Transcriptomic Signature of the Simulated Microgravity Response in Caenorhabditis elegans and Comparison to Spaceflight Experiments
Abstract
:1. Introduction
2. Materials and Methods
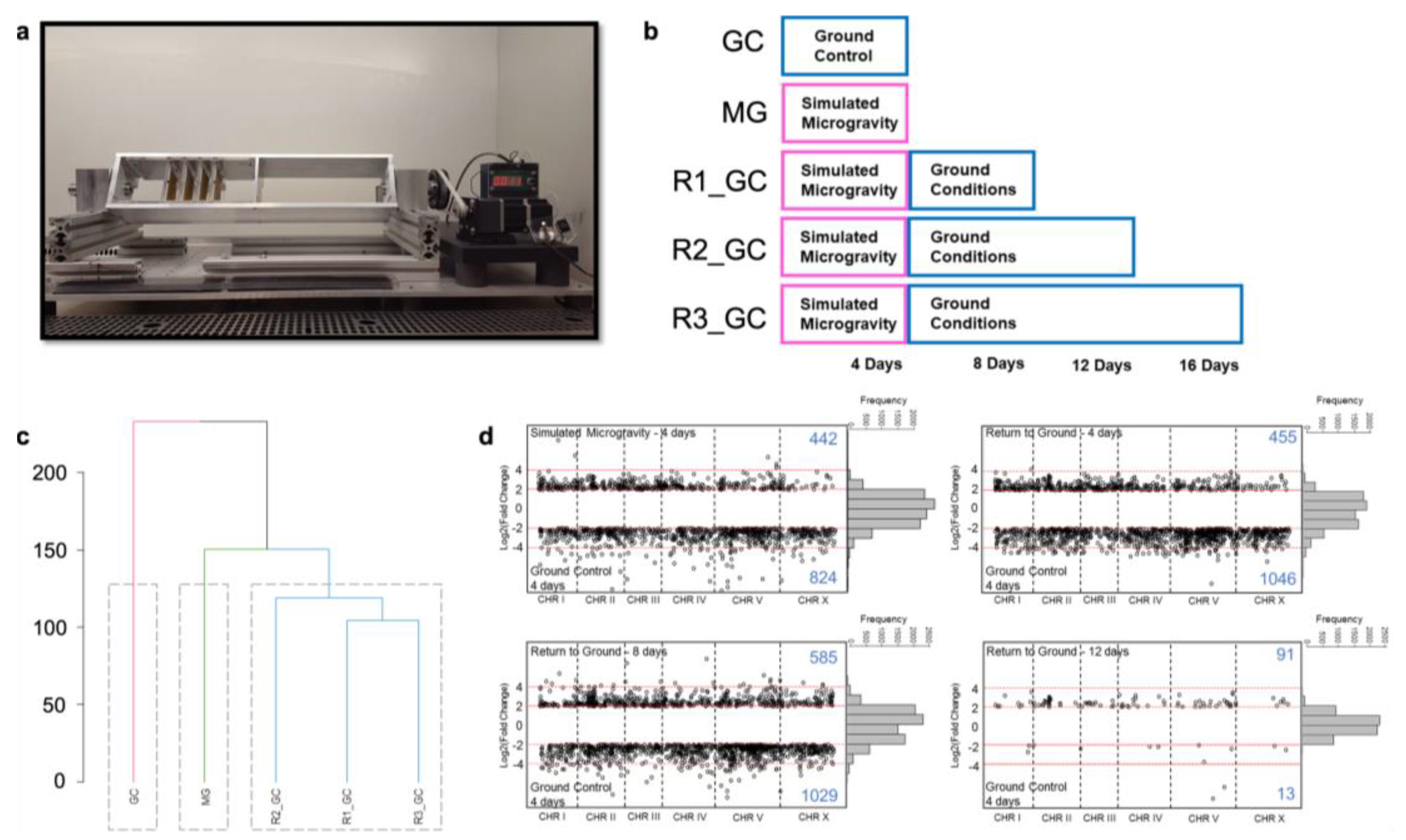
2.1. C. elegans Strain and Growth Conditions
2.2. RNA Isolation, Illumina Sequencing
2.3. Gene Expression Analysis
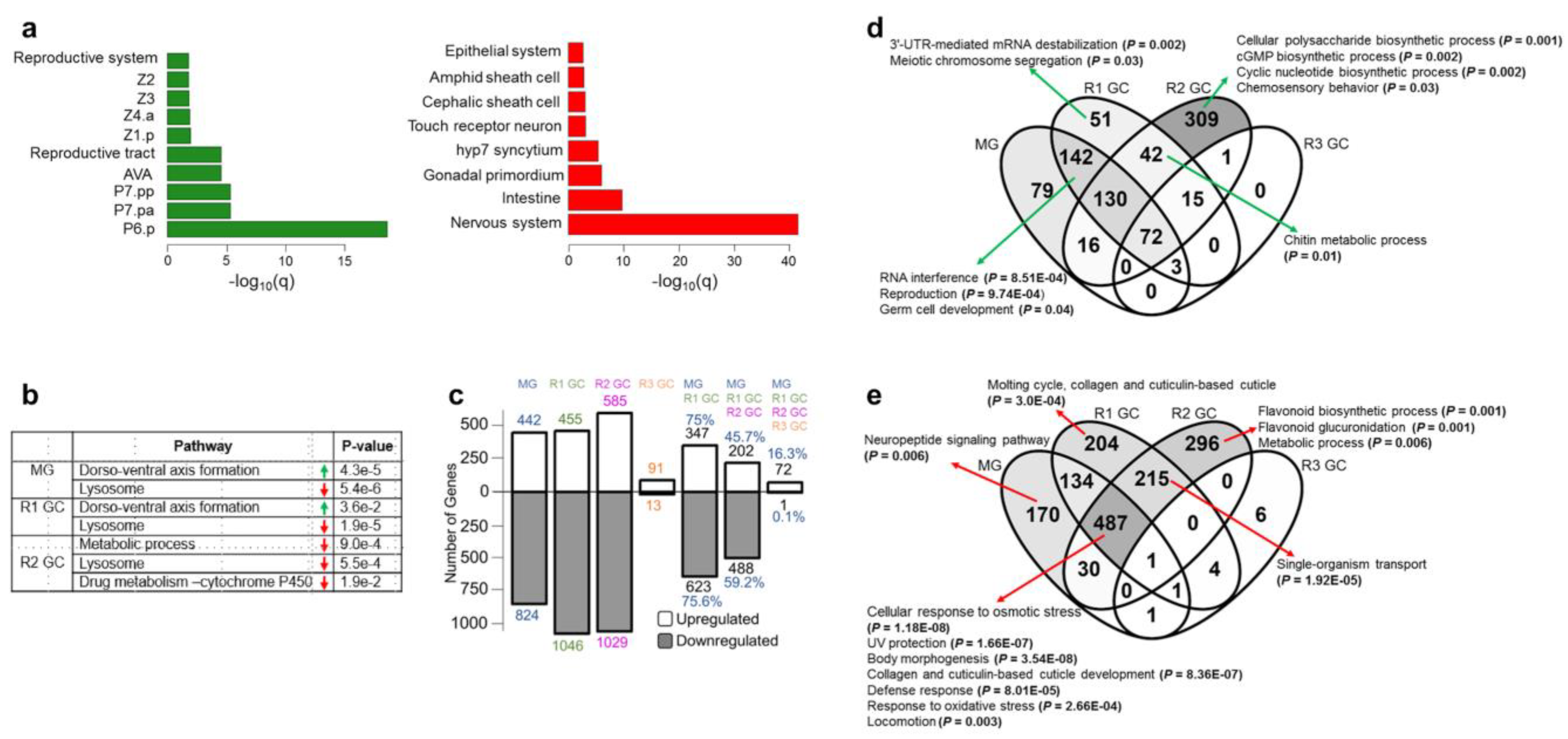
2.4. Functional Analysis of the Genes
2.5. Comparison of DEGs to the CERISE Four Days
2.6. Mass Spectrometry for Ceramides
2.7. Microgravity Simulation with Clinorotation
3. Results
3.1. Simulated Microgravity Triggers Differential Expression of Hundreds of Genes
3.2. Simulated Microgravity-Induced Gene Expression Differences Are Highly Maintained for Eight Days after Return to Ground Conditions
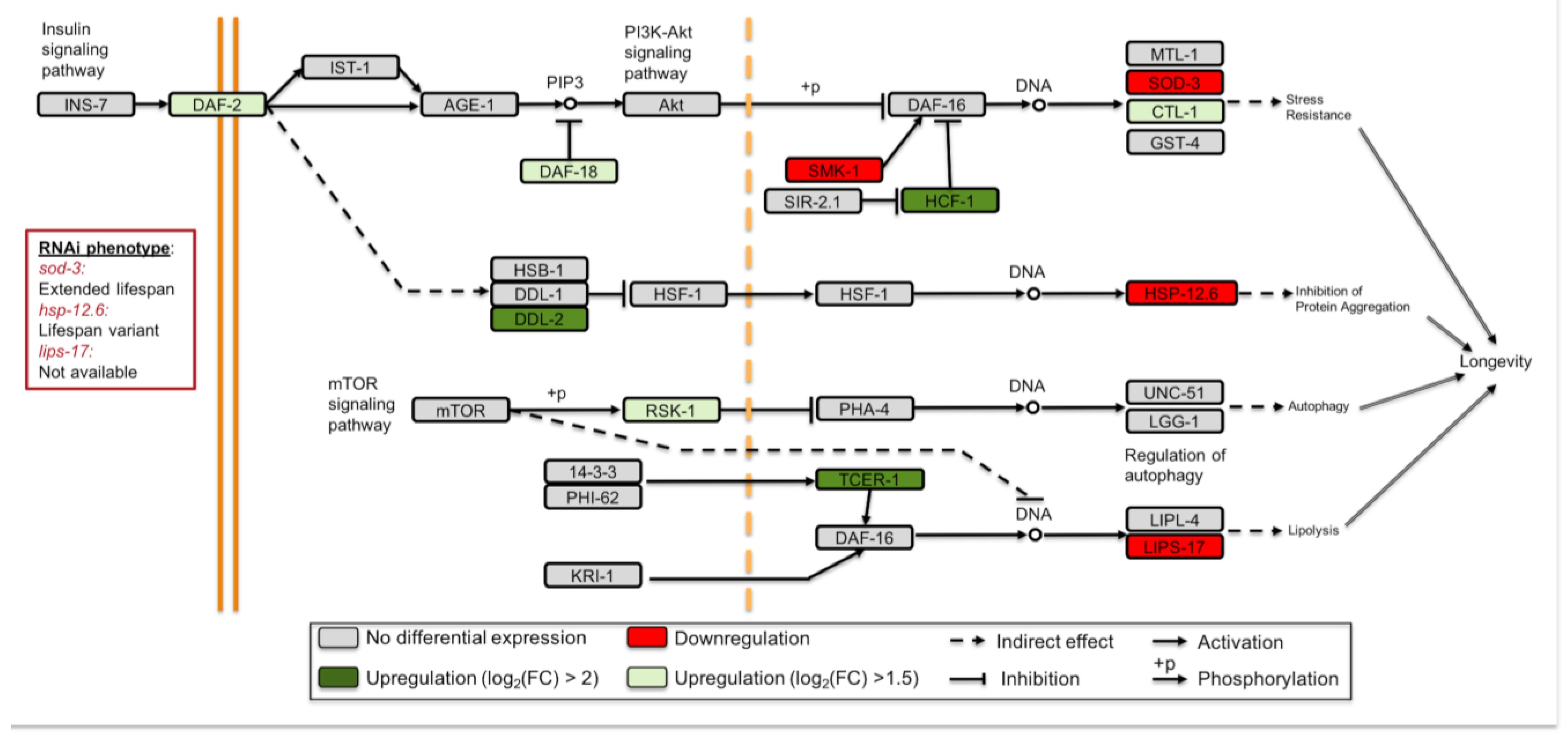
3.3. Longevity Regulating Pathways Are Affected under Simulated Microgravity
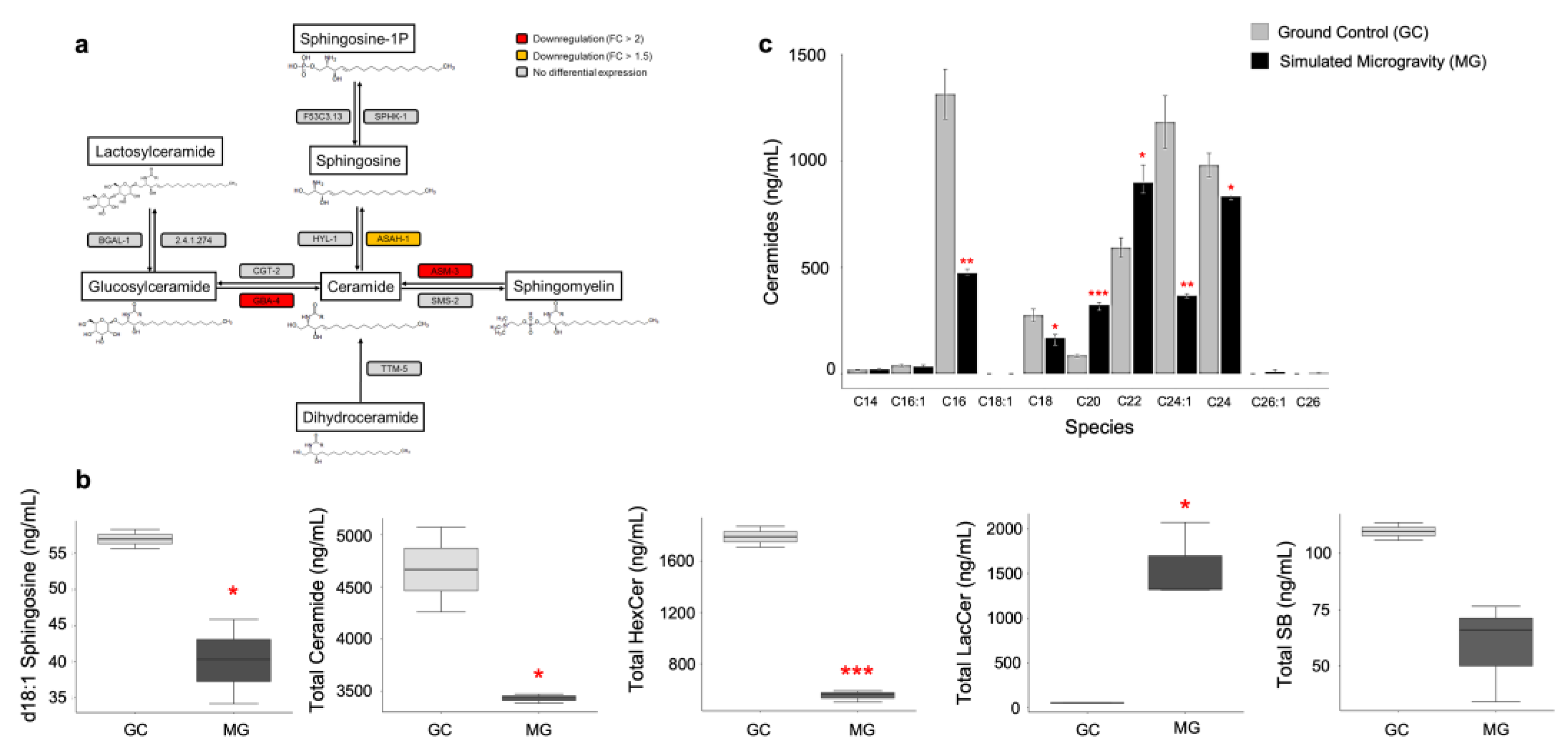
3.4. Sphingolipid Signaling Pathway Is Suppressed in Response to Simulated Microgravity
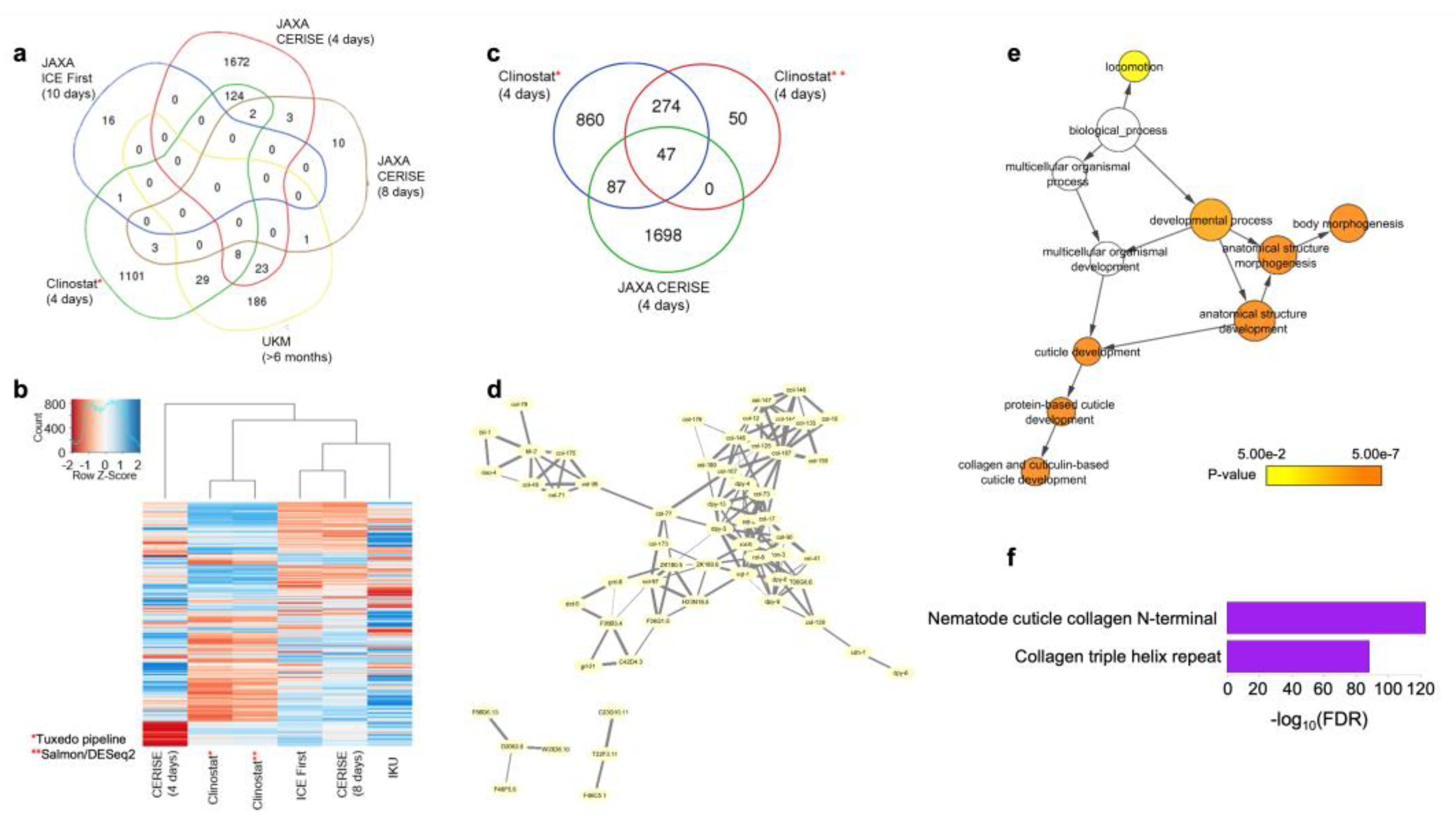
3.5. Identification of the Common Microgravity-Responsive Genes between Four-Day CERISE and Simulated Microgravity
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Stein, T.P. Weight, Muscle and Bone Loss during Space Flight: Another Perspective. Eur. J. Appl. Physiol. 2013, 113, 2171–2181. [Google Scholar] [CrossRef] [PubMed]
- Crucian, B.E.; Choukèr, A.; Simpson, R.J.; Mehta, S.; Marshall, G.; Smith, S.M.; Zwart, S.R.; Heer, M.; Ponomarev, S.; Whitmire, A.; et al. Immune System Dysregulation during Spaceflight: Potential Countermeasures for Deep Space Exploration Missions. Front. Immunol. 2018, 9, 1437. [Google Scholar] [CrossRef] [PubMed]
- Grimm, D.; Grosse, J.; Wehland, M.; Mann, V.; Reseland, J.E.; Sundaresan, A.; Corydon, T.J. The Impact of Microgravity on Bone in Humans. Bone 2016, 87, 44–56. [Google Scholar] [CrossRef]
- Sachs, J. Lectures on the Physiology of Plants; Clarendon Press: Oxford, UK, 1887. [Google Scholar]
- Kessler, J. The Internal Dynamics of Slowly Rotating Biological Systems. ASGSB Bull. 1992, 5, 11–21. [Google Scholar] [PubMed]
- Dedolph, R.R.; Oemick, D.A.; Wilson, B.R.; Smith, G.R. Causal Basis of Gravity Stimulus Nullification by Clinostat Rotation. Plant Physiol. 1967, 42, 1373–1383. [Google Scholar] [CrossRef] [Green Version]
- Gruener, R.; Hoeger, G. Vector-Averaged Gravity Alters Myocyte and Neuron Properties in Cell Culture. Aviat. Space. Environ. Med. 1991, 62, 1159–1165. [Google Scholar]
- Wuest, S.L.; Stern, P.; Casartelli, E.; Egli, M. Fluid Dynamics Appearing during Simulated Microgravity Using Random Positioning Machines. PLoS ONE 2017, 12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sievers, A.; Hejnowicz, Z. How Well Does the Clinostat Mimic the Effect of Microgravity on Plant Cells and Organs? ASGSB Bull. 1992, 5, 69–75. [Google Scholar]
- Brown, A.H.; Dahl, A.O.; Chapman, D.K. Limitation on the Use of the Horizontal Clinostat as a Gravity Compensator. PLANT Physiol. 1976, 58, 127–130. [Google Scholar] [CrossRef] [Green Version]
- Briegleb, W. Some Qualitative and Quantitative Aspects of the Fast-Rotating Clinostat as a Research Tool. ASGSB Bull. 1992, 5, 23–30. [Google Scholar]
- Hemmersbach, R.; Volkmann, D.; Häder, D.-P. Graviorientation in Protists and Plants. J. Plant Physiol. 1999, 154, 1–15. [Google Scholar] [CrossRef]
- Häder, D.-P.; Hemmersbach, R.; Lebert, M. Gravity and the Behavior of Unicellular Organisms; Cambridge University Press: Cambridge, UK, 2005; ISBN 9780511546211. [Google Scholar]
- Zhao, Y.; Johnsen, R.; Baillie, D.; Rose, A. Worms in Space? A Model Biological Dosimeter. Gravit. Space Biol. Bull 2005, 18, 11–16. [Google Scholar]
- Tissenbaum, H.A. Using C. elegans for Aging Research. Invertebr. Reprod. Dev. 2015, 59, 59–63. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Selch, F.; Higashibata, A.; Imamizo-Sato, M.; Higashitani, A.; Ishioka, N.; Szewczyk, N.J.; Conley, C.A. Genomic Response of the Nematode Caenorhabditis elegans to Spaceflight. Adv. Space Res. 2008, 41, 807–815. [Google Scholar] [CrossRef]
- Adenle, A.A.; Johnsen, B.; Szewczyk, N.J. Review of the Results from the International C. elegans First Experiment (ICE-FIRST). Adv. Sp. Res. 2009, 44, 210–216. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Higashibata, A.; Szewczyk, N.J.; Conley, C.A.; Imamizo-Sato, M.; Higashitani, A.; Ishioka, N. Decreased Expression of Myogenic Transcription Factors and Myosin Heavy Chains in Caenorhabditis elegans Muscles Developed during Spaceflight. J. Exp. Biol. 2006, 209, 3209–3218. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qiao, L.; Luo, S.; Liu, Y.; Li, X.; Wang, G.; Huang, Z. Reproductive and Locomotory Capacities of Caenorhabditis elegans Were Not Affected by Simulated Variable Gravities and Spaceflight during the Shenzhou-8 Mission. Astrobiology 2013, 13, 617–625. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Honda, Y.; Higashibata, A.; Matsunaga, Y.; Yonezawa, Y.; Kawano, T.; Higashitani, A.; Kuriyama, K.; Shimazu, T.; Tanaka, M.; Szewczyk, N.J.; et al. Genes Down-Regulated in Spaceflight Are Involved in the Control of Longevity in Caenorhabditis elegans. Sci. Rep. 2012, 2, 413–429. [Google Scholar] [CrossRef] [Green Version]
- Higashibata, A.; Hashizume, T.; Nemoto, K.; Higashitani, N.; Etheridge, T.; Mori, C.; Harada, S.; Sugimoto, T.; Szewczyk, N.J.; Baba, S.A.; et al. Microgravity Elicits Reproducible Alterations in Cytoskeletal and Metabolic Gene and Protein Expression in Space-Flown Caenorhabditis elegans. NPJ Microgravity 2016, 2, 15022. [Google Scholar] [CrossRef] [Green Version]
- Gao, Y.; Xu, D.; Zhao, L.; Zhang, M.; Sun, Y. Effects of Microgravity on DNA Damage Response in Caenorhabditis elegans during Shenzhou-8 Spaceflight. Int. J. Radiat. Biol. 2015, 91, 531–539. [Google Scholar] [CrossRef]
- Then, S.M.; Jusoh, N.F.; Harun, R.; Nathan, S.; Szewczyk, N.J.; Stodieck, L.; Jamal, R. Multi-Generational Culture of C. elegans on a Long-Term Space Flight Revealed Changes in Expression of Genes Involved in Longevity, DNA Repair, and Locomotion. Asia-Pac. J. Mol. Med. 2016, 4, 1. [Google Scholar]
- Honda, Y.; Honda, S.; Narici, M.; Szewczyk, N.J. Spaceflight and Ageing: Reflecting on Caenorhabditis elegans in Space. Gerontology 2014, 60, 138–142. [Google Scholar] [CrossRef]
- Ishioka, N.; Higashibata, A. Space Experiments Using C. elegans as a Model Organism. In Handbook of Space Pharmaceuticals; Springer: Cham, Switzerland, 2018. [Google Scholar]
- Çelen, I.İ.; Doh, J.H.; Sabanayagam, C.R. Effects of Liquid Cultivation on Gene Expression and Phenotype of C. elegans. BMC Genomics 2018, 19, 562. [Google Scholar] [CrossRef] [Green Version]
- Lev, I.; Bril, R.; Liu, Y.; Ceré, L.I.; Rechavi, O. Inter-Generational Consequences for Growing Caenorhabditis elegans in Liquid. Philos. Trans. R. Soc. B Biol. Sci. 2019, 374, 20180125. [Google Scholar] [CrossRef] [Green Version]
- Doh, J.H.; Moore, A.B.; Çelen, İ.; Moore, M.T.; Sabanayagam, C.R. ChIP and Chips: Introducing the WormPharm for Correlative Studies Employing Pharmacology and Genome-Wide Analyses in C. elegans. J. Biol. Methods 2016, 3, 1–11. [Google Scholar] [CrossRef]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data. Available online: www.Bioinformatics.Babraham.Ac.Uk/Projects/Fastqc/ (accessed on 1 January 2019).
- Trapnell, C.; Roberts, A.; Goff, L.; Pertea, G.; Kim, D.; Kelley, D.R.; Pimentel, H.; Salzberg, S.L.; Rinn, J.L.; Pachter, L. Differential Gene and Transcript Expression Analysis of RNA-Seq Experiments with TopHat and Cufflinks. Nat. Protoc. 2012, 7, 562–578. [Google Scholar] [CrossRef] [Green Version]
- Love, M.I.; Huber, W.; Anders, S. Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data with DESeq2. Genome Biol. 2014, 15. [Google Scholar] [CrossRef] [Green Version]
- Patro, R.; Duggal, G.; Love, M.I.; Irizarry, R.A.; Kingsford, C. Salmon Provides Fast and Bias-Aware Quantification of Transcript Expression. Nat. Methods 2017, 14, 417–419. [Google Scholar] [CrossRef] [Green Version]
- Cunningham, F.; Amode, M.R.; Barrell, D.; Beal, K.; Billis, K.; Brent, S.; Carvalho-Silva, D.; Clapham, P.; Coates, G.; Fitzgerald, S.; et al. Ensembl 2015. Nucleic Acids Res. 2014, 43, D662–D669. [Google Scholar] [CrossRef] [Green Version]
- Harris, T.W.; Baran, J.; Bieri, T.; Cabunoc, A.; Chan, J.; Chen, W.J.; Davis, P.; Done, J.; Grove, C.; Howe, K.; et al. WormBase 2014: New Views of Curated Biology. Nucleic Acids Res. 2014, 42, D789–D793. [Google Scholar] [CrossRef] [Green Version]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Systematic and Integrative Analysis of Large Gene Lists Using DAVID Bioinformatics Resources. Nat. Protoc. 2008, 4, 44–57. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Maere, S.; Heymans, K.; Kuiper, M. BiNGO: A Cytoscape Plugin to Assess Overrepresentation of Gene Ontology Categories in Biological Networks. Bioinformatics 2005, 21, 3448–3449. [Google Scholar] [CrossRef] [Green Version]
- Angeles-Albores, D.; Lee, R.Y.; Chan, J.; Sternberg, P.W. Tissue Enrichment Analysis for C. elegans Genomics. BMC Bioinform. 2016, 17, 366. [Google Scholar] [CrossRef] [Green Version]
- Szklarczyk, D.; Morris, J.H.; Cook, H.; Kuhn, M.; Wyder, S.; Simonovic, M.; Santos, A.; Doncheva, N.T.; Roth, A.; Bork, P.; et al. The STRING Database in 2017: Quality-Controlled Protein-Protein Association Networks, Made Broadly Accessible. Nucleic Acids Res. 2017, 45, D362–D368. [Google Scholar] [CrossRef]
- Kim, W.; Underwood, R.S.; Greenwald, I.; Shaye, D.D. OrthoList 2: A New Comparative Genomic Analysis of Human and Caenorhabditis elegans Genes. Genetics 2018, 210, 445–461. [Google Scholar] [CrossRef] [Green Version]
- Bligh, E.G.; Dyer, W.J. A Rapid Method of Total Lipid Extraction and Purification. Can. J. Biochem. Physiol. 1959, 37, 911–917. [Google Scholar] [CrossRef]
- Dedolph, R.R.; Dipert, M.H. The Physical Basis of Gravity Stimulus Nullification by Clinostat Rotation. Plant Physiol. 1971, 47, 756–764. [Google Scholar] [CrossRef]
- Lee Silver, I. The Dynamics of a Discrete Geotropic Sensor Subject to Rotation-Induced Gravity Compensation. J. Theor. Biol. 1976, 61, 353–362. [Google Scholar] [CrossRef]
- Zhao, S.; Fung-Leung, W.P.; Bittner, A.; Ngo, K.; Liu, X. Comparison of RNA-Seq and Microarray in Transcriptome Profiling of Activated T Cells. PLoS ONE 2014, 9. [Google Scholar] [CrossRef]
- Tee, L.F.; Neoh, H.; Then, S.M.; Murad, N.A.; Asillam, M.F.; Hashim, M.H.; Nathan, S.; Jamal, R. Effects of Simulated Microgravity on Gene Expression and Biological Phenotypes of a Single Generation Caenorhabditis elegans Cultured on 2 Different Media. Life Sci. Space Res. 2017, 15, 11–17. [Google Scholar] [CrossRef]
- Reinke, V.; Krause, M.; Okkema, P. Transcriptional Regulation of Gene Expression in C. elegans. WormBook 2013, 1–34. [Google Scholar] [CrossRef] [Green Version]
- Lin, K.; Hsin, H.; Libina, N.; Kenyon, C. Regulation of the Caenorhabditis elegans Longevity Protein DAF-16 by Insulin/IGF-1 and Germline Signaling. Nat. Genet. 2001, 28, 139–145. [Google Scholar] [CrossRef] [PubMed]
- Seung, W.O.; Mukhopadhyay, A.; Dixit, B.L.; Raha, T.; Green, M.R.; Tissenbaum, H.A. Identification of Direct DAF-16 Targets Controlling Longevity, Metabolism and Diapause by Chromatin Immunoprecipitation. Nat. Genet. 2006, 38, 251–257. [Google Scholar] [CrossRef]
- Murphy, C.T.; McCarroll, S.A.; Bargmann, C.I.; Fraser, A.; Kamath, R.S.; Ahringer, J.; Li, H.; Kenyon, C. Genes That Act Downstream of DAF-16 to Influence the Lifespan of Caenorhabditis elegans. Nature 2003, 424, 277–284. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.S.; Kennedy, S.; Tolonen, A.C.; Ruvkun, G. DAF-16 Target Genes That Control C. elegans Life-Span and Metabolism. Science 2003, 300, 644–647. [Google Scholar] [CrossRef] [Green Version]
- Tepper, R.G.; Ashraf, J.; Kaletsky, R.; Kleemann, G.; Murphy, C.T.; Bussemaker, H.J. PQM-1 Complements DAF-16 as a Key Transcriptional Regulator of DAF-2-Mediated Development and Longevity. Cell 2013, 154, 676–690. [Google Scholar] [CrossRef] [Green Version]
- Cui, M.; Wang, Y.; Cavaleri, J.; Kelson, T.; Teng, Y.; Han, M. Starvation-Induced Stress Response Is Critically Impacted by Ceramide Levels in Caenorhabditis elegans. Genetics 2017, 205, 775–785. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, Y.; Sun, H. ASM-3 Acid Sphingomyelinase Functions as a Positive Regulator of the DAF-2/AGE-1 Signaling Pathway and Serves as a Novel Anti-Aging Target. PLoS ONE 2012, 7, e45890. [Google Scholar] [CrossRef] [Green Version]
- Cutler, R.G.; Thompson, K.W.; Camandola, S.; Mack, K.T.; Mattson, M.P. Sphingolipid Metabolism Regulates Development and Lifespan in Caenorhabditis elegans. Mech. Ageing Dev. 2014, 143–144, 9–18. [Google Scholar] [CrossRef] [Green Version]
- Huang, X.; Withers, B.R.; Dickson, R.C. Sphingolipids and Lifespan Regulation. Biochim. Biophys Acta 2014, 1841, 657–664. [Google Scholar] [CrossRef] [Green Version]
- Hernández-Corbacho, M.J.; Jenkins, R.W.; Clarke, C.J.; Hannun, Y.A.; Obeid, L.M.; Snider, A.J.; Siskind, L.J. Accumulation of Long-Chain Glycosphingolipids during Aging Is Prevented by Caloric Restriction. PLoS ONE 2011, 6, e20411. [Google Scholar] [CrossRef] [Green Version]
- Straczkowski, M.; Kowalska, I. The Role of Skeletal Muscle Sphingolipids in the Development of Insulin Resistance. Rev. Diabet. Stud. 2008, 5, 13–24. [Google Scholar] [CrossRef] [Green Version]
- Bismuth, J.; Lin, P.; Yao, Q.; Chen, C. Ceramide: A Common Pathway for Atherosclerosis? Atherosclerosis 2008, 196, 497–504. [Google Scholar] [CrossRef] [Green Version]
- Chan, J.P.; Brown, J.; Hark, B.; Nolan, A.; Servello, D.; Hrobuchak, H.; Staab, T.A. Loss of Sphingosine Kinase Alters Life History Traits and Locomotor Function in Caenorhabditis elegans. Front. Genet. 2017, 8, 132. [Google Scholar] [CrossRef]
- Menuz, V.; Howell, K.S.; Gentina, S.; Epstein, S.; Riezman, I.; Fornallaz-Mulhauser, M.; Hengartner, M.O.; Gomez, M.; Riezman, H.; Martinou, J.C. Protection of C. elegans from Anoxia by HYL-2 Ceramide Synthase. Science 2009, 324, 381–384. [Google Scholar] [CrossRef] [Green Version]
- Deng, X.; Yin, X.; Allan, R.; Lu, D.D.; Maurer, C.W.; Haimovitz-Friedman, A.; Fuks, Z.; Shaham, S.; Kolesnick, R. Ceramide Biogenesis Is Required for Radiation-Induced Apoptosis in the Germ Line of C. elegans. Science 2008, 322, 110–115. [Google Scholar] [CrossRef] [Green Version]
- Gao, Y.; Arfat, Y.; Wang, H.; Goswami, N. Muscle Atrophy Induced by Mechanical Unloading: Mechanisms and Potential Countermeasures. Front. Physiol. 2018, 9, 235. [Google Scholar] [CrossRef] [Green Version]
- Salazar, J.J.; Michele, D.E.; Brooks, S.V. Inhibition of Calpain Prevents Muscle Weakness and Disruption of Sarcomere Structure during Hindlimb Suspension. J. Appl. Physiol. 2010, 108, 120–127. [Google Scholar] [CrossRef] [Green Version]
- Seale, P.; Bjork, B.; Yang, W.; Kajimura, S.; Chin, S.; Kuang, S.; Scimè, A.; Devarakonda, S.; Conroe, H.M.; Erdjument-Bromage, H.; et al. PRDM16 Controls a Brown Fat/Skeletal Muscle Switch. Nature 2008, 454, 961–967. [Google Scholar] [CrossRef] [Green Version]
- Cutler, R.G.; Pedersen, W.A.; Camandola, S.; Rothstein, J.D.; Mattson, M.P. Evidence That Accumulation of Ceramides and Cholesterol Esters Mediates Oxidative Stress—Induced Death of Motor Neurons in Amyotrophic Lateral Sclerosis. Ann. Neurol. 2002, 52, 448–457. [Google Scholar] [CrossRef]
- Graveley, A.; Vlasov, A.; Freeman, A.; Wu, K.; Szewczyk, N.J.; D’Cruz, R.; Batt, J. Levels of Acid Sphingomyelinase (ASM) in Caenorhabditis elegans in Microgravity. Gravit. Space Res. 2018, 6, 27–36. [Google Scholar] [CrossRef]
- Garrett-Bakelman, F.E.; Darshi, M.; Green, S.J.; Gur, R.C.; Lin, L.; Macias, B.R.; McKenna, M.J.; Meydan, C.; Mishra, T.; Nasrini, J.; et al. The NASA Twins Study: A Multidimensional Analysis of a Year-Long Human Spaceflight. Science 2019, 364, eaau8650. [Google Scholar] [CrossRef] [PubMed]
- Sahin, E.; Depinho, R.A. Linking Functional Decline of Telomeres, Mitochondria and Stem Cells during Ageing. Nature 2010, 464, 520–528. [Google Scholar] [CrossRef] [Green Version]
- Joeng, K.S.; Song, E.J.; Lee, K.J.; Lee, J. Long Lifespan in Worms with Long Telomeric DNA. Nat. Genet. 2004, 36, 607–611. [Google Scholar] [CrossRef]
- Monaghan, P. Telomeres and Life Histories: The Long and the Short of It. Ann. N. Y. Acad. Sci. 2010, 1206, 130–142. [Google Scholar] [CrossRef]
- Heidinger, B.J.; Blount, J.D.; Boner, W.; Griffiths, K.; Metcalfe, N.B.; Monaghan, P. Telomere Length in Early Life Predicts Lifespan. Proc. Natl. Acad. Sci. USA 2012, 109, 1743–1748. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saddoughi, S.A.; Song, P.; Ogretmen, B. Roles of Bioactive Sphingolipids in Cancer Biology and Therapeutics. In Lipids in Health and Disease; Springer: Dordrecht, The Netherlands, 2008; Volume 49, pp. 413–440. [Google Scholar]
- Taylor, P.W. Impact of Space Flight on Bacterial Virulence and Antibiotic Susceptibility. Infect. Drug Resist. 2015, 8, 249–262. [Google Scholar] [CrossRef] [Green Version]
- Szewczyk, N.J.; Tillman, J.; Conley, C.A.; Granger, L.; Segalat, L.; Higashitani, A.; Honda, S.; Honda, Y.; Kagawa, H.; Adachi, R.; et al. Description of International Caenorhabditis elegans Experiment First Flight (ICE-FIRST). Adv. Space Res. 2008, 42, 1072–1079. [Google Scholar] [CrossRef] [Green Version]
- Higashitani, A.; Hashizume, T.; Sugimoto, T.; Mori, C.; Nemoto, K.; Etheridge, T.; Higashitani, N.; Takanami, T.; Suzuki, H.; Fukui, K.; et al. C. elegans RNAi Space Experiment (CERISE) in Japanese Experiment Module KIBO. Biol. Sci. Sp. 2009, 23, 183–187. [Google Scholar] [CrossRef] [Green Version]
- Spencer, W.C.; Zeller, G.; Watson, J.D.; Henz, S.R.; Watkins, K.L.; McWhirter, R.D.; Petersen, S.; Sreedharan, V.T.; Widmer, C.; Jo, J.; et al. A Spatial and Temporal Map of C. elegans Gene Expression. Genome Res. 2011, 21, 325–341. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Çelen, İ.; Doh, J.H.; Sabanayagam, C. Genetic Adaptation of C. elegans to Environment Changes I: Multigenerational Analysis of the Transcriptome. bioRxiv 2017, 194506. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Çelen, İ.; Jayasinghe, A.; Doh, J.H.; Sabanayagam, C.R. Transcriptomic Signature of the Simulated Microgravity Response in Caenorhabditis elegans and Comparison to Spaceflight Experiments. Cells 2023, 12, 270. https://doi.org/10.3390/cells12020270
Çelen İ, Jayasinghe A, Doh JH, Sabanayagam CR. Transcriptomic Signature of the Simulated Microgravity Response in Caenorhabditis elegans and Comparison to Spaceflight Experiments. Cells. 2023; 12(2):270. https://doi.org/10.3390/cells12020270
Chicago/Turabian StyleÇelen, İrem, Aroshan Jayasinghe, Jung H. Doh, and Chandran R. Sabanayagam. 2023. "Transcriptomic Signature of the Simulated Microgravity Response in Caenorhabditis elegans and Comparison to Spaceflight Experiments" Cells 12, no. 2: 270. https://doi.org/10.3390/cells12020270
APA StyleÇelen, İ., Jayasinghe, A., Doh, J. H., & Sabanayagam, C. R. (2023). Transcriptomic Signature of the Simulated Microgravity Response in Caenorhabditis elegans and Comparison to Spaceflight Experiments. Cells, 12(2), 270. https://doi.org/10.3390/cells12020270




