Triphasic 3D In Vitro Model of Bone-Tendon-Muscle Interfaces to Study Their Regeneration
Abstract
:1. Introduction
2. Material and Methods
2.1. Preparation of Biphasic and Triphasic Composite Hydrogels
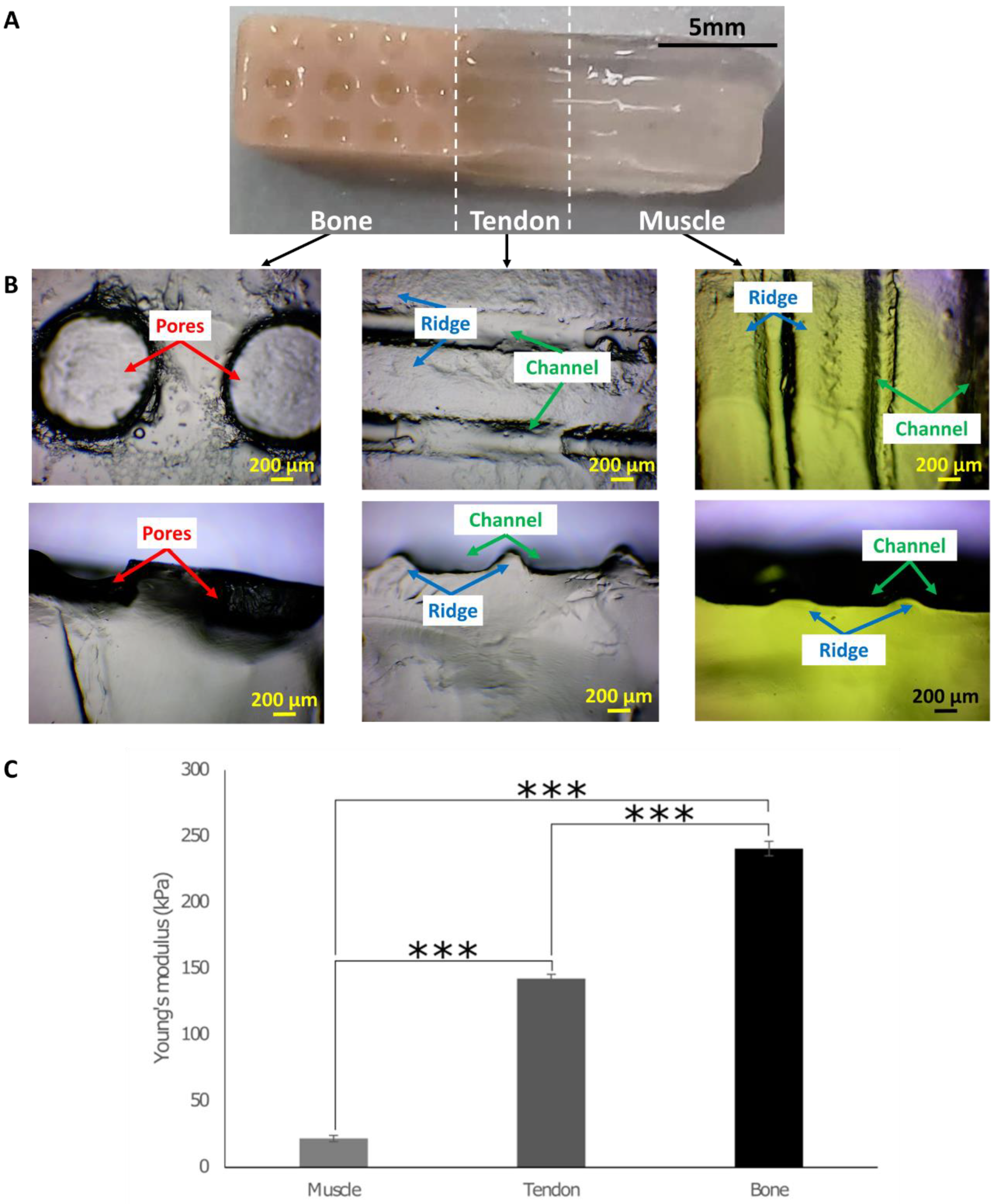
2.2. Determining Young’s Modulus from Compression Testing
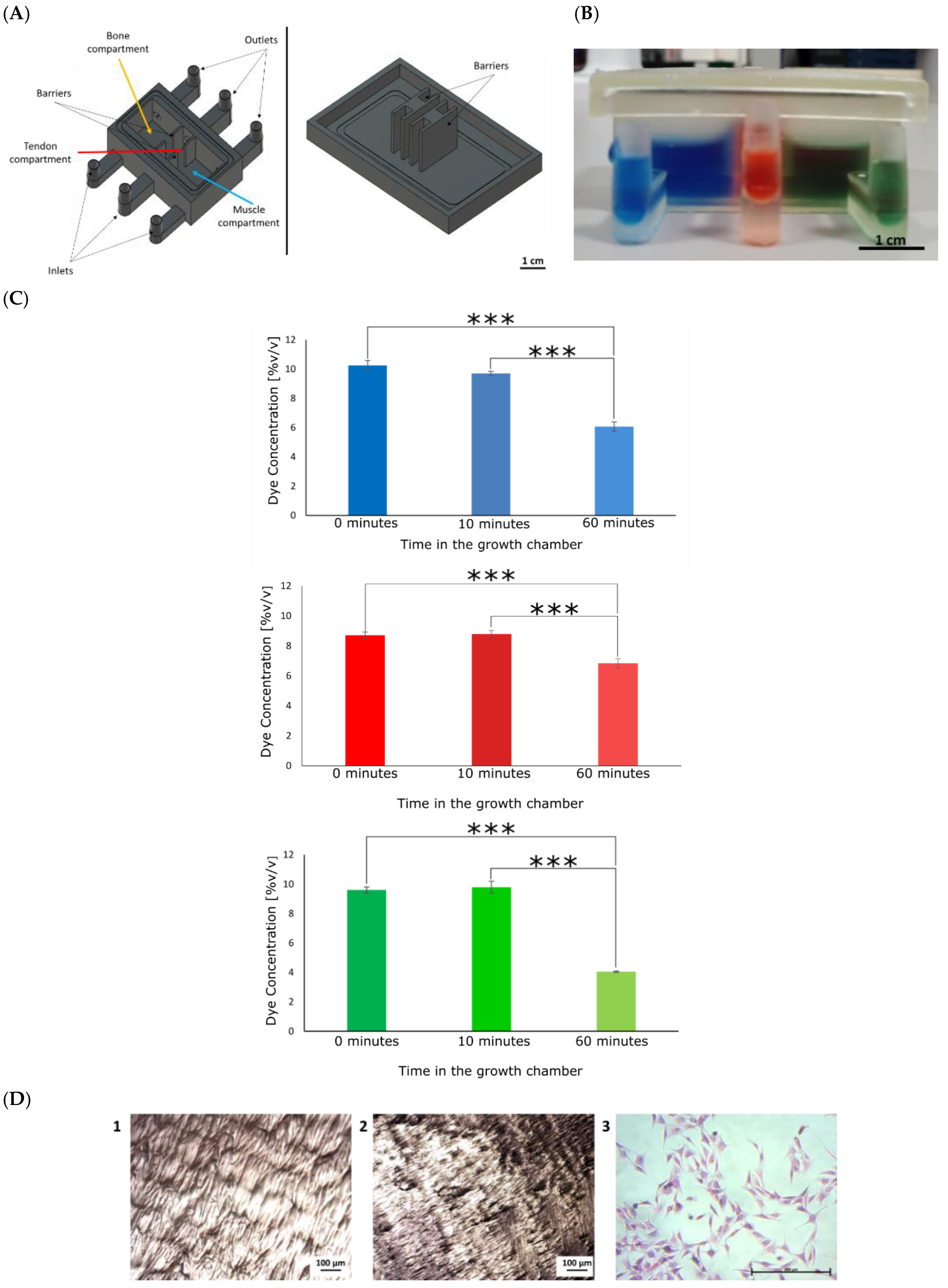
2.3. Design and Fabrication of the 3D In Vitro Model
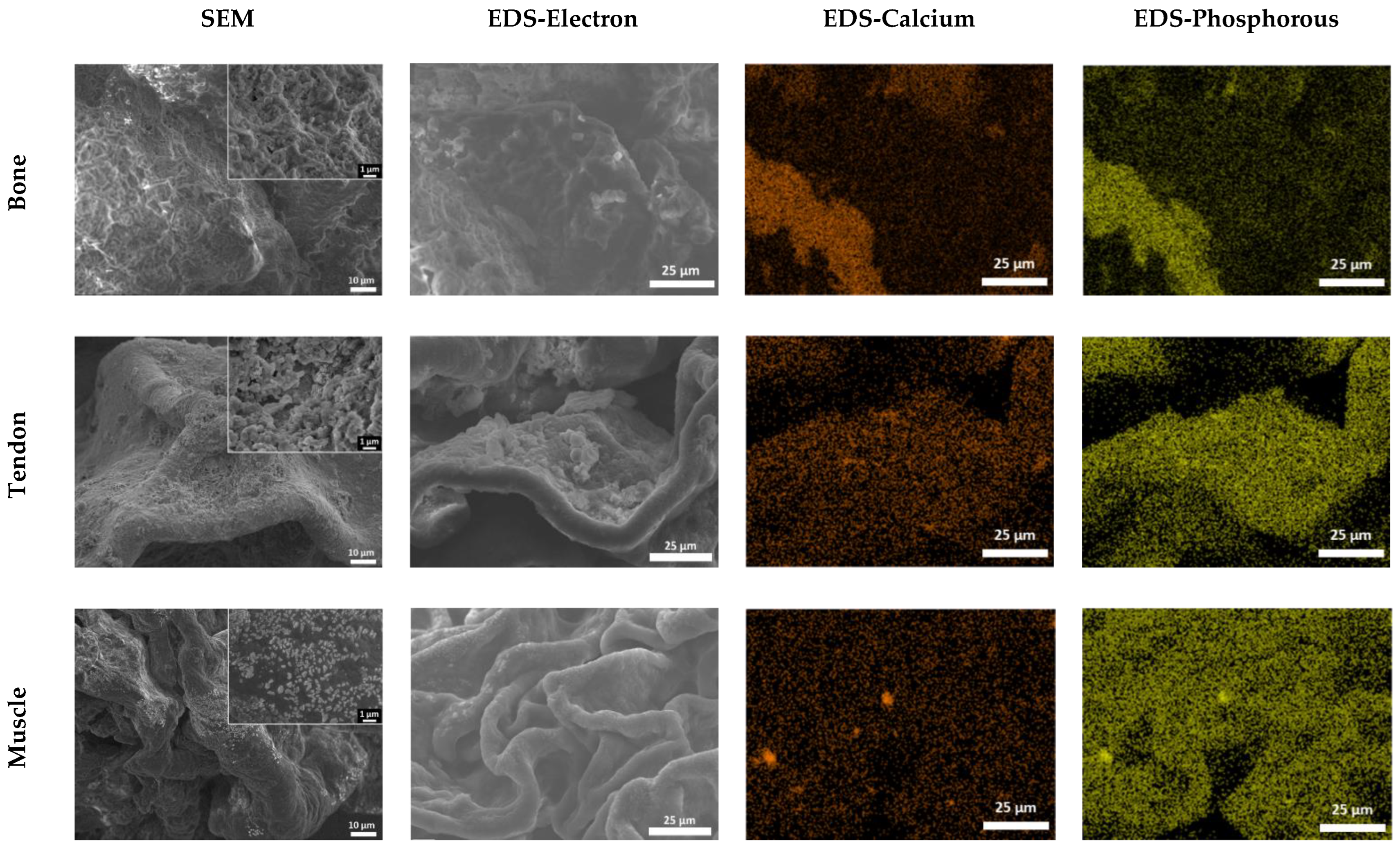
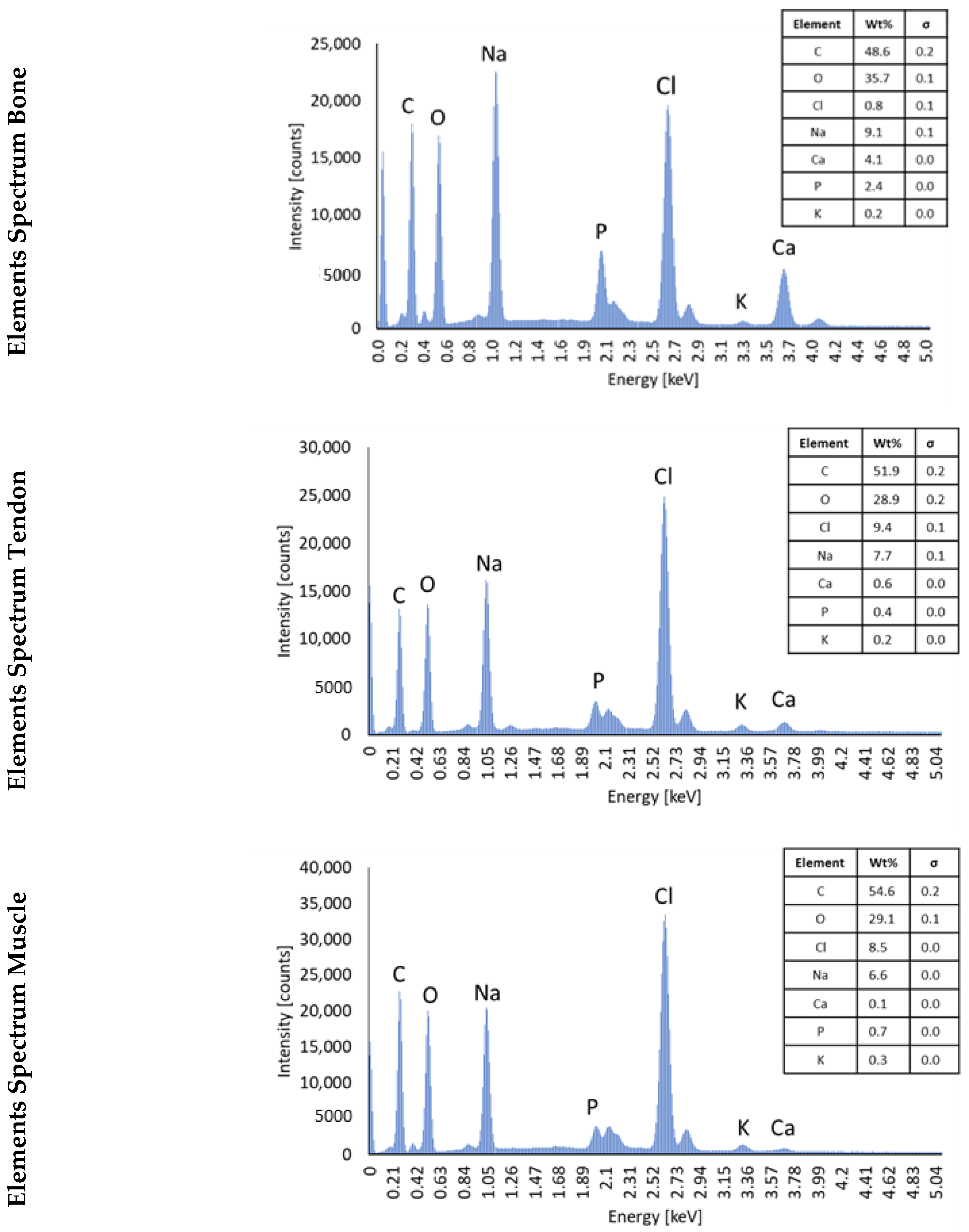
2.4. Energy Dispersive Spectroscopy (EDS) with Scanning Electron Microscope (SEM)
2.5. Fabrication of the Co-Culture Growth Chamber
2.6. Response of Cells Cultured on 3D Hydrogels
2.7. PicoGreen Assay
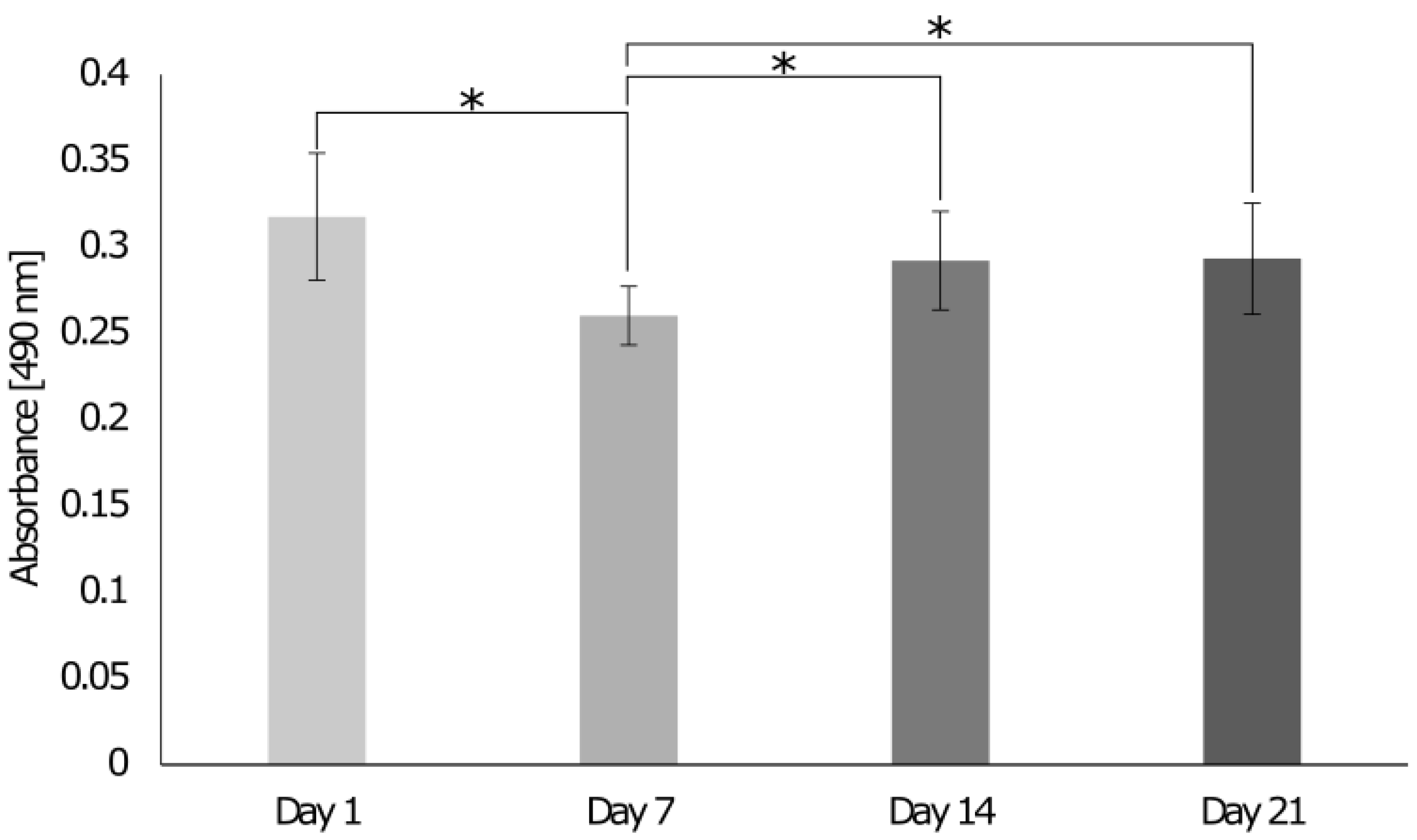
2.8. Alamar Blue Assay
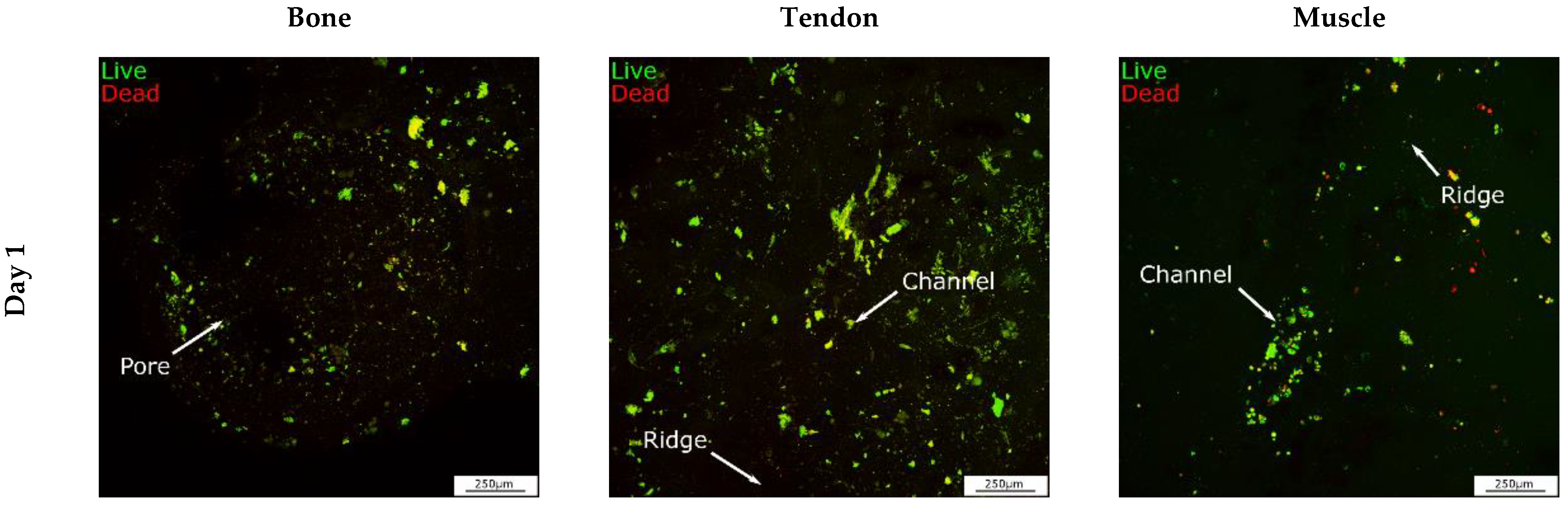
2.9. LIVE/DEAD™ Viability Assay
2.10. LDH Cytotoxicity Assay
2.11. Immunocytochemistry
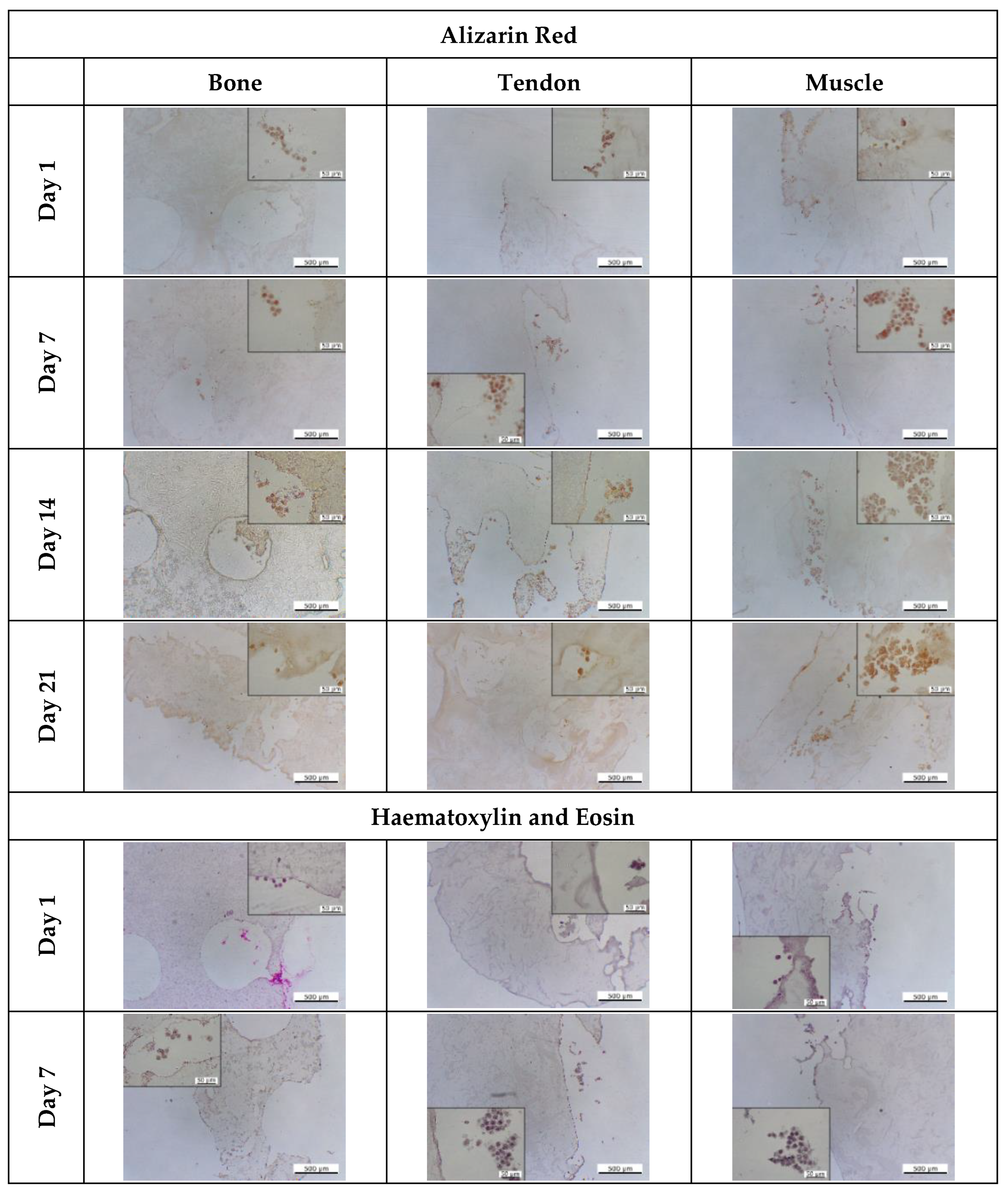
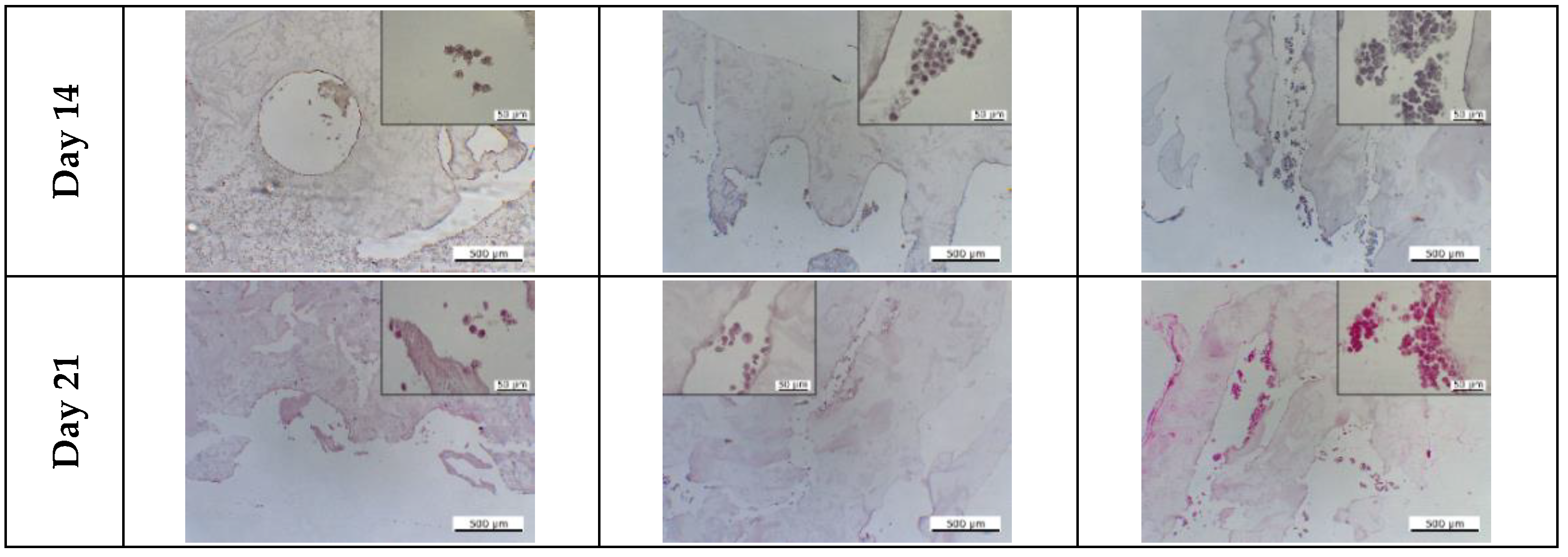
2.12. Histological Staining
2.13. Statistical Analysis
3. Results
3.1. Fabrication of the 3D In Vitro Model and the Growth Chamber
3.2. SEM/EDS Analysis to Assess the Composition of Hydrogels
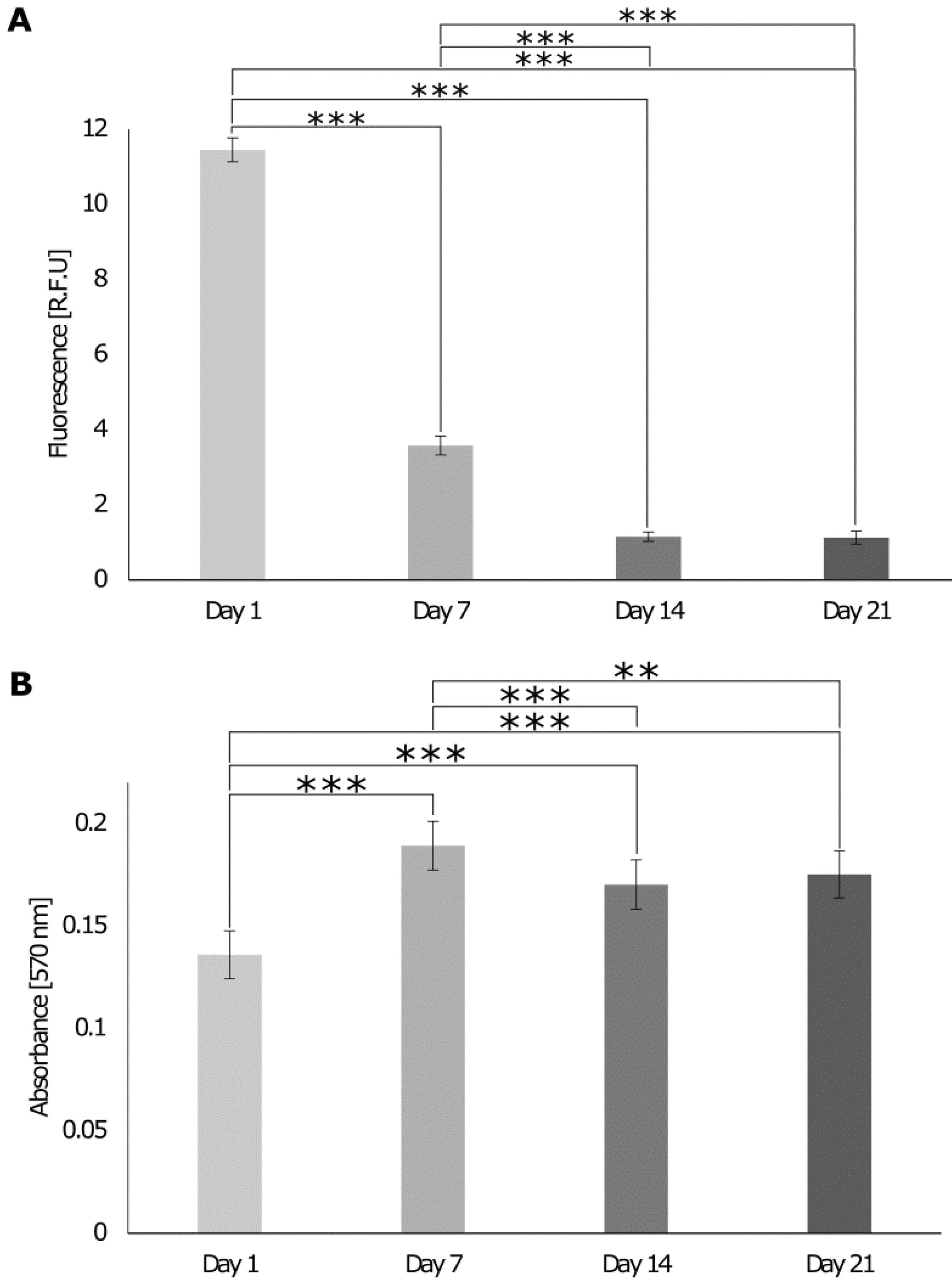
3.3. Identification of Cell Homeostasis
3.4. Biocompatibility of Biphasic and Triphasic Gels
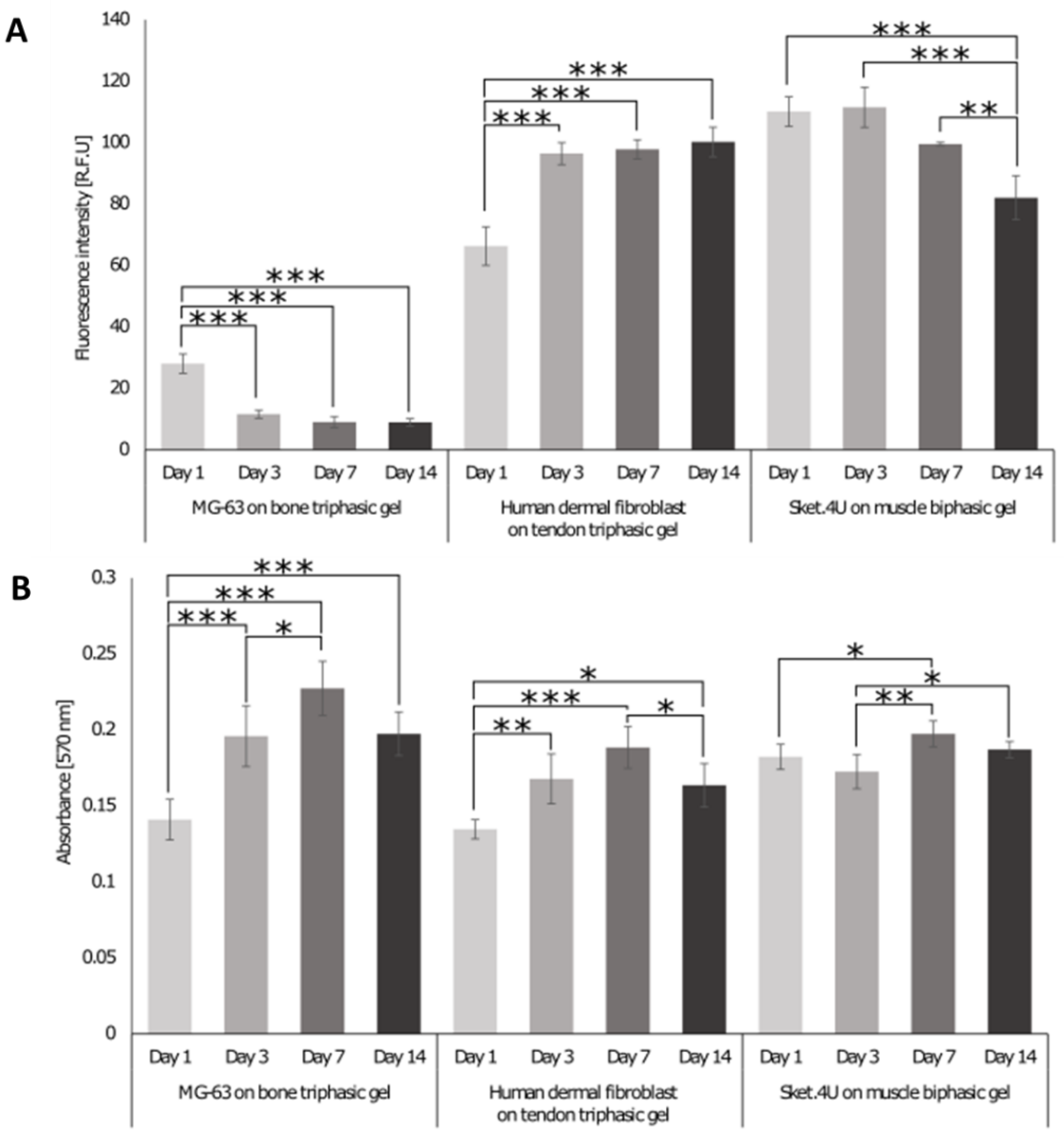
3.5. Biological Response of Cells Seeded on the 3D Interface Model
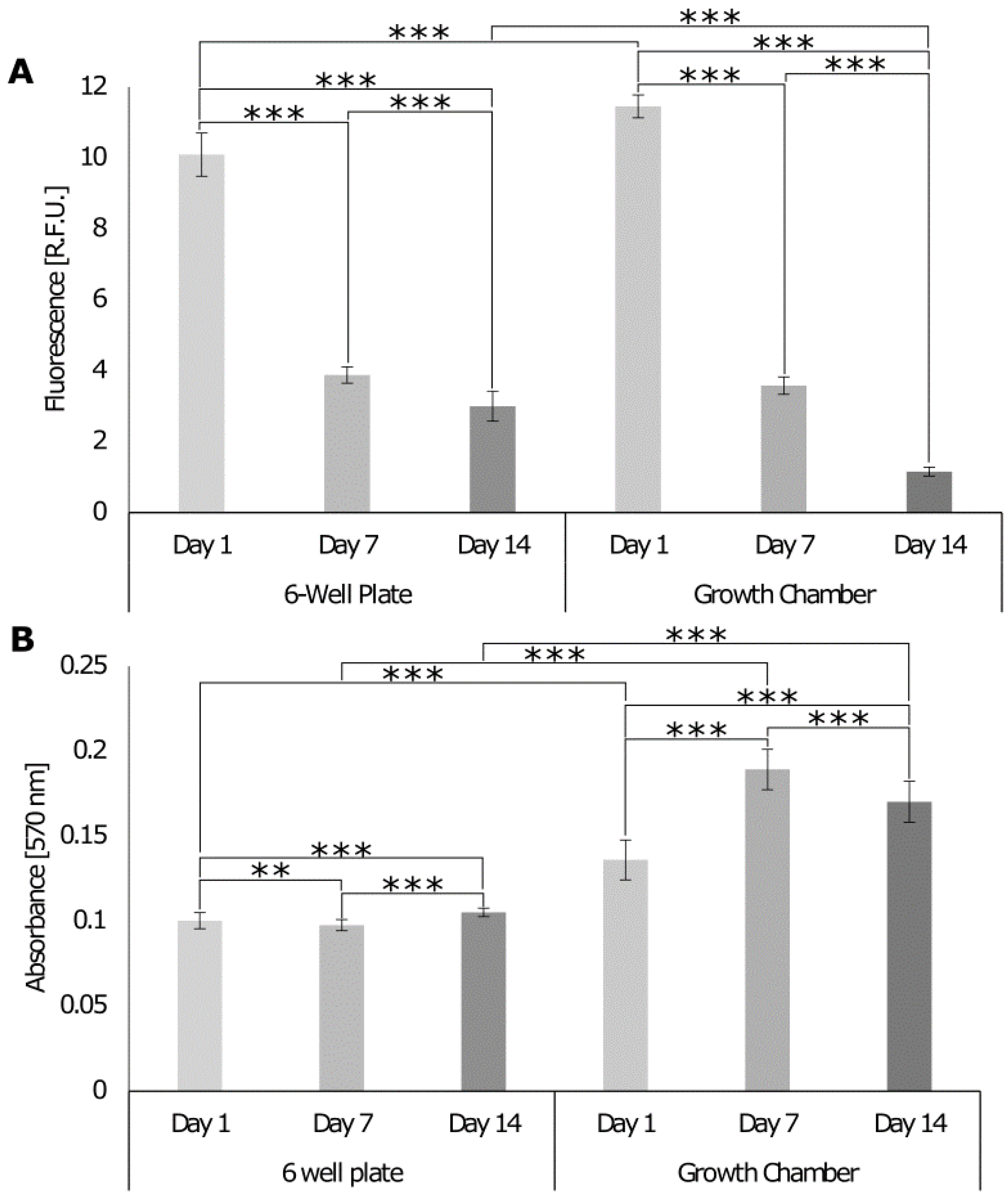
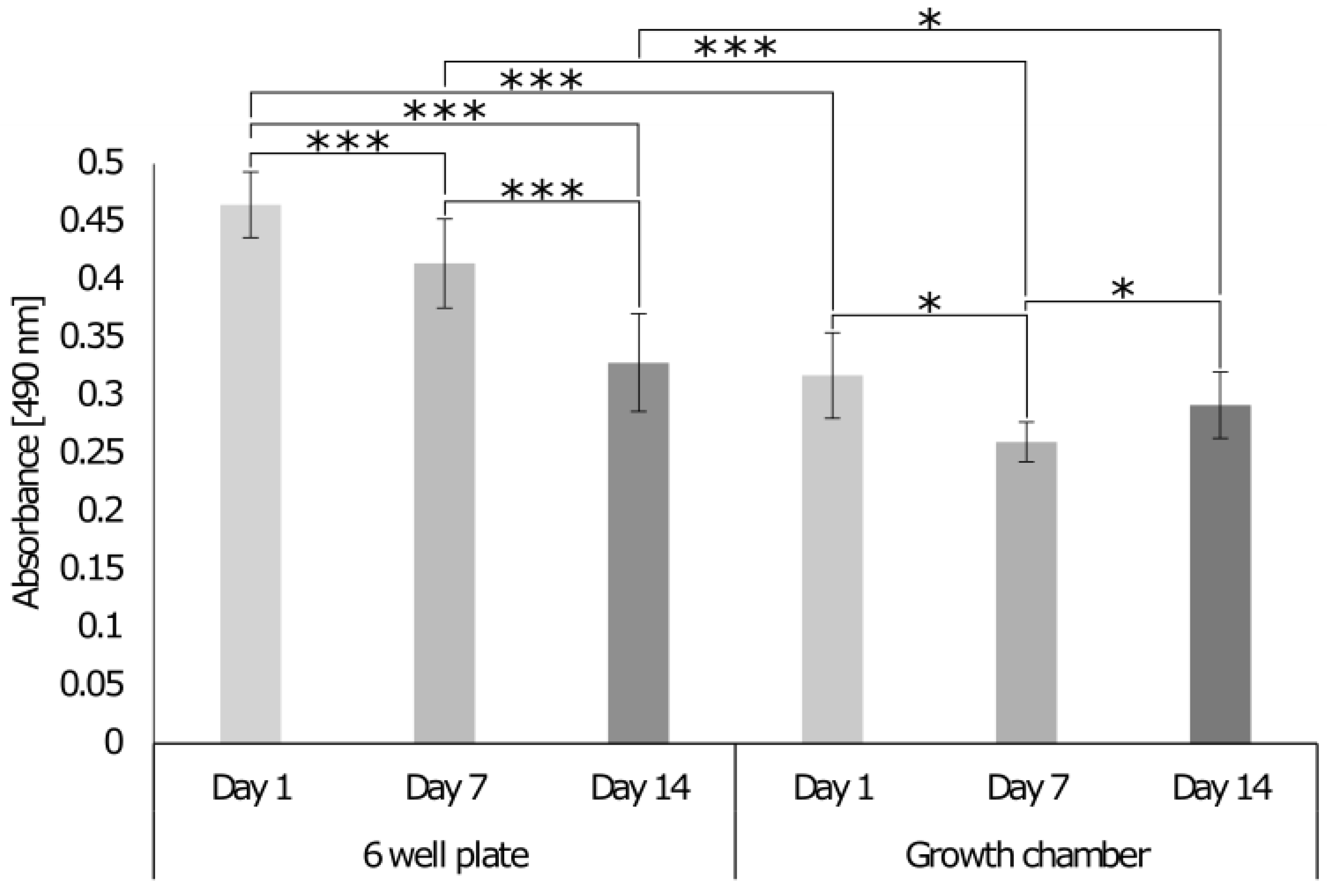
3.5.1. Comparison of the Growth Chamber versus the Standard Cell Culture Plate
3.5.2. Biological Response of Cells Cultures on the 3D In Vitro Interface Model
3.5.3. Assessment of Tissue Interface Development
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Yang, P.J.; Temenoff, J.S. Engineering orthopedic tissue interfaces. Tissue Eng. Part B Rev. 2009, 15, 127–141. [Google Scholar] [CrossRef] [Green Version]
- Lu, H.H.; Subramony, S.D.; Boushell, M.K.; Zhang, X. Tissue engineering strategies for the regeneration of orthopedic interfaces. Ann. Biomed. Eng. 2010, 38, 2142–2154. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Babatunde, O.O.; Jordan, J.L.; Van der Windt, D.A.; Hill, J.C.; Foster, N.E.; Protheroe, J. Effective treatment options for musculoskeletal pain in primary care: A systematic overview of current evidence. PLoS ONE 2017, 12, e0178621. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Balestri, M.W.; Morris, R.H.; Hunt, J.A.; Reinwald, Y. Current advances on the regeneration of musculoskeletal interfaces. Tissue Eng. Part B Rev. 2021, 27, 548–571. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Lipner, J.; Xie, J.; Manning, C.N.; Thomopoulos, S.; Xia, Y. Nanofiber scaffolds with gradients in mineral content for spatial control of osteogenesis. ACS Appl. Mater. Interfaces 2014, 6, 2842–2849. [Google Scholar] [CrossRef]
- Landis, W.J.; Hodgens, K.; Arena, J.; Song, M.; McEwen, B.F. Structural relations between collagen and mineral in bone as determined by high voltage electron microscopic tomography. Microsc. Res. Tech. 1996, 33, 192–202. [Google Scholar] [CrossRef]
- Baldino, L.; Cardea, S.; Maffulli, N.; Reverchon, E. Regeneration techniques for bone-to-tendon and muscle-to-tendon interfaces reconstruction. Br. Med. Bull. 2016, 117, 25–37. [Google Scholar] [CrossRef] [Green Version]
- Xiong, Y.; Yang, H.; Feng, J.; Shi, Z.; Wu, L. Effects of alendronate on the proliferation and osteogenic differentiation of MG-63 cells. J. Int. Med. Res. 2009, 37, 407–416. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, F.; Nerlich, M.; Docheva, D. Tendon injuries: Basic science and new repair proposals. EFORT Open Rev. 2017, 2, 332–342. [Google Scholar] [CrossRef] [PubMed]
- Qazi, T.H.; Mooney, D.J.; Pumberger, M.; Geißler, S.; Duda, G.N. Biomaterials based strategies for skeletal muscle tissue engineering: Existing technologies and future trends. Biomaterials 2015, 53, 502–521. [Google Scholar] [CrossRef]
- Gillies, A.R.; Lieber, R.L. Structure and function of the skeletal muscle extracellular matrix. Muscle Nerve 2011, 44, 318–331. [Google Scholar] [CrossRef] [Green Version]
- Bayrak, E.; Huri, P.Y. Engineering Musculoskeletal Tissue Interfaces. Front. Mater. 2018, 5, 24. [Google Scholar] [CrossRef] [Green Version]
- King, W.J.; Krebsbach, P.H. Multi-Tissue Interface Bioengineering. In Stem Cell Biology and Tissue Engineering in Dental Sciences; Elsevier Inc.: Amsterdam, The Netherlands, 2015; pp. 593–602. [Google Scholar] [CrossRef]
- Kruize, C.P.; Panahkhahi, S.; Putra, N.E.; Diaz-Payno, P.; van Osch, G.; Zadpoor, A.A.; Mirzaali, M.J. Biomimetic Approaches for the Design and Fabrication of Bone-to-Soft Tissue Interfaces. ACS Biomater. Sci. Eng. 2021. [Google Scholar] [CrossRef]
- Mosher, C.Z.; Spalazzi, J.P.; Lu, H.H. Stratified scaffold design for engineering composite tissues. Methods 2015, 84, 99–102. [Google Scholar] [CrossRef] [Green Version]
- Lipner, J.; Liu, W.; Liu, Y.; Boyle, J.; Genin, G.; Xia, Y.; Thomopoulos, S. The mechanics of PLGA nanofiber scaffolds with biomimetic gradients in mineral for tendon-to-bone repair. J. Mech. Behav. Biomed. Mater. 2014, 40, 59–68. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, H.H.; Cooper, J.A.; Manuel, S.; Freeman, J.W.; Attawia, M.A.; Ko, F.K.; Laurencin, C.T. Anterior cruciate ligament regeneration using braided biodegradable scaffolds: In vitro optimization studies. Biomaterials 2005, 26, 4805–4816. [Google Scholar] [CrossRef]
- Baudequin, T.; Gaut, L.; Mueller, M.; Huepkes, A.; Glasmacher, B.; Duprez, D.; Bedoui, F.; Legallais, C. The Osteogenic and Tenogenic Differentiation Potential of C3H10T1/2 (Mesenchymal Stem Cell Model) Cultured on PCL/PLA Electrospun Scaffolds in the Absence of Specific Differentiation Medium. Materials 2017, 10, 1387. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, P.; Han, F.; Zhang, P.; Sun, Y.; Lin, C.; Chen, J. Hydroxyapatite-doped polycaprolactone nanofiber membrane improves tendon–bone interface healing for anterior cruciate ligament reconstruction. Int. J. Nanomed. 2015, 10, 7333–7343. [Google Scholar] [CrossRef] [Green Version]
- Nowlin, J.; Bismi, M.A.; Delpech, B.; Dumas, P.; Zhou, Y.; Tan, G.Z. Engineering the hard–soft tissue interface with random-to-aligned nanofiber scaffolds. Nanobiomedicine 2018, 5, 1849543518803538. [Google Scholar] [CrossRef]
- Cooper, J.O.; Bumgardner, J.D.; Cole, J.A.; Smith, R.A.; Haggard, W.O. Co-cultured tissue-specific scaffolds for tendon/bone interface engineering. J. Tissue Eng. 2014, 5, 204173141454229. [Google Scholar] [CrossRef]
- Guo, J.; Su, W.; Jiang, J.; Ning, C.; Zhao, J.; Liu, X. Enhanced tendon to bone healing in rotator cuff tear by PLLA/CPS composite films prepared by a simple melt-pressing method: An in vitro and in vivo study. Compos. Part B: Eng. 2019, 165, 526–536. [Google Scholar] [CrossRef]
- Perikamana, S.K.M.; Lee, J.; Ahmad, T.; Kim, E.M.; Byun, H.; Lee, S.; Shin, H. Harnessing biochemical and structural cues for tenogenic differentiation of adipose derived stem cells (ADSCs) and development of an in vitro tissue interface mimicking tendon-bone insertion graft. Biomaterials 2018, 165, 79–93. [Google Scholar] [CrossRef]
- Kolluru, P.V.; Lipner, J.; Liu, W.; Xia, Y.; Thomopoulos, S.; Genin, G.M.; Chasiotis, I. Strong and tough mineralized PLGA nanofibers for tendon-to-bone scaffolds. Acta Biomater. 2013, 9, 9442–9450. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Merceron, T.K.; Burt, M.; Seol, Y.-J.; Kang, H.-W.; Lee, S.J.; Yoo, J.J.; Atala, A. A 3D bioprinted complex structure for engineering the muscle–tendon unit. Biofabrication 2015, 7, 035003. [Google Scholar] [CrossRef] [PubMed]
- Schweitzer, R.; Chyung, J.H.; Murtaugh, L.C.; Brent, A.E.; Rosen, V.; Olson, E.N.; Lassar, A.; Tabin, C.J. Analysis of the tendon cell fate using Scleraxis, a specific marker for tendons and ligaments. Development 2001, 128, 3855–3866. [Google Scholar] [CrossRef]
- O’Brien, F.J. Biomaterials & scaffolds for tissue engineering. Mater. Today 2011, 14, 88–95. [Google Scholar] [CrossRef]
- Goncalves, A.D.; Balestri, W.; Reinwald, Y. Biomedical Implants for Regenerative Therapies. In Biomaterials; IntechOpen: London, UK, 2020. [Google Scholar] [CrossRef] [Green Version]
- Fu, S.; Zhang, P. Surface modification of polylactic acid (PLA) and polyglycolic acid (PGA) monofilaments via the cold plasma method for acupoint catgut-embedding therapy applications. Text. Res. J. 2019, 89, 3839–3849. [Google Scholar] [CrossRef]
- Yuan, Z.; Memarzadeh, K.; Stephen, A.S.; Allaker, R.P.; Brown, R.A.; Huang, J. Development of a 3D Collagen Model for the In Vitro Evaluation of Magnetic-assisted Osteogenesis. Sci. Rep. 2018, 8, 16270. [Google Scholar] [CrossRef]
- Caliari, S.; Ramirez, M.A.; Harley, B.A. The development of collagen-GAG scaffold-membrane composites for tendon tissue engineering. Biomaterials 2011, 32, 8990–8998. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Farano, V.; Cresswell, M.; Gritsch, K.; Jackson, P.; Attik, N.; Grosgogeat, B.; Maurin, J.-C. Bioactivity evaluation of collagen-based scaffolds containing a series of Sr-doped melt-quench derived phosphate-based glasses. J. Mater. Sci. Mater. Med. 2018, 29, 101. [Google Scholar] [CrossRef]
- Shah, R.; Knowles, J.C.; Hunt, N.P.; Lewis, M.P. Development of a novel smart scaffold for human skeletal muscle regeneration. J. Tissue Eng. Regen. Med. 2013, 10, 162–171. [Google Scholar] [CrossRef] [PubMed]
- Marelli, B.; Achilli, M.; Alessandrino, A.; Freddi, G.; Tanzi, M.C.; Farè, S.; Mantovani, D. Collagen-Reinforced Electrospun Silk Fibroin Tubular Construct as Small Calibre Vascular Graft. Macromol. Biosci. 2012, 12, 1566–1574. [Google Scholar] [CrossRef] [PubMed]
- Achilli, M.; Lagueux, J.; Mantovani, D. On the Effects of UV-C and pH on the Mechanical Behavior, Molecular Conformation and Cell Viability of Collagen-Based Scaffold for Vascular Tissue Engineering. Macromol. Biosci. 2010, 10, 307–316. [Google Scholar] [CrossRef] [PubMed]
- Korpayev, S.; Kaygusuz, G.; Şen, M.; Orhan, K.; Oto, Ç.; Karakeçili, A. Chitosan/collagen based biomimetic osteochondral tissue constructs: A growth factor-free approach. Int. J. Biol. Macromol. 2020, 156, 681–690. [Google Scholar] [CrossRef] [PubMed]
- Köpf, M.; Campos, D.F.D.; Blaeser, A.; Sen, K.S.; Fischer, H. A tailored three-dimensionally printable agarose–collagen blend allows encapsulation, spreading, and attachment of human umbilical artery smooth muscle cells. Biofabrication 2016, 8, 025011. [Google Scholar] [CrossRef]
- Ulrich, T.A.; Jain, A.; Tanner, K.; MacKay, J.L.; Kumar, S. Probing cellular mechanobiology in three-dimensional culture with collagen–agarose matrices. Biomaterials 2010, 31, 1875–1884. [Google Scholar] [CrossRef]
- Kim, B.S.; Kim, E.J.; Choi, J.S.; Jeong, J.H.; Jo, C.H.; Cho, Y.W. Human collagen-based multilayer scaffolds for tendon-to-bone interface tissue engineering. J. Biomed. Mater. Res. Part A 2014, 102, 4044–4054. [Google Scholar] [CrossRef]
- Al-Munajjed, A.A.; Plunkett, N.A.; Gleeson, J.P.; Weber, T.; Jungreuthmayer, C.; Levingstone, T.; Hammer, J.; O’Brien, F.J. Development of a biomimetic collagen-hydroxyapatite scaffold for bone tissue engineering using a SBF immersion technique. J. Biomed. Mater. Res. Part B Appl. Biomater. 2009, 90B, 584–591. [Google Scholar] [CrossRef]
- Liu, L.; Zhang, L.; Ren, B.; Wang, F.; Zhang, Q. Preparation and characterization of collagen-hydroxyapatite composite used for bone tissue engineering scaffold. Artif. Cells Blood Substit. Biotechnol. 2003, 31, 435–448. [Google Scholar] [CrossRef]
- Rodrigues, S.C.; Salgado, C.L.; Sahu, A.; Garcia, M.P.; Fernandes, M.H.; Monteiro, F.J. Preparation and characterization of collagen-nanohydroxyapatite biocomposite scaffolds by cryogelation method for bone tissue engineering applications. J. Biomed. Mater. Res. Part A 2012, 101A, 1080–1094. [Google Scholar] [CrossRef]
- Ross, A.M.; Jiang, Z.; Bastmeyer, M.; Lahann, J. Physical Aspects of Cell Culture Substrates: Topography, Roughness, and Elasticity. Small 2011, 8, 336–355. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Janson, I.A.; Putnam, A.J. Extracellular matrix elasticity and topography: Material-based cues that affect cell function via conserved mechanisms. J. Biomed. Mater. Res. A 2014, 103, 1246–1258. [Google Scholar] [CrossRef] [Green Version]
- Bettinger, C.J.; Langer, R.; Borenstein, J.T. Engineering Substrate Topography at the Micro- and Nanoscale to Control Cell Function. Angew. Chem. Int. Ed. 2009, 48, 5406–5415. [Google Scholar] [CrossRef] [Green Version]
- Dalby, M.J.; Gadegaard, N.; Tare, R.; Andar, A.; Riehle, M.O.; Herzyk, P.; Wilkinson, C.D.W.; Oreffo, R.O.C. The control of human mesenchymal cell differentiation using nanoscale symmetry and disorder. Nat. Mater. 2007, 6, 997–1003. [Google Scholar] [CrossRef]
- Choi, H.J.; Lee, J.J.; Lee, J.B.; Sung, H.-J.; Shin, J.-W.; Shin, J.W.; Wu, Y.; Kim, J.K. MG-63 cells proliferation following various types of mechanical stimulation on cells by auxetic hybrid scaffolds. Biomater. Res. 2016, 20, 1–8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shor, L.; Güçeri, S.; Wen, X.; Gandhi, M.; Sun, W. Fabrication of three-dimensional polycaprolactone/hydroxyapatite tissue scaffolds and osteoblast-scaffold interactions in vitro. Biomaterials 2007, 28, 5291–5297. [Google Scholar] [CrossRef] [PubMed]
- Leong, W.S.; Wu, S.C.; Ng, K.; Tan, L.P. Electrospun 3D multi-scale fibrous scaffold for enhanced human dermal fibroblast infiltration. Int. J. Bioprinting 2016, 2, 81–92. [Google Scholar] [CrossRef]
- Wu, Y.; Wang, Z.; Fuh, J.Y.H.; Wong, Y.S.; Wang, W.; Thian, E.S. Direct E-jet printing of three-dimensional fibrous scaffold for tendon tissue engineering. J. Biomed. Mater. Res. B Appl. Biomater. 2015, 105, 616–627. [Google Scholar] [CrossRef]
- Chen, S.; Nakamoto, T.; Kawazoe, N.; Chen, G. Engineering multi-layered skeletal muscle tissue by using 3D microgrooved collagen scaffolds. Biomaterials 2015, 73, 23–31. [Google Scholar] [CrossRef]
- Cha, S.H.; Lee, H.J.; Koh, W.-G. Study of myoblast differentiation using multi-dimensional scaffolds consisting of nano and micropatterns. Biomater. Res. 2017, 21, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Ladd, M.R.; Lee, S.J.; Stitzel, J.D.; Atala, A.; Yoo, J.J. Co-electrospun dual scaffolding system with potential for muscle–tendon junction tissue engineering. Biomaterials 2011, 32, 1549–1559. [Google Scholar] [CrossRef] [PubMed]
- Vis, M.A.M.; Ito, K.; Hofmann, S. Impact of Culture Medium on Cellular Interactions in in vitro Co-culture Systems. Front. Bioeng. Biotechnol. 2020, 8, 911. [Google Scholar] [CrossRef]
- Harris, E.; Liu, Y.; Cunniffe, G.; Morrissey, D.; Carroll, S.; Mulhall, K.; Kelly, D.J. Biofabrication of soft tissue templates for engineering the bone-ligament interface. Biotechnol. Bioeng. 2017, 114, 2400–2411. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.-H.; Lin, F.-H.; Lin, C.-C.; Chou, C.-H.; Liu, H.-C. Cartilage tissue engineering on the surface of a novel gelatin-calcium-phosphate biphasic scaffold in a double-chamber bioreactor. J. Biomed. Mater. Res. B Appl. Biomater. 2004, 71B, 313–321. [Google Scholar] [CrossRef] [PubMed]
- Criscenti, G.; Longoni, A.; Di Luca, A.; De Maria, C.; Van Blitterswijk, C.A.; Vozzi, G.; Moroni, L. Triphasic scaffolds for the regeneration of the bone–ligament interface. Biofabrication 2016, 8, 015009. [Google Scholar] [CrossRef]
- Su, M.; Zhang, Q.; Zhu, Y.; Wang, S.; Lv, J.; Sun, J.; Qiu, P.; Fan, S.; Jin, K.; Chen, L.; et al. Preparation of Decellularized Triphasic Hierarchical Bone-Fibrocartilage-Tendon Composite Extracellular Matrix for Enthesis Regeneration. Adv. Healthc. Mater. 2019, 8, e1900831. [Google Scholar] [CrossRef]
- Moffat, K.L.; Kwei, A.S.-P.; Spalazzi, J.P.; Doty, S.B.; Levine, W.N.; Lu, H.H. Novel Nanofiber-Based Scaffold for Rotator Cuff Repair and Augmentation. Tissue Eng. Part A 2009, 15, 115–126. [Google Scholar] [CrossRef] [Green Version]
- Scaffaro, R.; Lopresti, F.; Botta, L.; Rigogliuso, S.; Ghersi, G. Preparation of three-layered porous PLA/PEG scaffold: Relationship between morphology, mechanical behavior and cell permeability. J. Mech. Behav. Biomed. Mater. 2016, 54, 8–20. [Google Scholar] [CrossRef]
- Cooper, J.A.; Lu, H.H.; Ko, F.K.; Freeman, J.W.; Laurencin, C.T. Fiber-based tissue-engineered scaffold for ligament replacement: Design considerations and in vitro evaluation. Biomaterials 2005, 26, 1523–1532. [Google Scholar] [CrossRef]
- Casanellas, I.; García-Lizarribar, A.; Lagunas, A.; Samitier, J. Producing 3D Biomimetic Nanomaterials for Musculoskeletal System Regeneration. Front. Bioeng. Biotechnol. 2018, 6, 128. [Google Scholar] [CrossRef]
- Cross, L.M.; Thakur, A.; Jalili, N.A.; Detamore, M.; Gaharwar, A.K. Nanoengineered biomaterials for repair and regeneration of orthopedic tissue interfaces. Acta Biomater. 2016, 42, 2–17. [Google Scholar] [CrossRef]
- Rossetti, L.; Kuntz, L.A.; Kunold, E.; Schock, J.; Müller, K.W.; Grabmayr, H.; Stolberg-Stolberg, J.; Pfeiffer, F.; Sieber, S.A.; Burgkart, R.; et al. The microstructure and micromechanics of the tendon–bone insertion. Nat. Mater. 2017, 16, 664–670. [Google Scholar] [CrossRef]
- Liu, C.-F.; Aschbacher-Smith, L.; Barthelery, N.J.; Dyment, N.; Butler, D.; Wylie, C. What We Should Know Before Using Tissue Engineering Techniques to Repair Injured Tendons: A Developmental Biology Perspective. Tissue Eng. Part B Rev. 2011, 17, 165–176. [Google Scholar] [CrossRef] [Green Version]
- Lee, Y.-S.; Arinzeh, T.L. Electrospun Nanofibrous Materials for Neural Tissue Engineering. Polymers 2011, 3, 413–426. [Google Scholar] [CrossRef] [Green Version]
- Liu, T.; Houle, J.D.; Xu, J.; Chan, B.P.; Chew, S.Y. Nanofibrous Collagen Nerve Conduits for Spinal Cord Repair. Tissue Eng. Part A 2012, 18, 1057–1066. [Google Scholar] [CrossRef] [Green Version]
- Boecker, A.H.; van Neerven, S.G.A.; Scheffel, J.; Tank, J.; Altinova, H.; Seidensticker, K.; Deumens, R.; Tolba, R.; Weis, J.; Brook, G.A.; et al. Pre-differentiation of mesenchymal stromal cells in combination with a microstructured nerve guide supports peripheral nerve regeneration in the rat sciatic nerve model. Eur. J. Neurosci. 2015, 43, 404–416. [Google Scholar] [CrossRef]
- Jorfi, M.; D’Avanzo, C.; Kim, D.Y.; Irimia, D. Three-Dimensional Models of the Human Brain Development and Diseases. Adv. Healthc. Mater. 2017, 7, 1700723. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Griffin, D.J.; Bonnevie, E.D.; Lachowsky, D.J.; Hart, J.C.; Sparks, H.D.; Moran, N.; Matthews, G.; Nixon, A.J.; Cohen, I.; Bonassar, L.J. Mechanical characterization of matrix-induced autologous chondrocyte implantation (MACI®) grafts in an equine model at 53 weeks. J. Biomech. 2015, 48, 1944–1949. [Google Scholar] [CrossRef] [PubMed]
- Muhonen, V.; Salonius, E.; Haaparanta, A.-M.; Järvinen, E.; Paatela, T.; Meller, A.; Hannula, M.; Björkman, M.; Pyhältö, T.; Ellä, V.; et al. Articular cartilage repair with recombinant human type II collagen/polylactide scaffold in a preliminary porcine study. J. Orthop. Res. 2015, 34, 745–753. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.; Wang, W.; Liu, S.; Wu, J.; Li, F.; Cao, L.; Liu, X.-D.; Mo, X.; Fan, C. Enhancement of chondrogenic differentiation of rabbit mesenchymal stem cells by oriented nanofiber yarn-collagen type I/hyaluronate hybrid. Mater. Sci. Eng. C 2016, 58, 1071–1076. [Google Scholar] [CrossRef]
- Ma, L.; Gao, C.Y.; Mao, Z.W.; Zhou, J.; Shen, J.C.; Hu, X.Q.; Han, C.M. Collagen/chitosan porous scaffolds with improved biostability for skin tissue engineering. Biomaterials 2003, 24, 4833–4841. [Google Scholar] [CrossRef] [PubMed]
- Rho, K.S.; Jeong, L.; Lee, G.; Seo, B.-M.; Park, Y.J.; Hong, S.-D.; Roh, S.; Cho, J.J.; Park, W.H.; Min, B.-M. Electrospinning of collagen nanofibers: Effects on the behavior of normal human keratinocytes and early-stage wound healing. Biomaterials 2006, 27, 1452–1461. [Google Scholar] [CrossRef]
- Niehues, H.; Bouwstra, J.A.; El Ghalbzouri, A.; Brandner, J.M.; Zeeuwen, P.L.J.M.; Van Den Bogaard, E.H. 3D skin models for 3R research: The potential of 3D reconstructed skin models to study skin barrier function. Exp. Dermatol. 2018, 27, 501–511. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Campbell, J.; Husmann, A.; Hume, R.; Biomaterials, C.W. Development of Three-Dimensional Collagen Scaffolds with Controlled Architecture for Cell Migration Studies Using Breast Cancer Cell Lines; Elsevier: Amsterdam, The Netherlands, 2017; Volume 114, pp. 34–43. [Google Scholar] [CrossRef] [Green Version]
- Sapudom, J.; Kalbitzer, L.; Wu, X.; Martin, S.; Kroy, K.; Pompe, T. Fibril bending stiffness of 3D collagen matrices instructs spreading and clustering of invasive and non-invasive breast cancer cells. Biomaterials 2019, 193, 47–57. [Google Scholar] [CrossRef] [PubMed]
- Wei, G.; Ma, P.X. Structure and properties of nano-hydroxyapatite/polymer composite scaffolds for bone tissue engineering. Biomaterials 2004, 25, 4749–4757. [Google Scholar] [CrossRef]
- Nocera, A.D.; Comín, R.; Salvatierra, N.A.; Cid, M.P. Development of 3D printed fibrillar collagen scaffold for tissue engineering. Biomed. Microdevices 2018, 20, 26. [Google Scholar] [CrossRef]
- Fuoco, C.; Petrilli, L.L.; Cannata, S.; Gargioli, C. Matrix scaffolding for stem cell guidance toward skeletal muscle tissue engineering. J. Orthop. Surg. Res. 2016, 11, 1–8. [Google Scholar] [CrossRef]
- Houben, A.; Van Hoorick, J.; Van Erps, J.; Thienpont, H.; Van Vlierberghe, S.; Dubruel, P. Indirect Rapid Prototyping: Opening Up Unprecedented Opportunities in Scaffold Design and Applications. Ann. Biomed. Eng. 2016, 45, 58–83. [Google Scholar] [CrossRef]
- Feng, X. Chemical and Biochemical Basis of Cell-Bone Matrix Interaction in Health and Disease. Curr. Chem. Biol. 2009, 3, 189–196. [Google Scholar] [CrossRef] [Green Version]
- Siu, P.M. Immediate Effects of 2 Different Whole-Body Vibration Frequencies on Muscle Peak Torque and Stiffness. Arch. Phys. Med. Rehabil. 2020, 91, 1608–1615. [Google Scholar] [CrossRef]
- Silldorff, M.D. Effect of Supraspinatus Tendon Injury on Supraspinatus and Infraspinatus Muscle Passive Tension and Associated Biochemistry. J. Bone Jt. Surg. 2014, 96, 175. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, B. Evaluation of normal cadaveric Achilles tendon and enthesis with ultrashort echo time (UTE) magnetic resonance imaging and indentation testing. NMR Biomed. 2019, 32, 4034. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maganaris, C.N.; Paul, J.P. In vivo human tendon mechanical properties. J. Physiol. 1999, 521, 307–313. [Google Scholar] [CrossRef]
- Augat, P.; Link, T.; Lang, T.F.; Lin, J.C.; Majumdar, S.; Genant, H.K. Anisotropy of the elastic modulus of trabecular bone specimens from different anatomical locations. Med. Eng. Phys. 1998, 20, 124–131. [Google Scholar] [CrossRef] [PubMed]
- Zysset, P.K.; Edward Guo, X.; Edward Hoffler, C.; Moore, K.E.; Goldstein, S.A. Elastic modulus and hardness of cortical and trabecular bone lamellae measured by nanoindentation in the human femur. J. Biomech. 1999, 32, 1005–1012. [Google Scholar] [CrossRef]
- Zhao, H.; Xu, K.; Zhu, P.; Wang, C.; Chi, Q. Smart hydrogels with high tunability of stiffness as a biomimetic cell carrier. Cell Biol. Int. 2019, 43, 84–97. [Google Scholar] [CrossRef]
- Stamenović, D.; Fredberg, J.J.; Wang, N.; Butler, J.P.; Ingber, D.E. A microstructural approach to cytoskeletal mechanics based on tensegrity. J. Theor. Biol. 1999, 181, 125–136. [Google Scholar] [CrossRef]
- Wang, N. Mechanical behavior in living cells consistent with the tensegrity model. Proc. Natl. Acad. Sci. USA 2001, 98, 7765–7770. [Google Scholar] [CrossRef] [Green Version]
- Billman, G.E. Homeostasis: The Underappreciated and Far Too Often Ignored Central Organizing Principle of Physiology. Front. Physiol. 2020, 11, 200. [Google Scholar] [CrossRef] [Green Version]
- Seidi, A.; Ramalingam, M.; Elloumi-Hannachi, I.; Ostrovidov, S.; Khademhosseini, A. Gradient biomaterials for soft-to-hard interface tissue engineering. Acta Biomater. 2011, 7, 1441–1451. [Google Scholar] [CrossRef]
- Frankfurt, O.S. Flow cytometric analysis of double-stranded RNA content distributions. J. Histochem. Cytochem. 1980, 28, 663–669. [Google Scholar] [CrossRef] [Green Version]
- Jones, L.J.; Gray, M.; Yue, S.T.; Haugland, R.P.; Singer, V.L. Sensitive determination of cell number using the CyQUANT® cell proliferation assay. J. Immunol. Methods 2001, 254, 85–98. [Google Scholar] [CrossRef] [PubMed]
- Langenbach, F.; Handschel, J. Effects of dexamethasone, ascorbic acid and β-glycerophosphate on the osteogenic differentiation of stem cells in vitro. Stem. Cell Res. Ther. 2013, 4, 1–7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Robertson, G.; Bushell, T.J.; Zagnoni, M. Chemically induced synaptic activity between mixed primary hippocampal co-cultures in a microfluidic system. Integr. Biol. 2014, 6, 636–644. [Google Scholar] [CrossRef] [Green Version]
- Sardelli, L.; Pacheco, D.P.; Zorzetto, L.; Rinoldi, C.; Święszkowski, W.; Petrini, P. Engineering biological gradients. J. Appl. Biomater. Funct. Mater. 2019, 17, 2280800019829023. [Google Scholar] [CrossRef] [PubMed]
- Oragui, E. The Role of Bioreactors in Tissue Engineering for Musculoskeletal Applications. Open Orthop. J. 2011, 5, 267–270. [Google Scholar] [CrossRef] [Green Version]
- Bar, A.; Ruvinov, E.; Cohen, S. Live imaging flow bioreactor for the simulation of articular cartilage regeneration after treatment with bioactive hydrogel. Biotechnol. Bioeng. 2018, 115, 2205–2216. [Google Scholar] [CrossRef]
- Reinwald, Y.; El Haj, A.J. Hydrostatic pressure in combination with topographical cues affects the fate of bone marrow-derived human mesenchymal stem cells for bone tissue regeneration. J. Biomed. Mater. Res. Part A 2017, 106, 629–640. [Google Scholar] [CrossRef] [Green Version]
- Zhao, F.; Ma, T. Perfusion bioreactor system for human mesenchymal stem cell tissue engineering: Dynamic cell seeding and construct development. Biotechnol. Bioeng. 2005, 91, 482–493. [Google Scholar] [CrossRef]
- Zhang, J.; Hu, Z.Q.; Turner, N.J.; Teng, S.F.; Cheng, W.Y.; Zhou, H.Y.; Zhang, L.; Hu, H.W.; Wang, Q.; Badylak, S.F. Perfusion-decellularized skeletal muscle as a three-dimensional scaffold with a vascular network template. Biomaterials 2016, 89, 114–126. [Google Scholar] [CrossRef]
- Garcia, A.G.; Labro, M.B.; Farhat, F.; Perot, J.-B.; Dermigny, Q.; Dufresne, M.; Grosset, J.-F.; Bedoui, F.; Legallais, C. Multi-scale approach to reconstruct a bioartificial system of system: The example of the bone-tendon-muscle continuum. In Proceedings of the 2018 13th System of Systems Engineering Conference, SoSE 2018, Paris, France, 19–22 June 2018; pp. 408–410. [Google Scholar] [CrossRef]
- Krafts, K.P. Tissue repair: The hidden drama. Organogenesis 2010, 6, 225–233. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roumeliotis, S.; Roumeliotis, A.; Dounousi, E.; Eleftheriadis, T.; Liakopoulos, V. Biomarkers of vascular calcification in serum. In Advances in Clinical Chemistry; Academic Press Inc.: Cambridge, MA, USA, 2020; Volume 98, pp. 91–147. [Google Scholar] [CrossRef]
- Salvatore, V.; Teti, G.; Focaroli, S.; Mazzotti, M.C.; Mazzotti, A.; Falconi, M. The tumor microenvironment promotes cancer progression and cell migration. Oncotarget 2017, 8, 9608–9616. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tschumperlin, D.J. Fibroblasts and the Ground They Walk On. Physiology 2013, 28, 380–390. [Google Scholar] [CrossRef] [PubMed]






| Type of Hydrogel | Collagen (mg/mL) | Agarose (% w/v) | Hydroxyapatite (% v/v) | Crosslinked |
|---|---|---|---|---|
| Biphasic (muscle) | 3 | 0.75 | 0 | No |
| Triphasic (tendon) | 3 | 0.75 | 0.2 | Yes |
| Triphasic (bone) | 3 | 0.75 | 40 | Yes |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Balestri, W.; Hickman, G.J.; Morris, R.H.; Hunt, J.A.; Reinwald, Y. Triphasic 3D In Vitro Model of Bone-Tendon-Muscle Interfaces to Study Their Regeneration. Cells 2023, 12, 313. https://doi.org/10.3390/cells12020313
Balestri W, Hickman GJ, Morris RH, Hunt JA, Reinwald Y. Triphasic 3D In Vitro Model of Bone-Tendon-Muscle Interfaces to Study Their Regeneration. Cells. 2023; 12(2):313. https://doi.org/10.3390/cells12020313
Chicago/Turabian StyleBalestri, Wendy, Graham J. Hickman, Robert H. Morris, John A. Hunt, and Yvonne Reinwald. 2023. "Triphasic 3D In Vitro Model of Bone-Tendon-Muscle Interfaces to Study Their Regeneration" Cells 12, no. 2: 313. https://doi.org/10.3390/cells12020313
APA StyleBalestri, W., Hickman, G. J., Morris, R. H., Hunt, J. A., & Reinwald, Y. (2023). Triphasic 3D In Vitro Model of Bone-Tendon-Muscle Interfaces to Study Their Regeneration. Cells, 12(2), 313. https://doi.org/10.3390/cells12020313








