J.A.; Aoyama, Y.; Kemp, B.; Comb, M.; Avruch, J. A Mitotic Cascade of NIMA Family Kinases. J. Biol. Chem. 2003, 278, 34897–34909. [Google Scholar] [CrossRef]
- Meirelles, G.V.; Lanza, D.C.F.; da Silva, J.C.; Bernachi, J.S.; Leme, A.F.P.; Kobarg, J. Characterization of hNek6 Interactome Reveals an Important Role for Its Short N-Terminal Domain and Colocalization with Proteins at the Centrosome. J. Proteome Res. 2010, 9, 6298–6316. [Google Scholar] [CrossRef]
- Fry, A.M.; O’Regan, L.; Sabir, S.; Bayliss, R. Cell cycle regulation by the NEK family of protein kinases. J. Cell Sci. 2012, 125, 4423–4433. [Google Scholar] [CrossRef] [PubMed]
- Melixetian, M.; Klein, D.K.; Sorensen, C.; Helin, K. NEK11 regulates CDC25A degradation and the IR-induced G2/M checkpoint. Nat. Cell Biol. 2009, 11, 1247–1253. [Google Scholar] [CrossRef] [PubMed]
- Schwab, M. Anchorage-Independent Cell Growth In Encyclopedia of Cancer; Springer: Berlin, Heidelberg, 2011; p. 173. [Google Scholar] [CrossRef]
- Park, J.; Lee, J.; Choi, C. Mitochondrial Network Determines Intracellular ROS Dynamics and Sensitivity to Oxidative Stress through Switching Inter-Mitochondrial Messengers. PLoS ONE 2011, 6, e23211. [Google Scholar] [CrossRef] [PubMed]
- Ďuračková, Z. Some Current Insights into Oxidative Stress. Physiol. Res. 2010, 59, 459–469. [Google Scholar] [CrossRef]
- Ighodaro, O.M.; Akinloye, O.A. First line defence antioxidants-superoxide dismutase (SOD), catalase (CAT) and glutathione peroxidase (GPX): Their fundamental role in the entire antioxidant defence grid. Alex. J. Med. 2018, 54, 287–293. [Google Scholar] [CrossRef]
- Nicolussi, A.; D’Inzeo, S.; Capalbo, C.; Giannini, G.; Coppa, A. The role of peroxiredoxins in cancer. Mol. Clin. Oncol. 2017, 6, 139–153. [Google Scholar] [CrossRef]
- Shi, Y.; Nikulenkov, F.; Zawacka-Pankau, J.; Li, H.; Gabdoulline, R.; Xu, J.; Eriksson, S.; Hedström, E.; Issaeva, N.; Kel, A.; et al. ROS-dependent activation of JNK converts p53 into an efficient inhibitor of oncogenes leading to robust apoptosis. Cell Death Differ. 2014, 21, 612–623. [Google Scholar] [CrossRef]
- Kozlov, S.V.; Waardenberg, A.J.; Engholm-Keller, K.; Arthur, J.W.; Graham, M.E.; Lavin, M. Reactive Oxygen Species (ROS)-Activated ATM-Dependent Phosphorylation of Cytoplasmic Substrates Identified by Large-Scale Phosphoproteomics Screen. Mol. Cell. Proteom. 2016, 15, 1032–1047. [Google Scholar] [CrossRef]
- Truong, T.H.; Carroll, K.S. Redox regulation of protein kinases. Crit. Rev. Biochem. Mol. Biol. 2013, 48, 332–356. [Google Scholar] [CrossRef]
- Ndombera, F.T. Anti-cancer agents and reactive oxygen species modulators that target cancer cell metabolism. Pure Appl. Chem. 2017, 89, 1333–1348. [Google Scholar] [CrossRef]
- Zuliani, T.; Denis, V.; Noblesse, E.; Schnebert, S.; Andre, P.; Dumas, M.; Ratinaud, M.-H. Hydrogen peroxide-induced cell death in normal human keratinocytes is differentiation dependent. Free. Radic. Biol. Med. 2005, 38, 307–316. [Google Scholar] [CrossRef] [PubMed]
- Marengo, B.; De Ciucis, C.; Ricciarelli, R.; Passalacqua, M.; Nitti, M.; Zingg, J.-M.; Marinari, U.M.; Pronzato, M.A.; Domenicotti, C. PKCδ Sensitizes Neuroblastoma Cells to L-Buthionine-Sulfoximine and Etoposide Inducing Reactive Oxygen Species Overproduction and DNA Damage. PLoS ONE 2011, 6, e14661. [Google Scholar] [CrossRef] [PubMed]
- Verzella, D.; Pescatore, A.; Capece, D.; Vecchiotti, D.; Ursini, M.V.; Franzoso, G.; Alesse, E.; Zazzeroni, F. Life, death, and autophagy in cancer: NF-κB turns up everywhere. Cell Death Dis. 2020, 11, 210. [Google Scholar] [CrossRef] [PubMed]
- Morgan, M.J.; Liu, Z.-G. Crosstalk of reactive oxygen species and NF-kappaκB signaling. Cell Res. 2011, 21, 103–115. [Google Scholar] [CrossRef] [PubMed]
- Djavaheri-Mergny, M.; Javelaud, D.; Wietzerbin, J.; Besançon, F. NF-κB activation prevents apoptotic oxidative stress via an increase of both thioredoxin and MnSOD levels in TNFα-treated Ewing sarcoma cells. FEBS Lett. 2004, 578, 111–115. [Google Scholar] [CrossRef]
- Kiningham, K.; Xu, Y.; Daosukho, C.; Popova, B.; Clair, D.S. Nuclear factor kappaB-dependent mechanisms coordinate the synergistic effect of PMA and cytokines on the induction of superoxide dismutase 2. Biochem. J. 2001, 353, 147–156. Available online: http://www.ncbi.nlm.nih.gov/pubmed/11115408 (accessed on 8 November 2022). [CrossRef]
- Rojo, A.I. Regulation of Cu/Zn-Superoxide Dismutase Expression via the Phosphatidylinositol 3 Kinase/Akt Pathway and Nuclear Factor-κB. J. Neurosci. 2004, 24, 7324–7334. [Google Scholar] [CrossRef]
- De Donato, M.; Righino, B.; Filippetti, F.; Battaglia, A.; Petrillo, M.; Pirolli, D.; Scambia, G.; De Rosa, M.C.; Gallo, D. Identification and antitumor activity of a novel inhibitor of the NIMA-related kinase NEK6. Sci. Rep. 2018, 8, 16047. [Google Scholar] [CrossRef]
- Das, T.P.; Suman, S.; Damodaran, C. Induction of reactive oxygen species generation inhibits epithelial-mesenchymal transition and promotes growth arrest in prostate cancer cells. Mol. Carcinog. 2013, 53, 537–547. [Google Scholar] [CrossRef] [PubMed]
- Sun, Z.-L.; Dong, J.-L.; Wu, J. Juglanin induces apoptosis and autophagy in human breast cancer progression via ROS/JNK promotion. Biomed. Pharmacother. 2017, 85, 303–312. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Chen, Y.; Zhao, J.; Shi, J.; Wang, M.; Qiu, S.; Hu, Y.; Xu, Y.; Cui, Y.; Liu, C.; et al. The Specific Inhibition of SOD1 Selectively Promotes Apoptosis of Cancer Cells via Regulation of the ROS Signaling Network. Oxidative Med. Cell. Longev. 2019, 2019, 9706792. [Google Scholar] [CrossRef] [PubMed]
- Zhu, S.; Jiang, L.; Wang, L.; Wang, L.; Zhang, C.; Ma, Y.; Huang, T. Identification of key genes and specific pathways potentially involved in androgen-independent, mitoxantrone-resistant prostate cancer. Cancer Manag. Res. 2019, 11, 419–430. [Google Scholar] [CrossRef] [PubMed]
- Papa, L.; Manfredi, G.; Germain, D. SOD1, an unexpected novel target for cancer therapy. Genes Cancer 2014, 5, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.W.; Sahm, H.; You, J.; Wang, M. Knock-down of superoxide dismutase 1 sensitizes cisplatin-resistant human ovarian cancer cells. Anticancer Res. 2010, 30, 2577–2581. Available online: http://www.ncbi.nlm.nih.gov/pubmed/20682985 (accessed on 8 November 2022).
- Ma, C.-S.; Lv, Q.-M.; Zhang, K.-R.; Tang, Y.-B.; Zhang, Y.-F.; Shen, Y.; Lei, H.-M.; Zhu, L. NRF2-GPX4/SOD2 axis imparts resistance to EGFR-tyrosine kinase inhibitors in non-small-cell lung cancer cells. Acta Pharmacol. Sin. 2020, 42, 613–623. [Google Scholar] [CrossRef]
- Zuo, J.; Zhao, M.; Liu, B.; Han, X.; Li, Y.; Wang, W.; Zhang, Q.; Lv, P.; Xing, L.; Shen, H.; et al. TNF-α-mediated upregulation of SOD-2 contributes to cell proliferation and cisplatin resistance in esophageal squamous cell carcinoma. Oncol. Rep. 2019, 42, 1497–1506. [Google Scholar] [CrossRef]
- Byun, J.M.; Kim, S.S.; Kim, K.T.; Kang, M.S.; Jeong, D.H.; Lee, D.S.; Jung, E.J.; Kim, Y.N.; Han, J.; Song, I.S.; et al. Overexpression of peroxiredoxin-3 and -5 is a potential biomarker for prognosis in endometrial cancer. Oncol. Lett. 2018, 15, 5111–5118. [Google Scholar] [CrossRef]
- Ramasamy, P.; Larkin, A.-M.; Linge, A.; Tiernan, D.; McAree, F.; Horgan, N.; Moriarty, P.; Beatty, S.; Murphy, C.; Clynes, M.; et al. PRDX3 is associated with metastasis and poor survival in uveal melanoma. J. Clin. Pathol. 2020, 73, 408–412. [Google Scholar] [CrossRef]
- Ummanni, R.; Barreto, F.; Venz, S.; Scharf, C.; Barett, C.; Mannsperger, H.A.; Brase, J.C.; Kuner, R.; Schlomm, T.; Sauter, G.; et al. Peroxiredoxins 3 and 4 Are Overexpressed in Prostate Cancer Tissue and Affect the Proliferation of Prostate Cancer Cells in Vitro. J. Proteome Res. 2012, 11, 2452–2466. [Google Scholar] [CrossRef] [PubMed]
- Whitaker, H.C.; Patel, D.; Howat, W.J.; Warren, A.Y.; Kay, J.D.; Sangan, T.; Marioni, J.C.; Mitchell, J.; Aldridge, S.; Luxton, H.J.; et al. Peroxiredoxin-3 is overexpressed in prostate cancer and promotes cancer cell survival by protecting cells from oxidative stress. Br. J. Cancer 2013, 109, 983–993. [Google Scholar] [CrossRef]
- Kim, Y.S.; Vallur, P.G.; Phaëton, R.; Mythreye, K.; Hempel, N. Insights into the Dichotomous Regulation of SOD2 in Cancer. Antioxidants 2017, 6, 86. [Google Scholar] [CrossRef] [PubMed]
- Miar, A.; Hevia, D.; Muñoz-Cimadevilla, H.; Astudillo, A.; Velasco, J.; Sainz, R.M.; Mayo, J.C. Manganese superoxide dismutase (SOD2/MnSOD)/catalase and SOD2/GPx1 ratios as biomarkers for tumor progression and metastasis in prostate, colon, and lung cancer. Free. Radic. Biol. Med. 2015, 85, 45–55. [Google Scholar] [CrossRef] [PubMed]
- Quiros-Gonzalez, I.; Gonzalez-Menendez, P.; Mayo, J.C.; Hevia, D.; Artime-Naveda, F.; Fernandez-Vega, S.; Fernandez-Fernandez, M.; Rodriguez-Gonzalez, P.; Garcia-Alonso, J.I.; Sainz, R.M. Androgen-Dependent Prostate Cancer Cells Reprogram Their Metabolic Signature upon GLUT1 Upregulation by Manganese Superoxide Dismutase. Antioxidants 2022, 11, 313. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Chen, Q.; Huang, C.; Rao, D.; Sang, C.; Zhu, S.; Gu, L.; Xie, C.; Tang, Z.; Xu, X. Transcription factor Nrf2 binds to circRNAPIBF1 to regulate SOD2 in lung adenocarcinoma progression. Mol. Carcinog. 2022, 61, 1161–1176. [Google Scholar] [CrossRef]
- Das, K.C.; Lewis-Molock, Y.; White, C.W. Activation of NF-kappa B and elevation of MnSOD gene expression by thiol reducing agents in lung adenocarcinoma (A549) cells. Am. J. Physiol. Cell. Mol. Physiol. 1995, 269, L588–L602. [Google Scholar] [CrossRef]
- Cao, Z.; Bhella, D.; Lindsay, J.G. Reconstitution of the Mitochondrial PrxIII Antioxidant Defence Pathway: General Properties and Factors Affecting PrxIII Activity and Oligomeric State. J. Mol. Biol. 2007, 372, 1022–1033. [Google Scholar] [CrossRef] [PubMed]
- Simon, H.-U.; Haj-Yehia, A.; Levi-Schaffer, F. Role of reactive oxygen species (ROS) in apoptosis induction. Apoptosis 2000, 5, 415–418. [Google Scholar] [CrossRef]
- de Feraudy, S.; Revet, I.; Bezrookove, V.; Feeney, L.; Cleaver, J.E. A minority of foci or pan-nuclear apoptotic staining of γH2AX in the S phase after UV damage contain DNA double-strand breaks. Proc. Natl. Acad. Sci. USA 2010, 107, 6870–6875. [Google Scholar] [CrossRef]
- Wang, X.; Olberding, K.E.; White, C.; Li, C. Bcl-2 proteins regulate ER membrane permeability to luminal proteins during ER stress-induced apoptosis. Cell Death Differ. 2010, 18, 38–47. [Google Scholar] [CrossRef]
- Kowaltowski, A.J.; Vercesi, A.E.; Fiskum, G. Bcl-2 prevents mitochondrial permeability transition and cytochrome c release via maintenance of reduced pyridine nucleotides. Cell Death Differ. 2000, 7, 903–910. [Google Scholar] [CrossRef]
- Lin, Y.; Fukuchi, J.; Hiipakka, R.A.; Kokontis, J.M.; Xiang, J. Up-regulation of Bcl-2 is required for the progression of prostate cancer cells from an androgen-dependent to an androgen-independent growth stage. Cell Res. 2007, 17, 531–536. [Google Scholar] [CrossRef] [PubMed]
- Marullo, R.; Werner, E.; Degtyareva, N.; Moore, B.; Altavilla, G.; Ramalingam, S.S.; Doetsch, P.W. Cisplatin Induces a Mitochondrial-ROS Response That Contributes to Cytotoxicity Depending on Mitochondrial Redox Status and Bioenergetic Functions. PLoS ONE 2013, 8, e81162. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pavan, I.C.B.; Basei, F.L.; Severino, M.B.; Rosa e Silva, I.; Issayama, L.K.; Mancini, M.C.S.; Góis, M.M.; da Silva, L.G.S.; Bezerra, R.M.N.; Simabuco, F.M.; et al. NEK6 Regulates Redox Balance and DNA Damage Response in DU-145 Prostate Cancer Cells. Cells 2023, 12, 256. https://doi.org/10.3390/cells12020256
Pavan ICB, Basei FL, Severino MB, Rosa e Silva I, Issayama LK, Mancini MCS, Góis MM, da Silva LGS, Bezerra RMN, Simabuco FM, et al. NEK6 Regulates Redox Balance and DNA Damage Response in DU-145 Prostate Cancer Cells. Cells. 2023; 12(2):256. https://doi.org/10.3390/cells12020256
Chicago/Turabian StylePavan, Isadora Carolina Betim, Fernanda Luisa Basei, Matheus Brandemarte Severino, Ivan Rosa e Silva, Luidy Kazuo Issayama, Mariana Camargo Silva Mancini, Mariana Marcela Góis, Luiz Guilherme Salvino da Silva, Rosangela Maria Neves Bezerra, Fernando Moreira Simabuco, and et al. 2023. "NEK6 Regulates Redox Balance and DNA Damage Response in DU-145 Prostate Cancer Cells" Cells 12, no. 2: 256. https://doi.org/10.3390/cells12020256
APA StylePavan, I. C. B., Basei, F. L., Severino, M. B., Rosa e Silva, I., Issayama, L. K., Mancini, M. C. S., Góis, M. M., da Silva, L. G. S., Bezerra, R. M. N., Simabuco, F. M., & Kobarg, J. (2023). NEK6 Regulates Redox Balance and DNA Damage Response in DU-145 Prostate Cancer Cells. Cells, 12(2), 256. https://doi.org/10.3390/cells12020256





