Preclinical Evaluation of a Novel Small Molecule LCC-21 to Suppress Colorectal Cancer Malignancy by Inhibiting Angiogenic and Metastatic Signatures
Abstract
1. Introduction
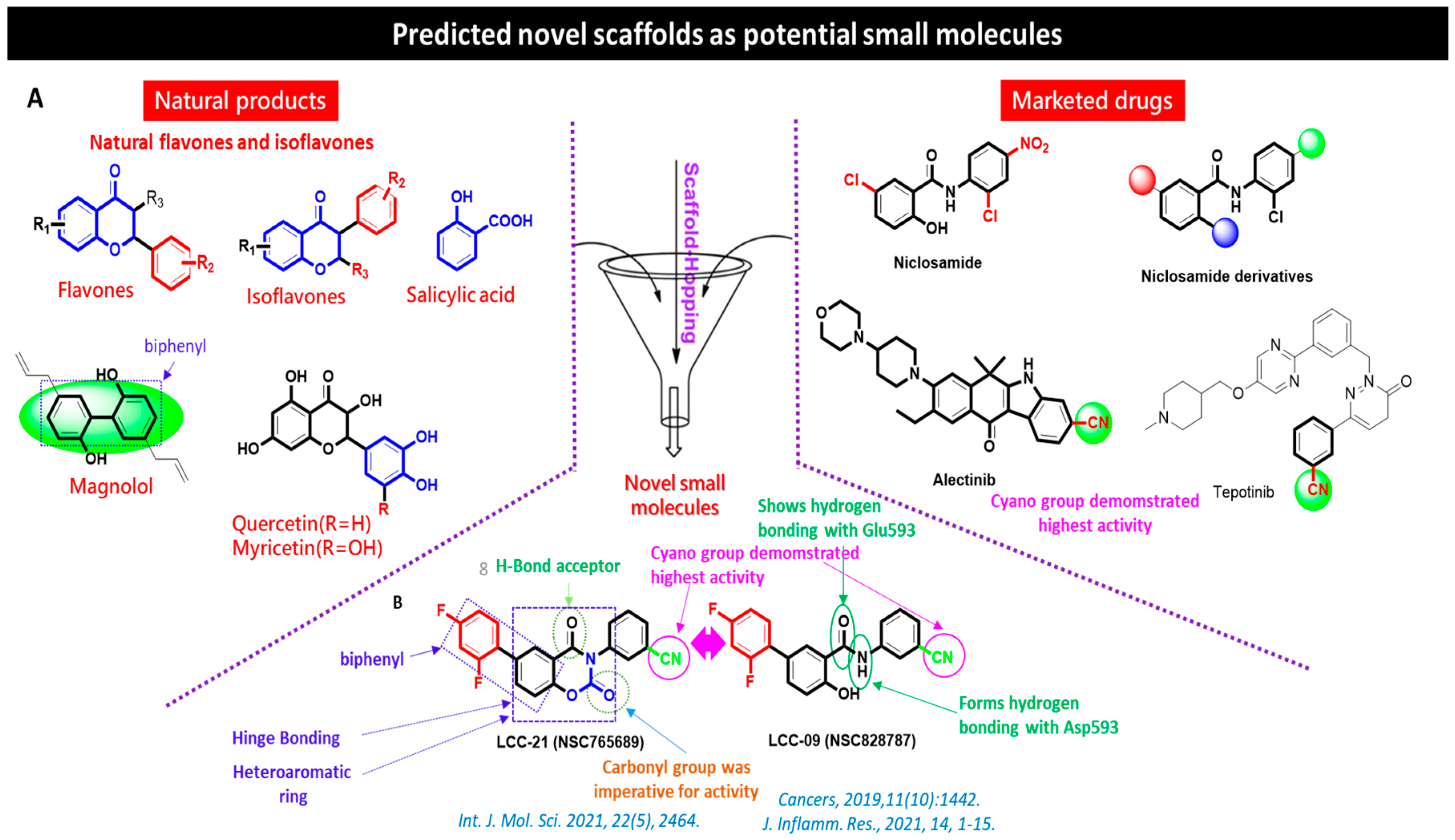
Chemical Synthesis of the Novel Multi-Target Small Molecules NSC828787 (LCC09) and NSC765689 (LCC-21)
2. Materials and Methods
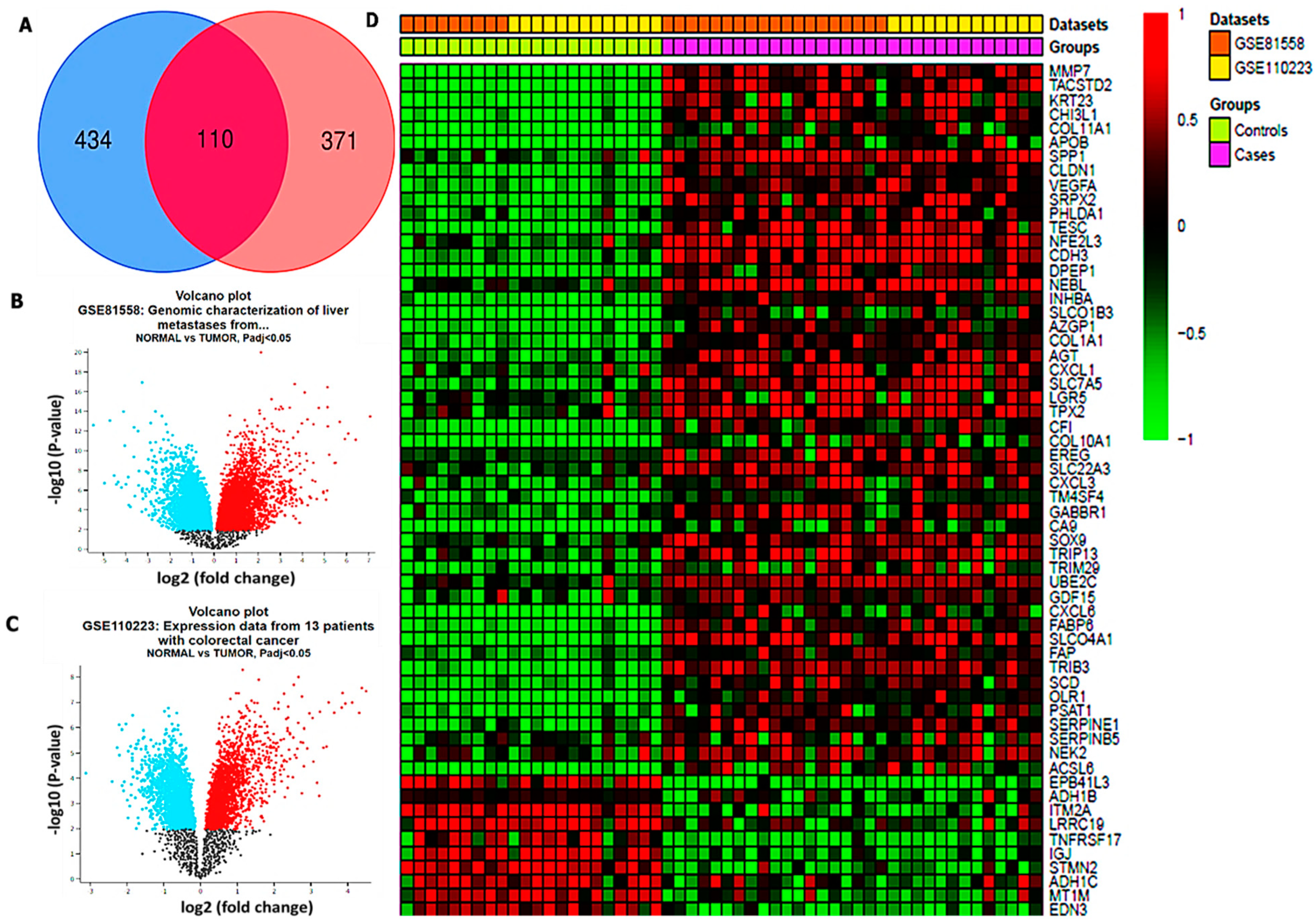
2.1. Microarray Dataset Extraction
2.2. Analysis of Pharmacokinetic (PK), Drug-Likeness, and Medicinal Chemical Properties of NSC765689 (LCC-21)
2.3. Validation of Differentially Expressed Genes (DEGs) in Colorectal Cancer Cohorts
2.4. Functional Enrichment Analysis
2.5. Analysis of Genomic Alterations of Targeted Genes and Immune infiltration in CRC
2.6. Bioinformatics Approaches to Single-Cell RNA-seq (scRNA-seq) Analysis
2.7. In Vitro Screening of LCC-21 against Full National Cancer Institute (NCI) Panels of Colorectal Tumor Cell Lines
2.8. Receptor–Ligand Interaction Analysis
2.9. Cell Culture and Reagents
2.10. Cell Viability Assay
2.11. Cell-Migration Assay
2.12. Colony-Formation Assays
2.13. Sodium Dodecylsulfate-Polyacrylamide Gel Electrophoresis (SDS-PAGE) and Immunoblot Analysis
2.14. Tumor-Sphere Formation
2.15. Data Analysis
3. Results
3.1. Identification of Differentially Expressed Genes (DEGs) in CRC
3.2. LCC-21 Successfully Meets Required Drug-Likeness Criteria
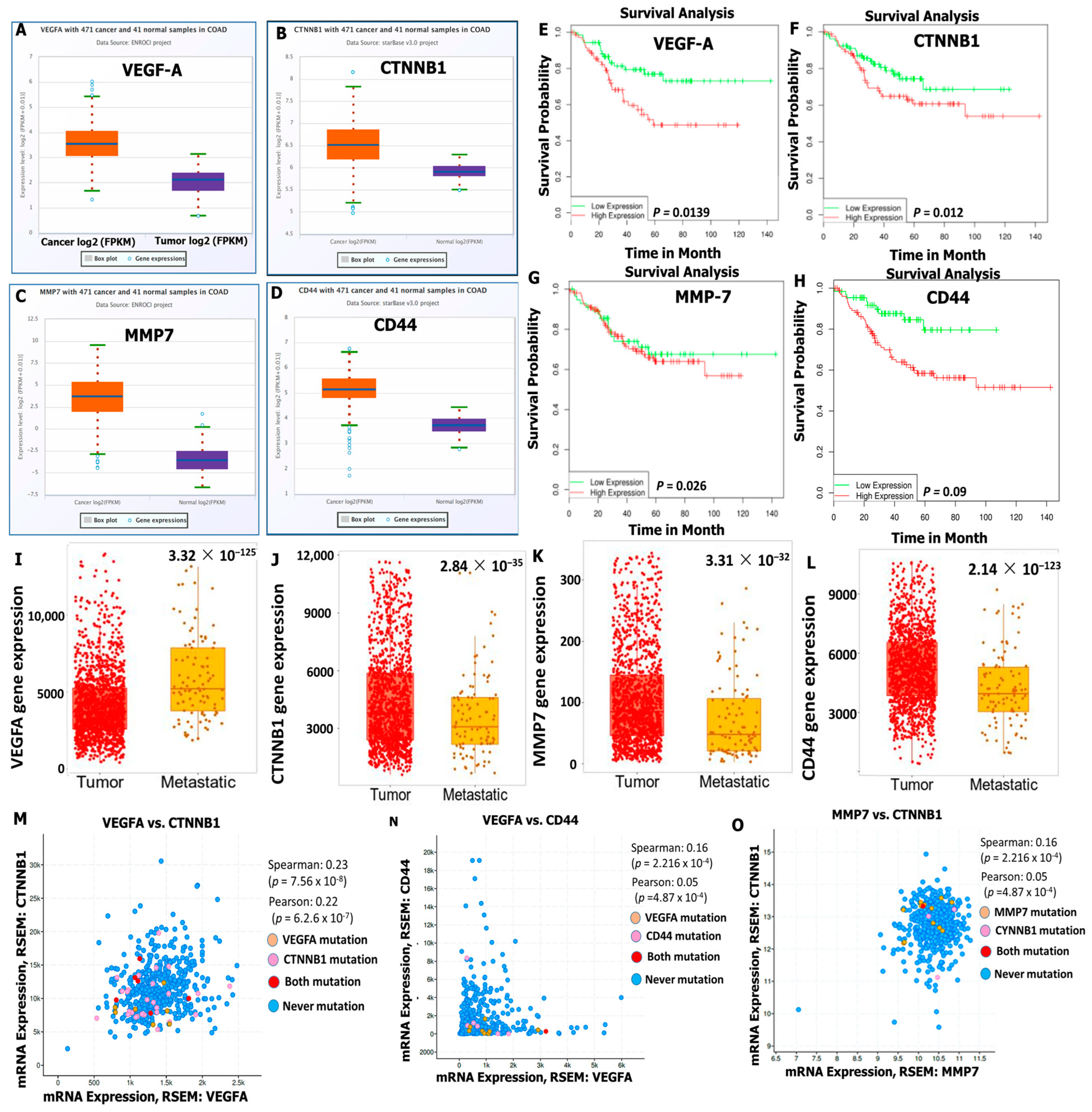
3.3. VEGFA/CTNNB1/MMP7/CD44 Oncogenic Signatures Are Overexpressed in CRC and Associated with a Poor Prognosis
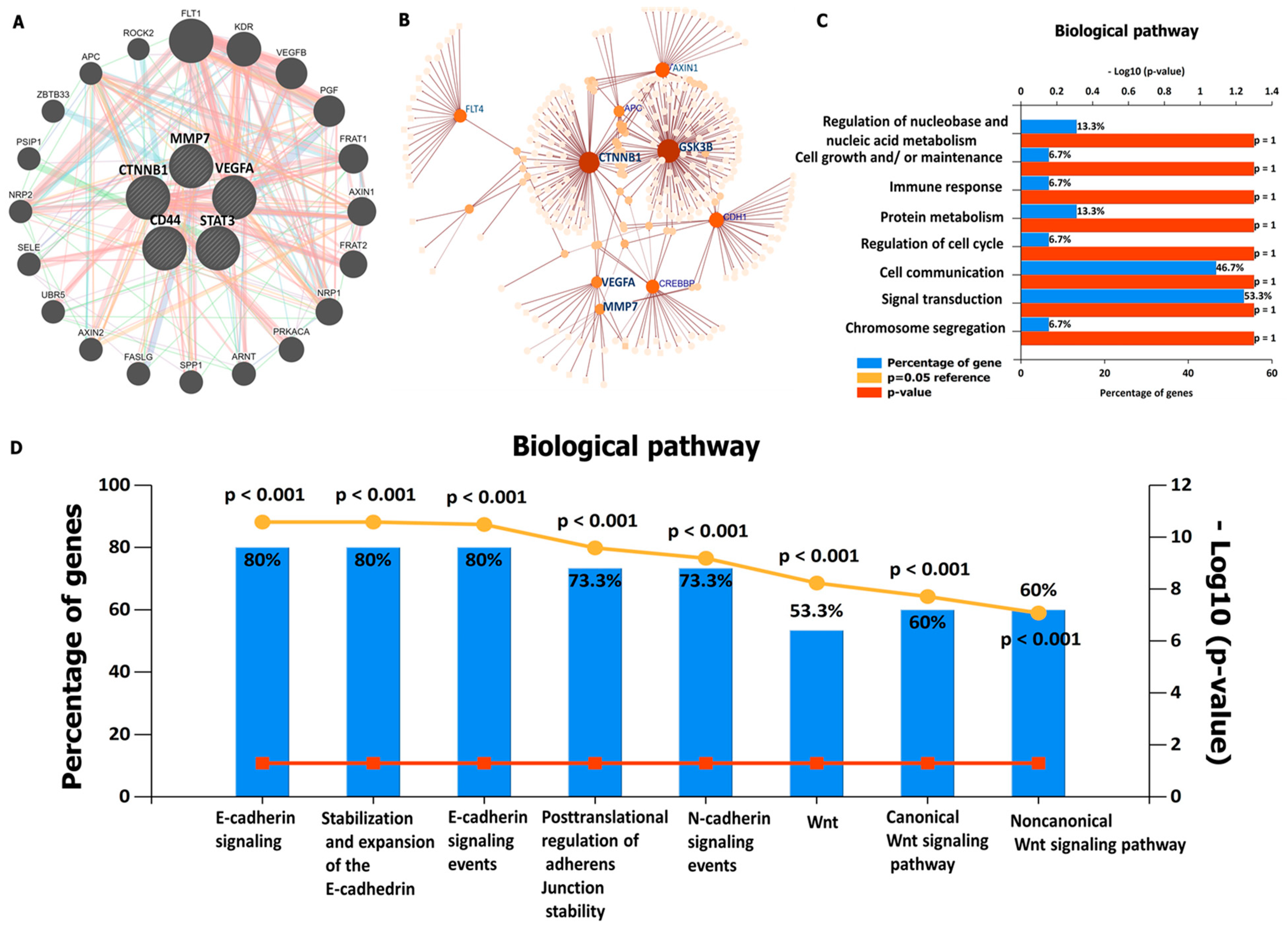
3.4. Protein–Protein Interaction (PPI) Network Construction and Functional Enrichment Analysis
3.5. VEGFA/CTNNB1/MMP7/CD44 Genes Are Altered in CRC Tissues and Immune Cells
3.6. VEGFA/CTNNB1CTNNB1/MMP7/CD44 Oncogene Expressions Are Correlated with Immune Cell Infiltration and Worse Prognosis in CRC
3.7. VEGFA/CTNNB1/MMP7/CD44 Gene Expression Influence the Immune Landscape within the TME of Colorectal Cancer
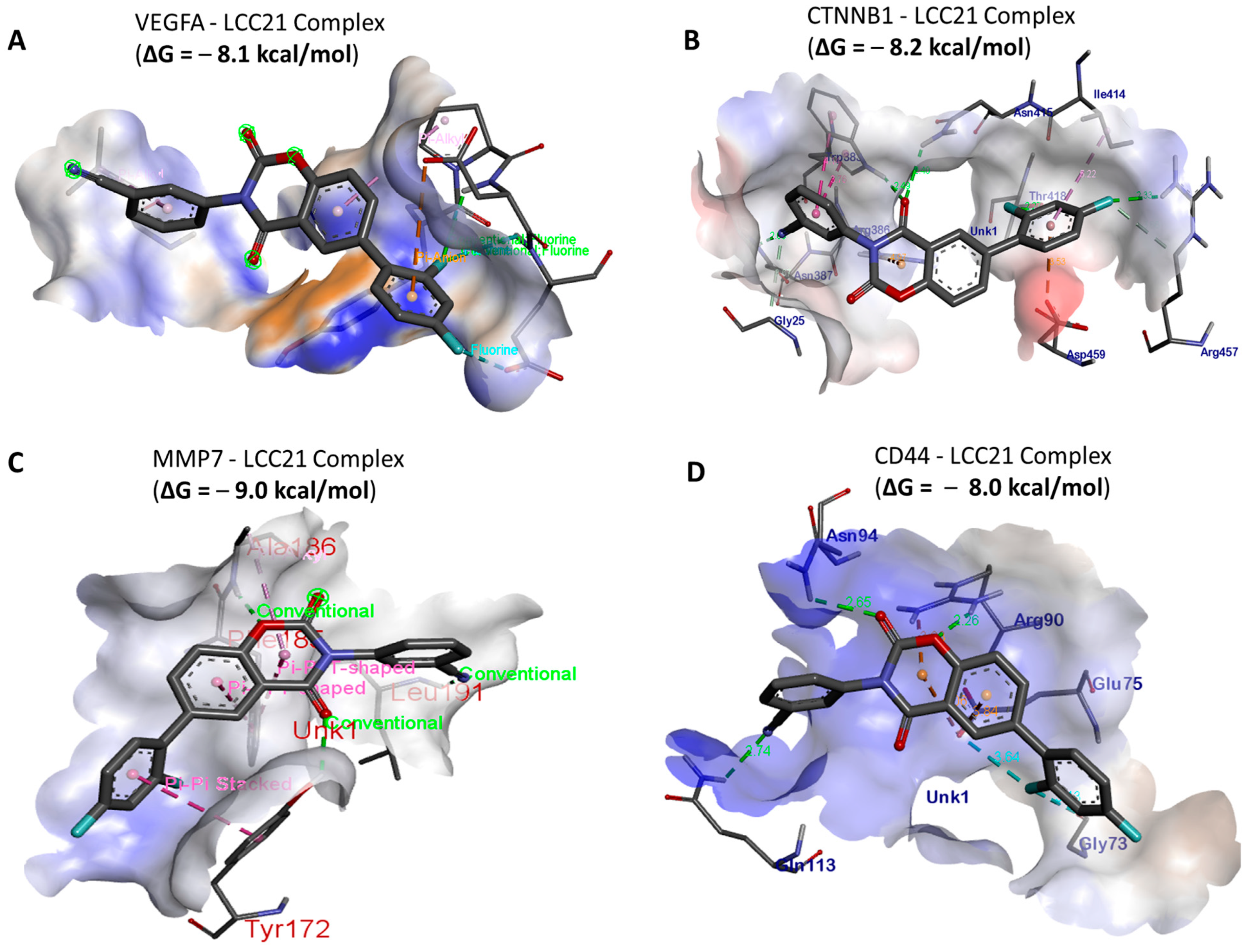
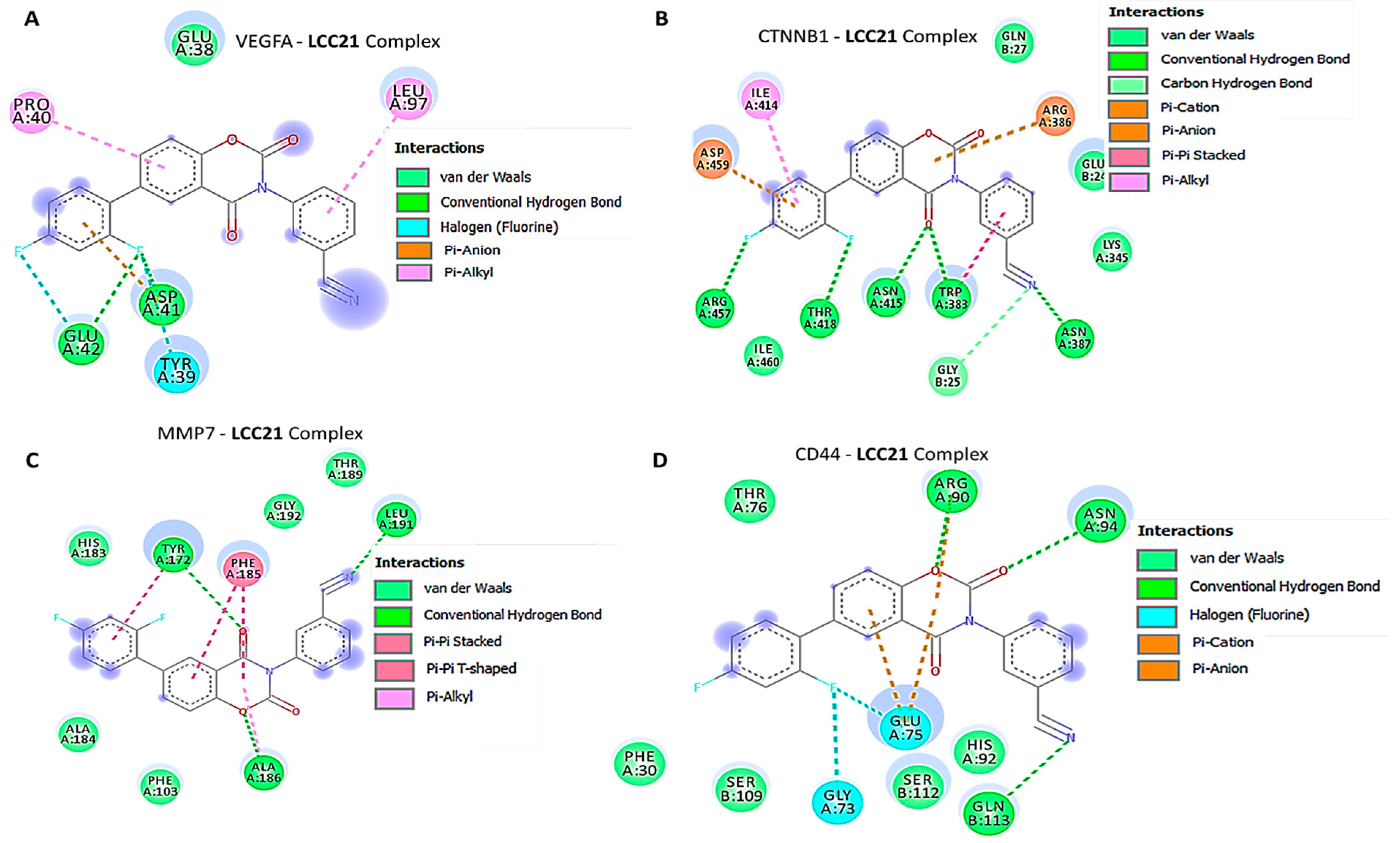
3.8. In Silico Molecular Docking Showed Putative Binding of (NSC765689) LCC-21 with VEGFA/CTNNB1/MMP7/CD44
3.9. Docking Analysis of VEGFA/CTNNB1/MMP7/CD44 with Their FDA Approved Inhibitors
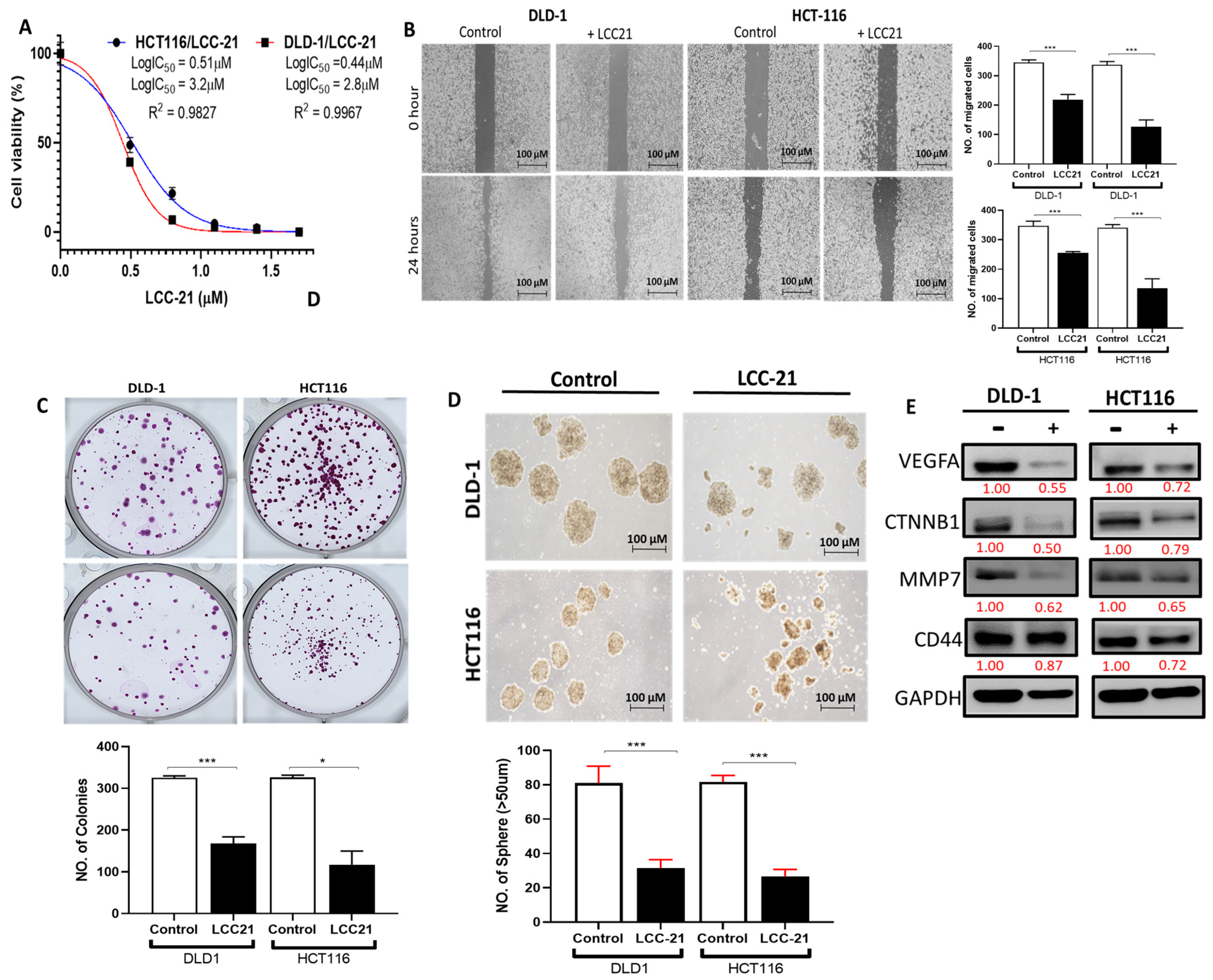
3.10. NSC765689 (LCC-21) Exhibited Anti-Proliferative and Cytotoxic Effects in NCI60 Human Colon Cancer Cell Lines
3.11. LCC-21 Decreases the Viability of CRC Cells through Modification of VEGFA/CTNNB1/MMP7/CD44 Oncogenic Signatures
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: Globocan Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Hauptman, N.; Boštjančič, E.; Žlajpah, M.; Ranković, B.; Zidar, N. Bioinformatics Analysis Reveals most Prominent Gene Candidates to Distinguish Colorectal Adenoma from Adenocarcinoma. Biomed. Res. Int. 2018, 2018, 9416515. [Google Scholar] [CrossRef] [PubMed]
- Linnekamp, J.F.; Wang, X.; Medema, J.P.; Vermeulen, L. Colorectal cancer heterogeneity and targeted therapy: A case for molecular disease subtypes. Cancer Res. 2015, 75, 245–249. [Google Scholar] [CrossRef] [PubMed]
- Cook, A.D.; Single, R.; McCahill, L.E. Surgical resection of primary tumors in patients who present with stage IV colorectal cancer: An analysis of surveillance, epidemiology, and end results data, 1988 to 2000. Ann. Surg. Oncol. 2005, 12, 637–645. [Google Scholar] [CrossRef] [PubMed]
- Isbister, W.H. Audit of definitive colorectal surgery in patients with early and advanced colorectal cancer. ANZ J. Surg. 2002, 72, 271–274. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R.; Desantis, C.; Jemal, A. Colorectal cancer statistics, 2014. CA Cancer J. Clin. 2014, 64, 104–117. [Google Scholar] [CrossRef] [PubMed]
- He, F.; Chen, H.; Yang, P.; Wu, Q.; Zhang, T.; Wang, C.; Wei, J.; Chen, Z.; Hu, H.; Li, W.; et al. Gankyrin sustains PI3K/GSK-3β/β-catenin signal activation and promotes colorectal cancer aggressiveness and progression. Oncotarget 2016, 7, 81156–81171. [Google Scholar] [CrossRef]
- Munro, M.J.; Wickremesekera, S.K.; Peng, L.; Tan, S.T.; Itinteang, T. Cancer stem cells in colorectal cancer: A review. J. Clin. Pathol. 2018, 71, 110–116. [Google Scholar] [CrossRef]
- Dienstmann, R.; Salazar, R.; Tabernero, J. Personalizing colon cancer adjuvant therapy: Selecting optimal treatments for individual patients. J. Clin. Oncol. 2015, 33, 1787–1796. [Google Scholar] [CrossRef]
- Dienstmann, R.; Vermeulen, L.; Guinney, J.; Kopetz, S.; Tejpar, S.; Tabernero, J. Consensus molecular subtypes and the evolution of precision medicine in colorectal cancer. Nat. Rev. Cancer 2017, 17, 79–92. [Google Scholar] [CrossRef]
- Gillies, R.J.; Verduzco, D.; Gatenby, R.A. Evolutionary dynamics of carcinogenesis and why targeted therapy does not work. Nat. Rev. Cancer 2012, 12, 487–493. [Google Scholar] [CrossRef] [PubMed]
- Kitajima, K.; Fujimori, T.; Fujii, S.; Takeda, J.; Ohkura, Y.; Kawamata, H.; Kumamoto, T.; Ishiguro, S.; Kato, Y.; Shimoda, T.; et al. Correlations between lymph node metastasis and depth of submucosal invasion in submucosal invasive colorectal carcinoma: A Japanese collaborative study. J. Gastroenterol. 2004, 39, 534–543. [Google Scholar] [CrossRef] [PubMed]
- George, M.L.; Tutton, M.G.; Janssen, F.; Arnaout, A.; Abulafi, A.M.; Eccles, S.A.; Swift, R.I. VEGF-A, VEGF-C, and VEGF-D in colorectal cancer progression. Neoplasia 2001, 3, 420–427. [Google Scholar] [CrossRef]
- Folkman, J. What is the evidence that tumors are angiogenesis dependent? J. Natl. Cancer Inst. 1990, 82, 4–6. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D.; Folkman, J. Patterns and emerging mechanisms of the angiogenic switch during tumorigenesis. Cell 1996, 86, 353–364. [Google Scholar] [CrossRef] [PubMed]
- Brown, L.F.; Berse, B.; Jackman, R.W.; Tognazzi, K.; Manseau, E.J.; Senger, D.R.; Dvorak, H.F. Expression of vascular permeability factor (vascular endothelial growth factor) and its receptors in adenocarcinomas of the gastrointestinal tract. Cancer Res. 1993, 53, 4727–4735. [Google Scholar]
- Kelland, L. The resurgence of platinum-based cancer chemotherapy. Nat. Rev. Cancer 2007, 7, 573–584. [Google Scholar] [CrossRef]
- Lajous, H.; Lelièvre, B.; Vauléon, E.; Lecomte, P.; Garcion, E. Rethinking Alkylating(-Like) Agents for Solid Tumor Management. Trends Pharmacol. Sci. 2019, 40, 342–357. [Google Scholar] [CrossRef]
- De Roock, W.; Claes, B.; Bernasconi, D.; De Schutter, J.; Biesmans, B.; Fountzilas, G.; Kalogeras, K.T.; Kotoula, V.; Papamichael, D.; Laurent-Puig, P.; et al. Effects of KRAS, BRAF, NRAS, and PIK3CA mutations on the efficacy of cetuximab plus chemotherapy in chemotherapy-refractory metastatic colorectal cancer: A retrospective consortium analysis. Lancet Oncol. 2010, 11, 753–762. [Google Scholar] [CrossRef]
- Chen, W.; Chen, M.; Barak, L.S. Development of small molecules targeting the Wnt pathway for the treatment of colon cancer: A high-throughput screening approach. Am. J. Physiol. Gastrointest. Liver Physiol. 2010, 299, G293–G300. [Google Scholar] [CrossRef]
- Hui, Y.F.; Reitz, J. Gemcitabine: A cytidine analogue active against solid tumors. Am. J. Health Syst. Pharm. 1997, 54, 162–170; quiz 197–168. [Google Scholar] [CrossRef] [PubMed]
- Van Cutsem, E.; Cervantes, A.; Nordlinger, B.; Arnold, D. Metastatic colorectal cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann. Oncol. 2014, 25 (Suppl. 3), iii1–iii9. [Google Scholar] [CrossRef] [PubMed]
- Willett, C.G.; Boucher, Y.; di Tomaso, E.; Duda, D.G.; Munn, L.L.; Tong, R.T.; Chung, D.C.; Sahani, D.V.; Kalva, S.P.; Kozin, S.V.; et al. Direct evidence that the VEGF-specific antibody bevacizumab has antivascular effects in human rectal cancer. Nat. Med. 2004, 10, 145–147. [Google Scholar] [CrossRef] [PubMed]
- Ferrara, N.; Hillan, K.J.; Gerber, H.P.; Novotny, W. Discovery and development of bevacizumab, an anti-VEGF antibody for treating cancer. Nat. Rev. Drug Discov. 2004, 3, 391–400. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Chen, X.; Li, W.; Ye, Y.; Du, X.; Sun, S.; Liu, L.; Zhang, H. Combination of Anti-EGFR and Anti-VEGF Drugs for the Treatment of Previously Treated Metastatic Colorectal Cancer: A Case Report and Literature Review. Front. Oncol. 2021, 11, 684309. [Google Scholar] [CrossRef]
- Hoeben, A.; Landuyt, B.; Highley, M.S.; Wildiers, H.; Van Oosterom, A.T.; De Bruijn, E.A. Vascular endothelial growth factor and angiogenesis. Pharmacol. Rev. 2004, 56, 549–580. [Google Scholar] [CrossRef]
- Keck, P.J.; Hauser, S.D.; Krivi, G.; Sanzo, K.; Warren, T.; Feder, J.; Connolly, D.T. Vascular permeability factor, an endothelial cell mitogen related to PDGF. Science 1989, 246, 1309–1312. [Google Scholar] [CrossRef]
- Senger, D.R.; Galli, S.J.; Dvorak, A.M.; Perruzzi, C.A.; Harvey, V.S.; Dvorak, H.F. Tumor cells secrete a vascular permeability factor that promotes accumulation of ascites fluid. Science 1983, 219, 983–985. [Google Scholar] [CrossRef]
- Joukov, V.; Pajusola, K.; Kaipainen, A.; Chilov, D.; Lahtinen, I.; Kukk, E.; Saksela, O.; Kalkkinen, N.; Alitalo, K. A novel vascular endothelial growth factor, VEGF-C, is a ligand for the Flt4 (VEGFR-3) and KDR (VEGFR-2) receptor tyrosine kinases. Embo J. 1996, 15, 290–298. [Google Scholar] [CrossRef]
- Achen, M.G.; Jeltsch, M.; Kukk, E.; Mäkinen, T.; Vitali, A.; Wilks, A.F.; Alitalo, K.; Stacker, S.A. Vascular endothelial growth factor D (VEGF-D) is a ligand for the tyrosine kinases VEGF receptor 2 (Flk1) and VEGF receptor 3 (Flt4). Proc. Natl. Acad. Sci. USA 1998, 95, 548–553. [Google Scholar] [CrossRef]
- Zhao, C.; Xiao, H.; Wu, X.; Li, C.; Liang, G.; Yang, S.; Lin, J. Rational combination of MEK inhibitor and the STAT3 pathway modulator for the therapy in K-Ras mutated pancreatic and colon cancer cells. Oncotarget 2015, 6, 14472–14487. [Google Scholar] [CrossRef] [PubMed]
- Cascio, S.; Ferla, R.; D’Andrea, A.; Gerbino, A.; Bazan, V.; Surmacz, E.; Russo, A. Expression of angiogenic regulators, VEGF and leptin, is regulated by the EGF/PI3K/STAT3 pathway in colorectal cancer cells. J. Cell Physiol. 2009, 221, 189–194. [Google Scholar] [CrossRef] [PubMed]
- Fan, F.; Wey, J.S.; McCarty, M.F.; Belcheva, A.; Liu, W.; Bauer, T.W.; Somcio, R.J.; Wu, Y.; Hooper, A.; Hicklin, D.J.; et al. Expression and function of vascular endothelial growth factor receptor-1 on human colorectal cancer cells. Oncogene 2005, 24, 2647–2653. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Kang, G.; Wang, T.; Huang, H. Tumor angiogenesis and anti-angiogenic gene therapy for cancer. Oncol. Lett. 2018, 16, 687–702. [Google Scholar] [CrossRef]
- Ellis, L.M.; Takahashi, Y.; Liu, W.; Shaheen, R.M. Vascular endothelial growth factor in human colon cancer: Biology and therapeutic implications. Oncologist 2000, 5 (Suppl. 1), 11–15. [Google Scholar] [CrossRef]
- Dvorak, H.F.; Nagy, J.A.; Feng, D.; Brown, L.F.; Dvorak, A.M. Vascular permeability factor/vascular endothelial growth factor and the significance of microvascular hyperpermeability in angiogenesis. Curr. Top Microbiol. Immunol. 1999, 237, 97–132. [Google Scholar] [CrossRef]
- Yoshida, N.; Kinugasa, T.; Ohshima, K.; Yuge, K.; Ohchi, T.; Fujino, S.; Shiraiwa, S.; Katagiri, M.; Akagi, Y. Analysis of Wnt and β-catenin Expression in Advanced Colorectal Cancer. Anticancer. Res. 2015, 35, 4403–4410. [Google Scholar]
- Lu, M.L.; Zhang, Y.; Li, J.; Fu, Y.; Li, W.H.; Zhao, G.F.; Li, X.H.; Wei, L.; Liu, G.B.; Huang, H. MicroRNA-124 inhibits colorectal cancer cell proliferation and suppresses tumor growth by interacting with PLCB1 and regulating Wnt/β-catenin signaling pathway. Eur. Rev. Med. Pharm. Sci. 2019, 23, 121–136. [Google Scholar] [CrossRef]
- Shan, Z.; An, N.; Qin, J.; Yang, J.; Sun, H.; Yang, W. Long non-coding RNA Linc00675 suppresses cell proliferation and metastasis in colorectal cancer via acting on miR-942 and Wnt/β-catenin signaling. Biomed. Pharm. 2018, 101, 769–776. [Google Scholar] [CrossRef]
- Kasprzak, A. Angiogenesis-Related Functions of Wnt Signaling in Colorectal Carcinogenesis. Cancers 2020, 12, 3601. [Google Scholar] [CrossRef]
- Calviello, G.; Resci, F.; Serini, S.; Piccioni, E.; Toesca, A.; Boninsegna, A.; Monego, G.; Ranelletti, F.O.; Palozza, P. Docosahexaenoic acid induces proteasome-dependent degradation of beta-catenin, down-regulation of survivin and apoptosis in human colorectal cancer cells not expressing COX-2. Carcinogenesis 2007, 28, 1202–1209. [Google Scholar] [CrossRef] [PubMed]
- Overall, C.M.; Kleifeld, O. Tumour microenvironment—Opinion: Validating matrix metalloproteinases as drug targets and anti-targets for cancer therapy. Nat. Rev. Cancer 2006, 6, 227–239. [Google Scholar] [CrossRef] [PubMed]
- Roy, R.; Zhang, B.; Moses, M.A. Making the cut: Protease-mediated regulation of angiogenesis. Exp. Cell Res. 2006, 312, 608–622. [Google Scholar] [CrossRef] [PubMed]
- Stetler-Stevenson, W.G.; Hewitt, R.; Corcoran, M. Matrix metalloproteinases and tumor invasion: From correlation and causality to the clinic. Semin. Cancer Biol. 1996, 7, 147–154. [Google Scholar] [CrossRef] [PubMed]
- Newell, K.J.; Witty, J.P.; Rodgers, W.H.; Matrisian, L.M. Expression and localization of matrix-degrading metalloproteinases during colorectal tumorigenesis. Mol. Carcinog. 1994, 10, 199–206. [Google Scholar] [CrossRef]
- Masaki, T.; Sugiyama, M.; Matsuoka, H.; Abe, N.; Izumisato, Y.; Sakamoto, A.; Atomi, Y. Matrix metalloproteinases may contribute compensationally to tumor invasion in T1 colorectal carcinomas. Anticancer. Res. 2003, 23, 4169–4173. [Google Scholar]
- Quintero-Fabián, S.; Arreola, R.; Becerril-Villanueva, E.; Torres-Romero, J.C.; Arana-Argáez, V.; Lara-Riegos, J.; Ramírez-Camacho, M.A.; Alvarez-Sánchez, M.E. Role of Matrix Metalloproteinases in Angiogenesis and Cancer. Front. Oncol. 2019, 9, 1370. [Google Scholar] [CrossRef]
- Brabletz, T.; Jung, A.; Dag, S.; Hlubek, F.; Kirchner, T. Beta-catenin regulates the expression of the matrix metalloproteinase-7 in human colorectal cancer. Am. J. Pathol. 1999, 155, 1033–1038. [Google Scholar] [CrossRef]
- Maurel, J.; Nadal, C.; Garcia-Albeniz, X.; Gallego, R.; Carcereny, E.; Almendro, V.; Mármol, M.; Gallardo, E.; Maria Augé, J.; Longarón, R.; et al. Serum matrix metalloproteinase 7 levels identifies poor prognosis advanced colorectal cancer patients. Int. J. Cancer 2007, 121, 1066–1071. [Google Scholar] [CrossRef]
- Ito, T.K.; Ishii, G.; Saito, S.; Yano, K.; Hoshino, A.; Suzuki, T.; Ochiai, A. Degradation of soluble VEGF receptor-1 by MMP-7 allows VEGF access to endothelial cells. Blood 2009, 113, 2363–2369. [Google Scholar] [CrossRef]
- Crawford, H.C.; Scoggins, C.R.; Washington, M.K.; Matrisian, L.M.; Leach, S.D. Matrix metalloproteinase-7 is expressed by pancreatic cancer precursors and regulates acinar-to-ductal metaplasia in exocrine pancreas. J. Clin. Investig. 2002, 109, 1437–1444. [Google Scholar] [CrossRef] [PubMed]
- Kurmyshkina, O.; Kovchur, P.; Schegoleva, L.; Volkova, T. Markers of Angiogenesis, Lymphangiogenesis, and Epithelial-Mesenchymal Transition (Plasticity) in CIN and Early Invasive Carcinoma of the Cervix: Exploring Putative Molecular Mechanisms Involved in Early Tumor Invasion. Int. J. Mol. Sci. 2020, 21, 6515. [Google Scholar] [CrossRef] [PubMed]
- Lizárraga-Verdugo, E.; Avendaño-Félix, M.; Bermúdez, M.; Ramos-Payán, R.; Pérez-Plasencia, C.; Aguilar-Medina, M. Cancer Stem Cells and Its Role in Angiogenesis and Vasculogenic Mimicry in Gastrointestinal Cancers. Front. Oncol. 2020, 10, 413. [Google Scholar] [CrossRef] [PubMed]
- Shangguan, W.; Fan, C.; Chen, X.; Lu, R.; Liu, Y.; Li, Y.; Shang, Y.; Yin, D.; Zhang, S.; Huang, Q.; et al. Endothelium originated from colorectal cancer stem cells constitute cancer blood vessels. Cancer Sci. 2017, 108, 1357–1367. [Google Scholar] [CrossRef]
- Gao, R.; Kim, C.; Sei, E.; Foukakis, T.; Crosetto, N.; Chan, L.K.; Srinivasan, M.; Zhang, H.; Meric-Bernstam, F.; Navin, N. Nanogrid single-nucleus RNA sequencing reveals phenotypic diversity in breast cancer. Nat. Commun. 2017, 8, 228. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.; Scurrah, C.R.; McKinley, E.T.; Simmons, A.J.; Ramirez-Solano, M.A.; Zhu, X.; Markham, N.O.; Heiser, C.N.; Vega, P.N.; Rolong, A.; et al. Differential pre-malignant programs and microenvironment chart distinct paths to malignancy in human colorectal polyps. Cell 2021, 184, 6262–6280.e26. [Google Scholar] [CrossRef]
- Mottaghi, S.; Abbaszadeh, H. Natural Lignans Honokiol and Magnolol as Potential Anticarcinogenic and Anticancer Agents. A Comprehensive Mechanistic Review. Nutr. Cancer 2022, 74, 761–778. [Google Scholar] [CrossRef]
- Newman, D.J.; Cragg, G.M. Natural Products as Sources of New Drugs over the nearly Four Decades from 01/1981 to 09/2019. J. Nat. Prod. 2020, 83, 770–803. [Google Scholar] [CrossRef]
- Xue, Z.; Yan, R.; Yang, B. Phenylethanoid glycosides and phenolic glycosides from stem bark of Magnolia officinalis. Phytochemistry 2016, 127, 50–62. [Google Scholar] [CrossRef]
- Kuo, W.L.; Chung, C.Y.; Hwang, T.L.; Chen, J.J. Biphenyl-type neolignans from Magnolia officinalis and their anti-inflammatory activities. Phytochemistry 2013, 85, 153–160. [Google Scholar] [CrossRef]
- Fu, Y.; Liu, B.; Zhang, N.; Liu, Z.; Liang, D.; Li, F.; Cao, Y.; Feng, X.; Zhang, X.; Yang, Z. Magnolol inhibits lipopolysaccharide-induced inflammatory response by interfering with TLR4 mediated NF-κB and MAPKs signaling pathways. J. Ethnopharmacol. 2013, 145, 193–199. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.C.; Liu, F.L.; Chen, C.L.; Chen, T.C.; Chang, D.M.; Huang, H.S. Discovery of 5-(2′,4′-difluorophenyl)-salicylanilides as new inhibitors of receptor activator of NF-κB ligand (RANKL)-induced osteoclastogenesis. Eur. J. Med. Chem. 2015, 98, 115–126. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Gharavi, R.; Pitta, M.; Gleichmann, M.; Mattson, M.P. Nicotinamide prevents NAD+ depletion and protects neurons against excitotoxicity and cerebral ischemia: NAD+ consumption by SIRT1 may endanger energetically compromised neurons. Neuromol. Med. 2009, 11, 28–42. [Google Scholar] [CrossRef] [PubMed]
- Ma, R.; Ma, Z.G.; Zhen, C.L.; Shen, X.; Li, S.L.; Li, L.; Zheng, Y.F.; Dong, D.L.; Sun, Z.J. Design, synthesis and characterization of poly (methacrylic acid-niclosamide) and its effect on arterial function. Mater. Sci. Eng. C Mater. Biol Appl. 2017, 77, 352–359. [Google Scholar] [CrossRef]
- Lee, C.C.; Liu, F.L.; Chen, C.L.; Chen, T.C.; Liu, F.C.; Ahmed Ali, A.A.; Chang, D.M.; Huang, H.S. Novel inhibitors of RANKL-induced osteoclastogenesis: Design, synthesis, and biological evaluation of 6-(2,4-difluorophenyl)-3-phenyl-2H-benzo[e][1,3]oxazine-2,4(3H)-diones. Bioorg. Med. Chem. 2015, 23, 4522–4532. [Google Scholar] [CrossRef]
- Kellam, P. Microarray gene expression database: Progress towards an international repository of gene expression data. Genome Biol. 2001, 2, 4011. [Google Scholar] [CrossRef]
- Edgar, R.; Domrachev, M.; Lash, A.E. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002, 30, 207–210. [Google Scholar] [CrossRef]
- Sayagués, J.M.; Corchete, L.A.; Gutiérrez, M.L.; Sarasquete, M.E.; Del Mar Abad, M.; Bengoechea, O.; Fermiñán, E.; Anduaga, M.F.; Del Carmen, S.; Iglesias, M.; et al. Genomic characterization of liver metastases from colorectal cancer patients. Oncotarget 2016, 7, 72908–72922. [Google Scholar] [CrossRef]
- Vlachavas, E.I.; Pilalis, E.; Papadodima, O.; Koczan, D.; Willis, S.; Klippel, S.; Cheng, C.; Pan, L.; Sachpekidis, C.; Pintzas, A.; et al. Radiogenomic Analysis of F-18-Fluorodeoxyglucose Positron Emission Tomography and Gene Expression Data Elucidates the Epidemiological Complexity of Colorectal Cancer Landscape. Comput. Struct. Biotechnol. J. 2019, 17, 177–185. [Google Scholar] [CrossRef]
- Daina, A.; Michielin, O.; Zoete, V. Swiss ADME: A free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017, 7, 42717. [Google Scholar] [CrossRef]
- Lipinski, C.A.; Lombardo, F.; Dominy, B.W.; Feeney, P.J. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev. 2001, 46, 3–26. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Wang, L.; Lv, M.; Pei, R.; Li, P.; Pei, Z.; Wang, Y.; Su, W.; Xie, X.Q. Alz Platform: An Alzheimer’s disease domain-specific chemogenomics knowledgebase for polypharmacology and target identification research. J. Chem. Inf. Model. 2014, 54, 1050–1060. [Google Scholar] [CrossRef] [PubMed]
- Gfeller, D.; Grosdidier, A.; Wirth, M.; Daina, A.; Michielin, O.; Zoete, V. Swiss Target Prediction: A web server for target prediction of bioactive small molecules. Nucleic Acids Res. 2014, 42, W32–W38. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Zhou, Z.; Yang, Q.; Yang, K. Differential expression of miR-30a-5p in post stroke depression and bioinformatics analysis of the possible mechanism. Nan Fang Yi Ke Da Xue Xue Bao 2020, 40, 922–929. [Google Scholar] [CrossRef]
- Huang, D.W.; Sherman, B.T.; Tan, Q.; Collins, J.R.; Alvord, W.G.; Roayaei, J.; Stephens, R.; Baseler, M.W.; Lane, H.C.; Lempicki, R.A. The DAVID Gene Functional Classification Tool: A novel biological module-centric algorithm to functionally analyze large gene lists. Genome Biol. 2007, 8, R183. [Google Scholar] [CrossRef]
- Pathan, M.; Keerthikumar, S.; Ang, C.S.; Gangoda, L.; Quek, C.Y.; Williamson, N.A.; Mouradov, D.; Sieber, O.M.; Simpson, R.J.; Salim, A.; et al. Fun Rich: An open access standalone functional enrichment and interaction network analysis tool. Proteomics 2015, 15, 2597–2601. [Google Scholar] [CrossRef]
- Zhou, G.; Soufan, O.; Ewald, J.; Hancock, R.E.W.; Basu, N.; Xia, J. Network Analyst 3.0: A visual analytics platform for comprehensive gene expression profiling and meta-analysis. Nucleic Acids Res. 2019, 47, W234–W241. [Google Scholar] [CrossRef]
- Xia, J.; Benner, M.J.; Hancock, R.E. Network Analyst—Integrative approaches for protein-protein interaction network analysis and visual exploration. Nucleic Acids Res. 2014, 42, W167–W174. [Google Scholar] [CrossRef]
- Li, T.; Fan, J.; Wang, B.; Traugh, N.; Chen, Q.; Liu, J.S.; Li, B.; Liu, X.S. TIMER: A Web Server for Comprehensive Analysis of Tumor-Infiltrating Immune Cells. Cancer Res. 2017, 77, e108–e110. [Google Scholar] [CrossRef]
- Tang, R.; Zhang, Y.; Liang, C.; Xu, J.; Meng, Q.; Hua, J.; Liu, J.; Zhang, B.; Yu, X.; Shi, S. The role of m6A-related genes in the prognosis and immune microenvironment of pancreatic adenocarcinoma. PeerJ 2020, 8, e9602. [Google Scholar] [CrossRef]
- Lee, H.O.; Hong, Y.; Etlioglu, H.E.; Cho, Y.B.; Pomella, V.; Van den Bosch, B.; Vanhecke, J.; Verbandt, S.; Hong, H.; Min, J.W.; et al. Lineage-dependent gene expression programs influence the immune landscape of colorectal cancer. Nat. Genet. 2020, 52, 594–603. [Google Scholar] [CrossRef] [PubMed]
- Mokgautsi, N.; Wen, Y.T.; Lawal, B.; Khedkar, H.; Sumitra, M.R.; Wu, A.T.H.; Huang, H.S. An Integrated Bioinformatics Study of a Novel Niclosamide Derivative, NSC765689, a Potential GSK3β/β-Catenin/STAT3/CD44 Suppressor with Anti-Glioblastoma Properties. Int. J. Mol. Sci. 2021, 22, 2464. [Google Scholar] [CrossRef] [PubMed]
- Shoemaker, R.H. The NCI60 human tumour cell line anticancer drug screen. Nat. Rev. Cancer 2006, 6, 813–823. [Google Scholar] [CrossRef] [PubMed]
- Rehman, S.K.; Haynes, J.; Collignon, E.; Brown, K.R.; Wang, Y.; Nixon, A.M.L.; Bruce, J.P.; Wintersinger, J.A.; Singh Mer, A.; Lo, E.B.L.; et al. Colorectal Cancer Cells Enter a Diapause-like DTP State to Survive Chemotherapy. Cell 2021, 184, 226–242.e21. [Google Scholar] [CrossRef] [PubMed]
- Mokgautsi, N.; Wang, Y.C.; Lawal, B.; Khedkar, H.; Sumitra, M.R.; Wu, A.T.H.; Huang, H.S. Network Pharmacological Analysis through a Bioinformatics Approach of Novel NSC765600 and NSC765691 Compounds as Potential Inhibitors of CCND1/CDK4/PLK1/CD44 in Cancer Types. Cancers 2021, 13, 2523. [Google Scholar] [CrossRef]
- Cousins, K.R. Computer review of Chem Draw Ultra 12.0. J. Am. Chem. Soc. 2011, 133, 8388. [Google Scholar] [CrossRef]
- Seeliger, D.; de Groot, B.L. Ligand docking and binding site analysis with PyMOL and Autodock/Vina. J. Comput. Aided. Mol. Des. 2010, 24, 417–422. [Google Scholar] [CrossRef]
- Dotse, E.; Bian, Y. Isolation of colorectal cancer stem-like cells. Cytotechnology 2016, 68, 609–619. [Google Scholar] [CrossRef]
- Brenner, H.; Kloor, M.; Pox, C.P. Colorectal cancer. Lancet 2014, 383, 1490–1502. [Google Scholar] [CrossRef]
- Azcárate-Peril, M.A.; Sikes, M.; Bruno-Bárcena, J.M. The intestinal microbiota, gastrointestinal environment and colorectal cancer: A putative role for probiotics in prevention of colorectal cancer? Am. J. Physiol. Gastrointest Liver Physiol. 2011, 301, G401–G424. [Google Scholar] [CrossRef]
- Zugazagoitia, J.; Guedes, C.; Ponce, S.; Ferrer, I.; Molina-Pinelo, S.; Paz-Ares, L. Current Challenges in Cancer Treatment. Clin. Ther. 2016, 38, 1551–1566. [Google Scholar] [CrossRef] [PubMed]
- Zeuner, A.; Todaro, M.; Stassi, G.; De Maria, R. Colorectal cancer stem cells: From the crypt to the clinic. Cell Stem. Cell 2014, 15, 692–705. [Google Scholar] [CrossRef] [PubMed]
- Marijnen, C.A.; Kapiteijn, E.; van de Velde, C.J.; Martijn, H.; Steup, W.H.; Wiggers, T.; Kranenbarg, E.K.; Leer, J.W. Acute side effects and complications after short-term preoperative radiotherapy combined with total mesorectal excision in primary rectal cancer: Report of a multicenter randomized trial. J. Clin. Oncol. 2002, 20, 817–825. [Google Scholar] [CrossRef] [PubMed]
- McDermott, D.F.; Huseni, M.A.; Atkins, M.B.; Motzer, R.J.; Rini, B.I.; Escudier, B.; Fong, L.; Joseph, R.W.; Pal, S.K.; Reeves, J.A.; et al. Clinical activity and molecular correlates of response to atezolizumab alone or in combination with bevacizumab versus sunitinib in renal cell carcinoma. Nat. Med. 2018, 24, 749–757. [Google Scholar] [CrossRef]
- Schmieder, R.; Hoffmann, J.; Becker, M.; Bhargava, A.; Müller, T.; Kahmann, N.; Ellinghaus, P.; Adams, R.; Rosenthal, A.; Thierauch, K.H.; et al. Regorafenib (BAY 73-4506): Antitumor and antimetastatic activities in preclinical models of colorectal cancer. Int. J. Cancer 2014, 135, 1487–1496. [Google Scholar] [CrossRef]
- Fernando, N.H.; Hurwitz, H.I. Inhibition of vascular endothelial growth factor in the treatment of colorectal cancer. Semin. Oncol. 2003, 30, 39–50. [Google Scholar] [CrossRef]
- Ferrara, N. VEGF and the quest for tumour angiogenesis factors. Nat. Rev. Cancer 2002, 2, 795–803. [Google Scholar] [CrossRef]
- Hu, T.H.; Yao, Y.; Yu, S.; Han, L.L.; Wang, W.J.; Guo, H.; Tian, T.; Ruan, Z.P.; Kang, X.M.; Wang, J.; et al. SDF-1/CXCR4 promotes epithelial-mesenchymal transition and progression of colorectal cancer by activation of the Wnt/β-catenin signaling pathway. Cancer Lett. 2014, 354, 417–426. [Google Scholar] [CrossRef]
- Woenne, E.C.; Lederle, W.; Zwick, S.; Palmowski, M.; Krell, H.; Semmler, W.; Mueller, M.M.; Kiessling, F. MMP inhibition blocks fibroblast-dependent skin cancer invasion, reduces vascularization and alters VEGF-A and PDGF-BB expression. Anticancer 2010, 30, 703–711. [Google Scholar]
- Deryugina, E.I.; Quigley, J.P. Tumor angiogenesis: MMP-mediated induction of intravasation- and metastasis-sustaining neovasculature. Matrix Biol. 2015, 44–46, 94–112. [Google Scholar] [CrossRef]
- Kumar, A.; Cherukumilli, M.; Mahmoudpour, S.H.; Brand, K.; Bandapalli, O.R. ShRNA-mediated knock-down of CXCL8 inhibits tumor growth in colorectal liver metastasis. Biochem. Biophys. Res. Commun. 2018, 500, 731–737. [Google Scholar] [CrossRef] [PubMed]
- Takada, K.; Zhu, D.; Bird, G.H.; Sukhdeo, K.; Zhao, J.J.; Mani, M.; Lemieux, M.; Carrasco, D.E.; Ryan, J.; Horst, D.; et al. Targeted disruption of the BCL9/β-catenin complex inhibits oncogenic Wnt signaling. Sci. Transl. Med. 2012, 4, 148ra117. [Google Scholar] [CrossRef] [PubMed]
- Cheng, X.; Xu, X.; Chen, D.; Zhao, F.; Wang, W. Therapeutic potential of targeting the Wnt/β-catenin signaling pathway in colorectal cancer. Biomed. Pharm. 2019, 110, 473–481. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.H.; Chen, Y.X.; Fang, J.Y. Comprehensive review of targeted therapy for colorectal cancer. Signal. Transduct Target. Ther. 2020, 5, 22. [Google Scholar] [CrossRef] [PubMed]
- White, B.D.; Chien, A.J.; Dawson, D.W. Dysregulation of Wnt/β-catenin signaling in gastrointestinal cancers. Gastroenterology 2012, 142, 219–232. [Google Scholar] [CrossRef] [PubMed]
- Wilson, C.L.; Matrisian, L.M. Matrilysin: An epithelial matrix metalloproteinase with potentially novel functions. Int. J. Biochem. Cell Biol. 1996, 28, 123–136. [Google Scholar] [CrossRef]
- Adachi, Y.; Yamamoto, H.; Itoh, F.; Hinoda, Y.; Okada, Y.; Imai, K. Contribution of matrilysin (MMP-7) to the metastatic pathway of human colorectal cancers. Gut 1999, 45, 252–258. [Google Scholar] [CrossRef]
- Jeffery, N.; McLean, M.H.; El-Omar, E.M.; Murray, G.I. The matrix metalloproteinase/tissue inhibitor of matrix metalloproteinase profile in colorectal polyp cancers. Histopathology 2009, 54, 820–828. [Google Scholar] [CrossRef]
- Fang, Y.J.; Lu, Z.H.; Wang, G.Q.; Pan, Z.Z.; Zhou, Z.W.; Yun, J.P.; Zhang, M.F.; Wan, D.S. Elevated expressions of MMP7, TROP2, and survivin are associated with survival, disease recurrence, and liver metastasis of colon cancer. Int. J. Color. Dis. 2009, 24, 875–884. [Google Scholar] [CrossRef]
- Sharma, P.; Allison, J.P. Immune checkpoint targeting in cancer therapy: Toward combination strategies with curative potential. Cell 2015, 161, 205–214. [Google Scholar] [CrossRef]
- De Simone, M.; Arrigoni, A.; Rossetti, G.; Gruarin, P.; Ranzani, V.; Politano, C.; Bonnal, R.J.P.; Provasi, E.; Sarnicola, M.L.; Panzeri, I.; et al. Transcriptional Landscape of Human Tissue Lymphocytes Unveils Uniqueness of Tumor-Infiltrating T Regulatory Cells. Immunity 2016, 45, 1135–1147. [Google Scholar] [CrossRef] [PubMed]







| Target | Common Name | Uniprot ID | ChEMBL ID | Target Class |
|---|---|---|---|---|
| Cyclin-dependent kinase 9 | CDK9 | P50750 | CHEMBL3116 | Kinase |
| Tyrosine-protein kinase JAK3 | JAK3 | P52333 | CHEMBL2148 | Kinase |
| Vascular endothelial growth factor-α | VEGFA | P00533 | CHEMBL203 | Kinase |
| Glycogen synthase kinase-3β | GSK3β | P49841 | CHEMBL262 | Kinase |
| Mitogen-activated protein kinase | MAPK14 | Q16539 | CHEMBL260 | Kinase |
| Signal transducer and activator of transcription 3 | STAT3 | P40763 | CHEMBL4026 | Transcription factor |
| Cyclin-dependent kinase 1 Catenin β1 | CDK1 CTNNB1 | P06493 P36861 | CHEMBL308 CHEMBL309 | Kinase Kinase |
| MYC proto-oncogene | MYC | P35557 | CHEMBL3820 | Enzyme |
| Matrix metalloproteinase 7 | MMP7 | P14780 | CHEMBL321 | Enzyme |
| VEGFA-LCC-21 Complex | (Δ = −8.1 kcal/mol) | CTNNB1-LCC-21 Complex | (Δ = −8.2 kcal/mol) |
|---|---|---|---|
| Type of interactions and number of bonds | distance of interacting Amino acids | Type of interactions and number of bonds | distance of interacting Amino acids |
| Conventional Hydrogen bond (3) | LEU (2.48 Å), CYS61 (2.04 Å), ASP63 (2.39 Å) | Conventional Hydrogen bond (5) | ARG457 (1.17 Å), THR418 (2.12 Å), ASN415 (1.87 Å), TRP383 ASN387 (3.17 Å) TRP383 (2.17 Å) |
| Van der Waals Forces | VAL33, LEU33, SER50, GLU64, ILE48, LYS48, ASN62 | Van der Waals Forces | ILE460, GLU24, LYS348 |
| Halogen | GLY59 | Carbon hydrogen bond | GLY25 |
| Pi–pi anion | ASP34 | Pi-cation | ASP387 |
| Amide pi-stacked | ASP63 | Pi-anion | ARG386 |
| Pi-Alkyl | CYS51, CYS60 | ||
| MMP7-LCC-21 Complex | (Δ = −9.0 kcal/mol) | CD44-LCC-21 Complex | (Δ = −8.0 kcal/mol) |
| Type of interactions and number of bonds | distance of interacting Amino acids | Type of interactions and number of bonds | distance of interacting Amino acids |
| Conventional Hydrogen bond (3) | LEU19 (2.24 Å),TRY172 (1.29 Å), ALA186 (2.00 Å) | Conventional Hydrogen bond (3) | ARG90 (3.64 Å), ASN94 (2.39 Å), GLN113 (3.00 Å) |
| Van der Waals Forces | HIS183, ALA184, GLY192, GLY190, THR189 | Van der Waals Forces | CYS77, THR76, PHE30, SER109, HIS92, SER112 |
| Pi–pi cation | PHE185 | Halogens | GLU75, GLY73 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mokgautsi, N.; Kuo, Y.-C.; Huang, Y.-J.; Chen, C.-H.; Mukhopadhyay, D.; Wu, A.T.H.; Huang, H.-S. Preclinical Evaluation of a Novel Small Molecule LCC-21 to Suppress Colorectal Cancer Malignancy by Inhibiting Angiogenic and Metastatic Signatures. Cells 2023, 12, 266. https://doi.org/10.3390/cells12020266
Mokgautsi N, Kuo Y-C, Huang Y-J, Chen C-H, Mukhopadhyay D, Wu ATH, Huang H-S. Preclinical Evaluation of a Novel Small Molecule LCC-21 to Suppress Colorectal Cancer Malignancy by Inhibiting Angiogenic and Metastatic Signatures. Cells. 2023; 12(2):266. https://doi.org/10.3390/cells12020266
Chicago/Turabian StyleMokgautsi, Ntlotlang, Yu-Cheng Kuo, Yan-Jiun Huang, Chien-Hsin Chen, Debabrata Mukhopadhyay, Alexander T. H. Wu, and Hsu-Shan Huang. 2023. "Preclinical Evaluation of a Novel Small Molecule LCC-21 to Suppress Colorectal Cancer Malignancy by Inhibiting Angiogenic and Metastatic Signatures" Cells 12, no. 2: 266. https://doi.org/10.3390/cells12020266
APA StyleMokgautsi, N., Kuo, Y.-C., Huang, Y.-J., Chen, C.-H., Mukhopadhyay, D., Wu, A. T. H., & Huang, H.-S. (2023). Preclinical Evaluation of a Novel Small Molecule LCC-21 to Suppress Colorectal Cancer Malignancy by Inhibiting Angiogenic and Metastatic Signatures. Cells, 12(2), 266. https://doi.org/10.3390/cells12020266








