Development of New Genome Editing Tools for the Treatment of Hyperlipidemia
Abstract
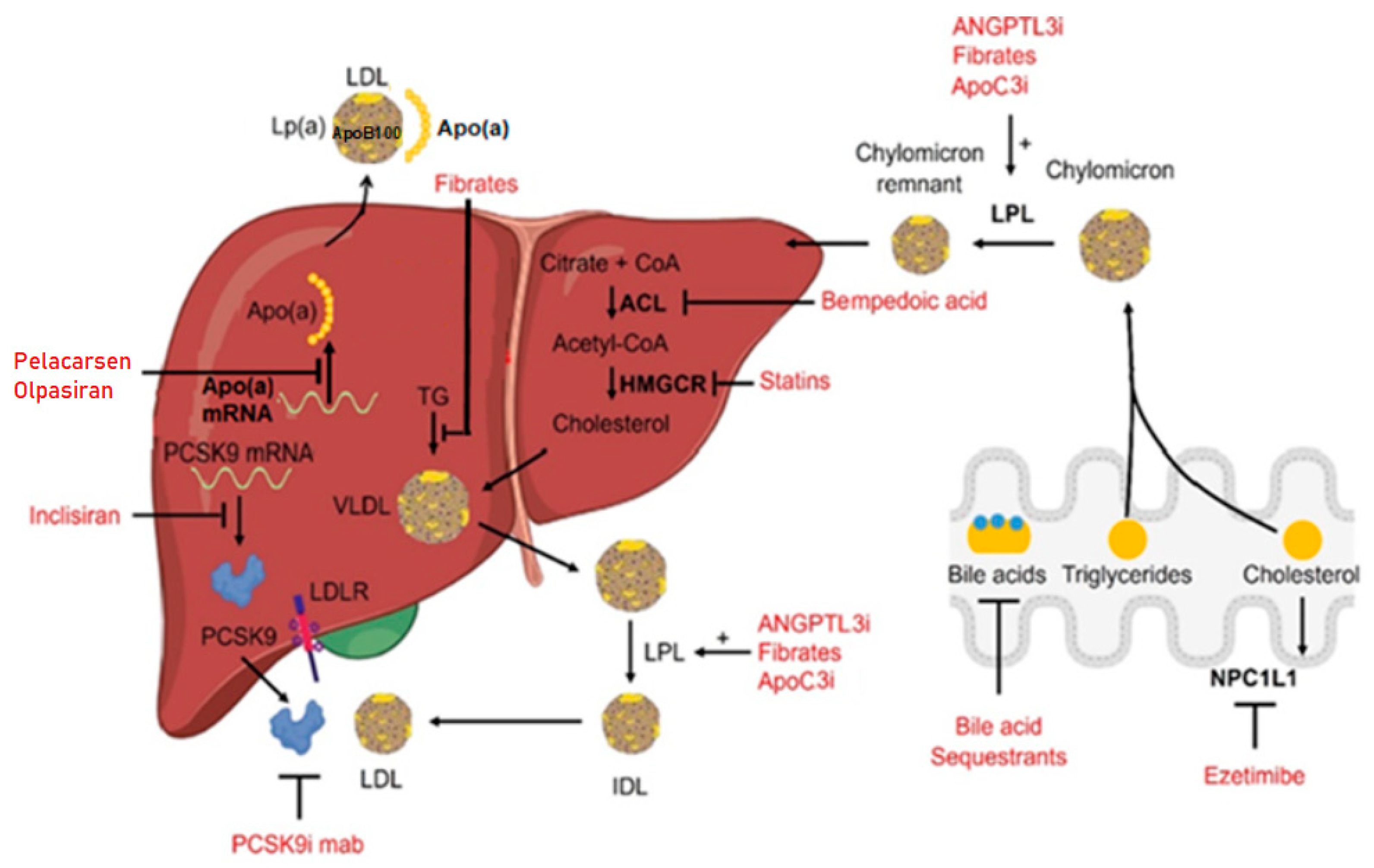
:1. Introduction
2. PCSK9 Gene Editing Strategies in Mice
3. PCSK9 Gene Editing Strategies in Non-Human Primates
4. ANGPTL3 Gene Editing Strategies in Mice
5. ANGPTL3 Gene Editing Strategies in Non-Human Primates
6. LDLR Gene Editing Strategies in Mice
7. LDLR Gene Editing Strategies in Other Animal Models
8. Apolipoproteins Gene Editing Strategies in Mice
9. Apolipoproteins Gene Editing Strategies in Other Animal Models
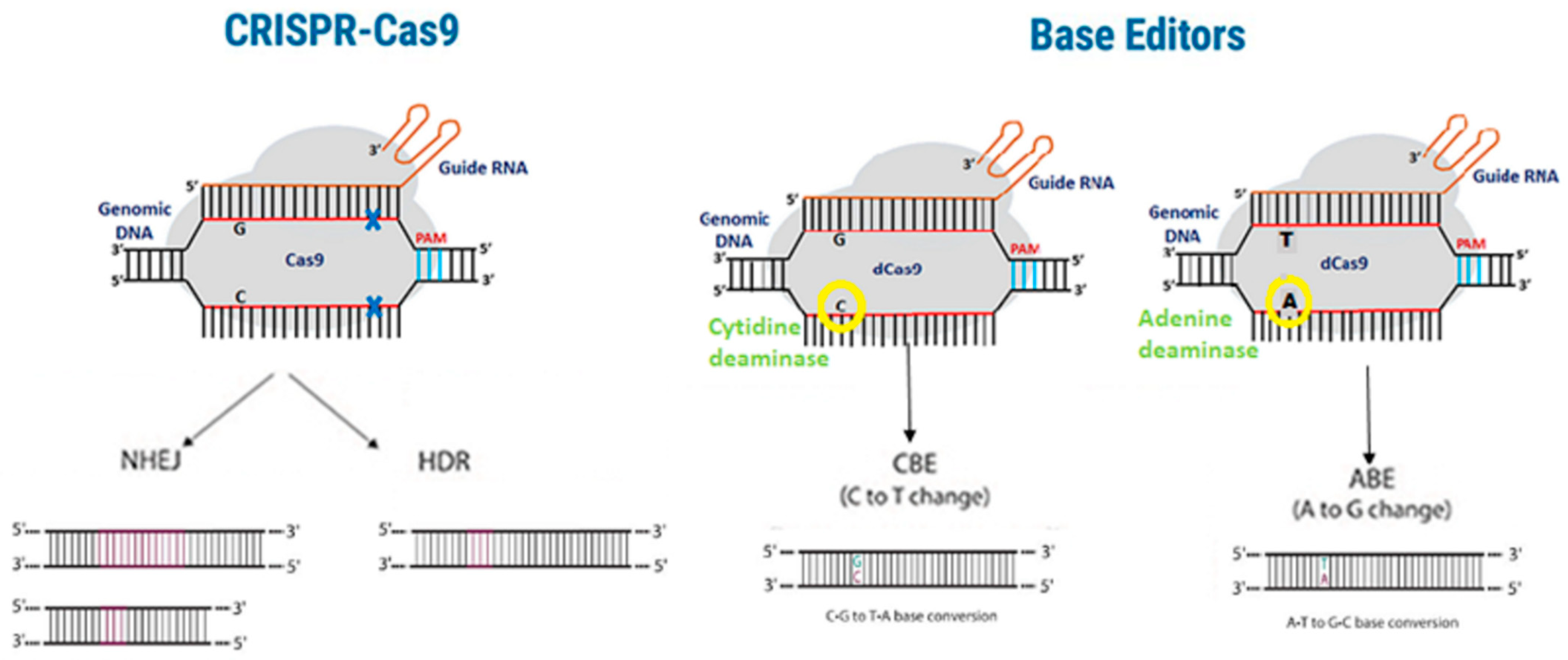
10. Off-Target Effects in CRISPR-Cas9 Gene Editing
11. Conclusions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AAV: | Adeno-associated virus |
| ABE: | Adenine base editor |
| ALT: | Alanine aminotransferase |
| ANGPTL3: | Angiopoietin like 3 |
| Apo(a): | Apoliporotein (a) |
| ApoA1: | Apolipoprotein A1 |
| ApoB: | Apolipoprotein B |
| ApoC3: | Apolipoprotein C3 |
| ASGPR: | Asialoglycoprotein receptor |
| AST: | Aspartate aminotransferase |
| Cas9: | CRISPR-associated protein 9 |
| CRISPR: | Clustered regularly interspaced short palindromic repeats |
| CVD: | Cardiovascular diseases |
| ER: | Endoplasmic reticulum |
| FDA: | Food and Drug Administration |
| FH: | Familial hypercholesterolemia |
| GalNac: | Multi-valent N-AcetylGalactosamine |
| gRNA: | guide RNA |
| HDL: | High-density lipoprotein |
| HMG-CoA: | β-Hydroxy β-methylglutaryl-CoA |
| HoFH: | Homozygous familial hypercholesterolemia |
| KO: | Knockout |
| LDL: | Low-density lipoprotein |
| LDLR: | Low-density lipoprotein receptor |
| LLN: | Lipid-like nanoparticle |
| LNP: | Lipid nanoparticle |
| Lp(a): | Lipoprotein A |
| NHP: | Non-human primates |
| PCSK9: | Proprotein convertase subtilisin/kexin type 9 |
| PPAR-alpha: | Peroxisome proliferator-activated receptor alpha |
| RNP: | Ribonucleoprotein |
| RXR: | Retinoid X receptor |
| siRNA: | Small interfering RNA |
| TC: | Total cholesterol |
| TG: | Triglycerides |
| VLDL: | Very low density lipoprotein |
| WT: | Wild-type |
References
- Jinek, M.; Chylinski, K.; Fonfara, I.; Hauer, M.; Doudna, J.A.; Charpentier, E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 2012, 337, 816–821. [Google Scholar] [CrossRef]
- Karimian, A.; Azizian, K.; Parsian, H.; Rafieian, S.; Shafiei-Irannejad, V.; Kheyrollah, M.; Yousefi, M.; Majidinia, M.; Yousefi, B. CRISPR/Cas9 technology as a potent molecular tool for gene therapy. J. Cell. Physiol. 2019, 234, 12267–12277. [Google Scholar] [CrossRef]
- Jarrett, K.E.; Lee, C.M.; Yeh, Y.-H.; Hsu, R.H.; Gupta, R.; Zhang, M.; Rodriguez, P.J.; Lee, C.S.; Gillard, B.K.; Bissig, K.-D.; et al. Somatic genome editing with CRISPR/Cas9 generates and corrects a metabolic disease. Sci. Rep. 2017, 7, 44624. [Google Scholar] [CrossRef]
- Yin, H.; Song, C.-Q.; Dorkin, J.R.; Zhu, L.J.; Li, Y.; Wu, Q.; Park, A.; Yang, J.; Suresh, S.; Bizhanova, A.; et al. Therapeutic genome editing by combined viral and non-viral delivery of CRISPR system components in vivo. Nat. Biotechnol. 2016, 34, 328–333. [Google Scholar] [CrossRef]
- Akinc, A.; Querbes, W.; De, S.; Qin, J.; Frank-Kamenetsky, M.; Jayaprakash, K.N.; Jayaraman, M.; Rajeev, K.G.; Cantley, W.L.; Dorkin, J.R.; et al. Targeted delivery of RNAi therapeutics with endogenous and exogenous ligand-based mechanisms. Mol. Ther. 2010, 18, 1357–1364. [Google Scholar] [CrossRef]
- Ahn, C.H.; Choi, S.H. New drugs for treating dyslipidemia: Beyond statins. Diabetes Metab. J. 2015, 39, 87–94. [Google Scholar] [CrossRef]
- Brautbar, A.; Ballantyne, C.M. Pharmacological strategies for lowering LDL cholesterol: Statins and beyond. Nat. Rev. Cardiol. 2011, 8, 253–265. [Google Scholar] [CrossRef]
- Muscoli, S.; Ifrim, M.; Russo, M.; Candido, F.; Sanseviero, A.; Milite, M.; Di Luozzo, M.; Marchei, M.; Sangiorgi, G.M. Current Options and Future Perspectives in the Treatment of Dyslipidemia. J. Clin. Med. 2022, 11, 4716. [Google Scholar] [CrossRef]
- Nurmohamed, N.S.; Stroes, E.S.G. Working towards full eradication of lipid-driven cardiovascular risk? Neth. Heart J. 2022, 30, 15–24. [Google Scholar] [CrossRef]
- Schirris, T.J.J.; Renkema, G.H.; Ritschel, T.; Voermans, N.C.; Bilos, A.; van Engelen, B.G.M.; Brandt, U.; Koopman, W.J.H.; Beyrath, J.D.; Rodenburg, R.J.; et al. Statin-Induced Myopathy Is Associated with Mitochondrial Complex III Inhibition. Cell Metab. 2015, 22, 399–407. [Google Scholar] [CrossRef]
- Lim, G.B. Dyslipidaemia: Balancing the benefits and risks of statin therapy. Nat. Rev. Cardiol. 2016, 13, 632–633. [Google Scholar] [CrossRef]
- Bardolia, C.; Amin, N.S.; Turgeon, J. Emerging Non-statin Treatment Options for Lowering Low-Density Lipoprotein Cholesterol. Front. Cardiovasc. Med. 2021, 8, 789931. [Google Scholar] [CrossRef]
- Garcia-Calvo, M.; Lisnock, J.; Bull, H.G.; Hawes, B.E.; Burnett, D.A.; Braun, M.P.; Crona, J.H.; Davis, H.R.J.; Dean, D.C.; Detmers, P.A.; et al. The target of ezetimibe is Niemann-Pick C1-Like 1 (NPC1L1). Proc. Natl. Acad. Sci. USA 2005, 102, 8132–8137. [Google Scholar] [CrossRef]
- Pandor, A.; Ara, R.M.; Tumur, I.; Wilkinson, A.J.; Paisley, S.; Duenas, A.; Durrington, P.N.; Chilcott, J. Ezetimibe monotherapy for cholesterol lowering in 2,722 people: Systematic review and meta-analysis of randomized controlled trials. J. Intern. Med. 2009, 265, 568–580. [Google Scholar] [CrossRef] [PubMed]
- Nodari, S.; Rocca, P.; Saporetti, A.; Bettari, L.; Foresti, A.L.; Tanghetti, E.; Metra, M.; Dei Cas, L. The combination of Ezetimibe and Statin: A new treatment for hypercholesterolemia. Heart Int. 2007, 3, 12. [Google Scholar] [CrossRef] [PubMed]
- Descamps, O.; Tomassini, J.E.; Lin, J.; Polis, A.B.; Shah, A.; Brudi, P.; Hanson, M.E.; Tershakovec, A.M. Variability of the LDL-C lowering response to ezetimibe and ezetimibe + statin therapy in hypercholesterolemic patients. Atherosclerosis 2015, 240, 482–489. [Google Scholar] [CrossRef] [PubMed]
- Ghasempour, G.; Zamani-Garmsiri, F.; Shaikhnia, F.; Soleimani, A.A.; Fard, S.R.H.; Leila, J.; Teimuri, S.; Parvaz, N.; Mohammadi, P.; Najafi, M. Efficacy and Safety of Alirocumab and Evolocumab as Proprotein Convertase Subtilisin/Kexin Type 9 (PCSK9) Inhibitors in Familial Hypercholesterolemia: A Systematic Review and Meta-Analysis. Curr. Med. Chem. 2023, 31, 223–241. [Google Scholar] [CrossRef] [PubMed]
- McDonagh, M.; Peterson, K.; Holzhammer, B.; Fazio, S. A Systematic Review of PCSK9 Inhibitors Alirocumab and Evolocumab. J. Manag. Care Spec. Pharm. 2016, 22, 641q–653q. [Google Scholar] [CrossRef]
- Kuzmich, N.; Andresyuk, E.; Porozov, Y.; Tarasov, V.; Samsonov, M.; Preferanskaya, N.; Veselov, V.; Alyautdin, R. PCSK9 as a Target for Development of a New Generation of Hypolipidemic Drugs. Molecules 2022, 27, 434. [Google Scholar] [CrossRef]
- Lagace, T.A. PCSK9 and LDLR degradation: Regulatory mechanisms in circulation and in cells. Curr. Opin. Lipidol. 2014, 25, 387–393. [Google Scholar] [CrossRef]
- Choi, J.Y.; Na, J.O. Pharmacological Strategies beyond Statins: Ezetimibe and PCSK9 Inhibitors. J. Lipid. Atheroscler. 2019, 8, 183–191. [Google Scholar] [CrossRef] [PubMed]
- Ray, K.K.; Wright, R.S.; Kallend, D.; Koenig, W.; Leiter, L.A.; Raal, F.J.; Bisch, J.A.; Richardson, T.; Jaros, M.; Wijngaard, P.L.; et al. Two Phase 3 Trials of Inclisiran in Patients with Elevated LDL Cholesterol. N. Engl. J. Med. 2020, 382, 1507–1519. [Google Scholar] [CrossRef]
- Katsiki, N.; Vrablik, M.; Banach, M.; Gouni-Berthold, I. Inclisiran, Low-Density Lipoprotein Cholesterol and Lipoprotein (a). Pharmaceuticals 2023, 16, 577. [Google Scholar] [CrossRef]
- Kosmas, C.E.; Bousvarou, M.D.; Sourlas, A.; Papakonstantinou, E.J.; Peña Genao, E.; Echavarria Uceta, R.; Guzman, E. Angiopoietin-Like Protein 3 (ANGPTL3) Inhibitors in the Management of Refractory Hypercholesterolemia. Clin. Pharmacol. 2022, 14, 49–59. [Google Scholar] [CrossRef] [PubMed]
- Karwatowska-Prokopczuk, E.; Tardif, J.-C.; Gaudet, D.; Ballantyne, C.M.; Shapiro, M.D.; Moriarty, P.M.; Baum, S.J.; Amour, E.S.; Alexander, V.J.; Xia, S.; et al. Effect of olezarsen targeting APOC-III on lipoprotein size and particle number measured by NMR in patients with hypertriglyceridemia. J. Clin. Lipidol. 2022, 16, 617–625. [Google Scholar] [CrossRef] [PubMed]
- Tardif, J.-C.; Karwatowska-Prokopczuk, E.; Amour, E.S.; Ballantyne, C.M.; Shapiro, M.D.; Moriarty, P.M.; Baum, S.J.; Hurh, E.; Bartlett, V.J.; Kingsbury, J.; et al. Apolipoprotein C-III reduction in subjects with moderate hypertriglyceridaemia and at high cardiovascular risk. Eur. Heart J. 2022, 43, 1401–1412. [Google Scholar] [CrossRef]
- Tsimikas, S.; Viney, N.J.; Hughes, S.G.; Singleton, W.; Graham, M.J.; Baker, B.F.; Burkey, J.L.; Yang, Q.; Marcovina, S.M.; Geary, R.S.; et al. Antisense therapy targeting apolipoprotein(a): A randomised, double-blind, placebo-controlled phase 1 study. Lancet 2015, 386, 1472–1483. [Google Scholar] [CrossRef]
- Viney, N.J.; van Capelleveen, J.C.; Geary, R.S.; Xia, S.; Tami, J.A.; Yu, R.Z.; Marcovina, S.M.; Hughes, S.G.; Graham, M.J.; Crooke, R.M.; et al. Antisense oligonucleotides targeting apolipoprotein(a) in people with raised lipoprotein(a): Two randomised, double-blind, placebo-controlled, dose-ranging trials. Lancet 2016, 388, 2239–2253. [Google Scholar] [CrossRef]
- Koren, M.J.; Moriarty, P.M.; Baum, S.J.; Neutel, J.; Hernandez-Illas, M.; Weintraub, H.S.; Florio, M.; Kassahun, H.; Melquist, S.; Varrieur, T.; et al. Preclinical development and phase 1 trial of a novel siRNA targeting lipoprotein(a). Nat. Med. 2022, 28, 96–103. [Google Scholar] [CrossRef]
- Sohn, W.; Winkle, P.; Neutel, J.; Wu, Y.; Jabari, F.; Terrio, C.; Varrieur, T.; Wang, J.; Hellawell, J. Pharmacokinetics, Pharmacodynamics, and Tolerability of Olpasiran in Healthy Japanese and Non-Japanese Participants: Results from a Phase I, Single-dose, Open-label Study. Clin. Ther. 2022, 44, 1237–1247. [Google Scholar] [CrossRef]
- Rider, D.A.; Eisermann, M.; Löffler, K.; Aleku, M.; Swerdlow, D.I.; Dames, S.; Hauptmann, J.; Morrison, E.; Lindholm, M.W.; Schubert, S.; et al. Pre-clinical assessment of SLN360, a novel siRNA targeting LPA, developed to address elevated lipoprotein (a) in cardiovascular disease. Atherosclerosis 2022, 349, 240–247. [Google Scholar] [CrossRef] [PubMed]
- Ganji, S.H.; Kamanna, V.S.; Kashyap, M.L. Niacin and cholesterol: Role in cardiovascular disease (review). J. Nutr. Biochem. 2003, 14, 298–305. [Google Scholar] [CrossRef] [PubMed]
- The HPS2-THRIVE Collaborative Group. Effects of extended-release niacin with laropiprant in high-risk patients. N. Engl. J. Med. 2014, 371, 203–212. [Google Scholar] [CrossRef]
- The AIM-HIGH Investigators. Niacin in patients with low HDL cholesterol levels receiving intensive statin therapy. N. Engl. J. Med. 2011, 365, 2255–2267. [Google Scholar] [CrossRef] [PubMed]
- Zeman, M.; Vecka, M.; Perlík, F.; Hromádka, R.; Staňková, B.; Tvrzická, E. Niacin in the Treatment of Hyperlipidemias in Light of New Clinical Trials: Has Niacin Lost its Place? Med. Sci. Monit. 2015, 21, 2156–2162. [Google Scholar] [PubMed]
- Hansen, M.; Sonne, D.P.; Knop, F.K. Bile acid sequestrants: Glucose-lowering mechanisms and efficacy in type 2 diabetes. Curr. Diab. Rep. 2014, 14, 482. [Google Scholar] [CrossRef]
- Lefebvre, P.; Cariou, B.; Lien, F.; Kuipers, F.; Staels, B. Role of bile acids and bile acid receptors in metabolic regulation. Physiol. Rev. 2009, 89, 147–191. [Google Scholar] [CrossRef]
- Feng, Y.; Li, Q.; Ou, G.; Yang, M.; Du, L. Bile acid sequestrants: A review of mechanism and design. J. Pharm. Pharmacol. 2021, 73, 855–861. [Google Scholar] [CrossRef]
- Abourbih, S.; Filion, K.B.; Joseph, L.; Schiffrin, E.L.; Rinfret, S.; Poirier, P.; Pilote, L.; Genest, J.; Eisenberg, M.J. Effect of fibrates on lipid profiles and cardiovascular outcomes: A systematic review. Am. J. Med. 2009, 122, 962.E1–962.E8. [Google Scholar] [CrossRef]
- Kim, N.H.; Kim, S.G. Fibrates Revisited: Potential Role in Cardiovascular Risk Reduction. Diabetes Metab. J. 2020, 44, 213–221. [Google Scholar] [CrossRef]
- Fazio, S.; Linton, M.F. The role of fibrates in managing hyperlipidemia: Mechanisms of action and clinical efficacy. Curr. Atheroscler. Rep. 2004, 6, 148–157. [Google Scholar] [CrossRef] [PubMed]
- Staels, B.; Dallongeville, J.; Auwerx, J.; Schoonjans, K.; Leitersdorf, E.; Fruchart, J.-C. Mechanism of action of fibrates on lipid and lipoprotein metabolism. Circulation 1998, 98, 2088–2093. [Google Scholar] [CrossRef] [PubMed]
- Bilen, O.; Ballantyne, C.M. Bempedoic Acid (ETC-1002): An Investigational Inhibitor of ATP Citrate Lyase. Curr. Atheroscler. Rep. 2016, 18, 61. [Google Scholar] [CrossRef]
- Ray, K.K.; Bays, H.E.; Catapano, A.L.; Lalwani, N.D.; Bloedon, L.T.; Sterling, L.R.; Robinson, P.L.; Ballantyne, C.M. Safety and Efficacy of Bempedoic Acid to Reduce LDL Cholesterol. N. Engl. J. Med. 2019, 380, 1022–1032. [Google Scholar] [CrossRef]
- Nissen, S.E.; Lincoff, A.M.; Brennan, D.; Ray, K.K.; Mason, D.; Kastelein, J.J.; Thompson, P.D.; Libby, P.; Cho, L.; Plutzky, J.; et al. Bempedoic Acid and Cardiovascular Outcomes in Statin-Intolerant Patients. N. Engl. J. Med. 2023, 388, 1353–1364. [Google Scholar] [CrossRef]
- Sundararaman, S.S.; Döring, Y.; van Der Vorst, E.P. PCSK9: A Multi-Faceted Protein That Is Involved in Cardiovascular Biology. Biomedicines 2021, 9, 793. [Google Scholar] [CrossRef]
- Xia, X.-D.; Peng, Z.-S.; Gu, H.-M.; Wang, M.; Wang, G.-Q.; Zhang, D.-W. Regulation of PCSK9 Expression and Function: Mechanisms and Therapeutic Implications. Front. Cardiovasc. Med. 2021, 8, 764038. [Google Scholar] [CrossRef]
- Horton, J.D.; Cohen, J.C.; Hobbs, H.H. Molecular biology of PCSK9: Its role in LDL metabolism. Trends Biochem. Sci. 2007, 32, 71–77. [Google Scholar] [CrossRef]
- Defesche, J.C.; Gidding, S.S.; Harada-Shiba, M.; Hegele, R.A.; Santos, R.D.; Wierzbicki, A.S. Familial hypercholesterolaemia. Nat. Rev. Dis. Primers 2017, 3, 17093. [Google Scholar] [CrossRef] [PubMed]
- Di Taranto, M.D.; Benito-Vicente, A.; Giacobbe, C.; Uribe, K.B.; Rubba, P.; Etxebarria, A.; Guardamagna, O.; Gentile, M.; Martín, C.; Fortunato, G. Identification and in vitro characterization of two new PCSK9 Gain of Function variants found in patients with Familial Hypercholesterolemia. Sci. Rep. 2017, 7, 15282. [Google Scholar] [CrossRef] [PubMed]
- Leren, T.P.; Berge, K.E. Identification of mutations in the apolipoprotein B-100 gene and in the PCSK9 gene as the cause of hypocholesterolemia. Clin. Chim. Acta 2008, 397, 92–95. [Google Scholar] [CrossRef] [PubMed]
- Berge, K.E.; Ose, L.; Leren, T.P. Missense mutations in the PCSK9 gene are associated with hypocholesterolemia and possibly increased response to statin therapy. Arterioscler. Thromb. Vasc. Biol. 2006, 26, 1094–1100. [Google Scholar] [CrossRef]
- Garvie, C.W.; Fraley, C.V.; Elowe, N.H.; Culyba, E.K.; Lemke, C.T.; Hubbard, B.K.; Kaushik, V.K.; Daniels, D.S. Point mutations at the catalytic site of PCSK9 inhibit folding, autoprocessing, and interaction with the LDL receptor. Protein Sci. 2016, 25, 2018–2027. [Google Scholar] [CrossRef] [PubMed]
- Kudo, T.; Sasaki, K.; Tada, H. Familial hypobetalipoproteinemia caused by homozygous loss-of-function mutations in PCSK9: A case report. J. Clin. Lipidol. 2022, 16, 596–600. [Google Scholar] [CrossRef]
- Katzmann, J.L.; Cupido, A.J.; Laufs, U. Gene Therapy Targeting PCSK9. Metabolites 2022, 12, 70. [Google Scholar] [CrossRef] [PubMed]
- Stankov, S.; Cuchel, M. Gene editing for dyslipidemias: New tools to “cut” lipids. Atherosclerosis 2023, 368, 14–24. [Google Scholar] [CrossRef]
- Ding, Q.; Strong, A.; Patel, K.M.; Ng, S.-L.; Gosis, B.S.; Regan, S.N.; Cowan, C.A.; Rader, D.J.; Musunuru, K. Permanent alteration of PCSK9 with in vivo CRISPR-Cas9 genome editing. Circ. Res. 2014, 115, 488–492. [Google Scholar] [CrossRef]
- Wang, X.; Raghavan, A.; Chen, T.; Qiao, L.; Zhang, Y.; Ding, Q.; Musunuru, K. CRISPR-Cas9 Targeting of PCSK9 in Human Hepatocytes In Vivo-Brief Report. Arterioscler. Thromb. Vasc. Biol. 2016, 36, 783–786. [Google Scholar] [CrossRef]
- Zhu, J.; Huang, X.; Yang, Y. Innate immune response to adenoviral vectors is mediated by both Toll-like receptor-dependent and -independent pathways. J. Virol. 2007, 81, 3170–3180. [Google Scholar] [CrossRef]
- Mingozzi, F.; High, K.A. Immune responses to AAV vectors: Overcoming barriers to successful gene therapy. Blood 2013, 122, 23–36. [Google Scholar] [CrossRef]
- Mingozzi, F.; High, K.A. Therapeutic in vivo gene transfer for genetic disease using AAV: Progress and challenges. Nat. Rev. Genet. 2011, 12, 341–355. [Google Scholar] [CrossRef] [PubMed]
- Kotterman, M.A.; Chalberg, T.W.; Schaffer, D.V. Viral Vectors for Gene Therapy: Translational and Clinical Outlook. Annu. Rev. Biomed. Eng. 2015, 17, 63–89. [Google Scholar] [CrossRef] [PubMed]
- Ran, F.A.; Cong, L.; Yan, W.X.; Scott, D.A.; Gootenberg, J.S.; Kriz, A.J.; Zetsche, B.; Shalem, O.; Wu, X.; Makarova, K.S.; et al. In vivo genome editing using Staphylococcus aureus Cas9. Nature 2015, 520, 186–191. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Su, J.; Liu, Y.; Jin, X.; Zhong, X.; Mo, L.; Wang, Q.; Deng, H.; Yang, Y. In vivo PCSK9 gene editing using an all-in-one self-cleavage AAV-CRISPR system. Mol. Ther. Methods Clin. Dev. 2021, 20, 652–659. [Google Scholar] [CrossRef]
- Jiang, C.; Mei, M.; Li, B.; Zhu, X.; Zu, W.; Tian, Y.; Wang, Q.; Guo, Y.; Dong, Y.; Tan, X. A non-viral CRISPR/Cas9 delivery system for therapeutically targeting HBV DNA and pcsk9 in vivo. Cell Res. 2017, 27, 440–443. [Google Scholar] [CrossRef]
- Li, B.; Luo, X.; Deng, B.; Wang, J.; McComb, D.W.; Shi, Y.; Gaensler, K.M.L.; Tan, X.; Dunn, A.L.; Kerlin, B.A.; et al. An Orthogonal Array Optimization of Lipid-like Nanoparticles for mRNA Delivery in Vivo. Nano Lett. 2015, 15, 8099–8107. [Google Scholar] [CrossRef]
- Cheng, Q.; Wei, T.; Farbiak, L.; Johnson, L.T.; Dilliard, S.A.; Siegwart, D.J. Selective organ targeting (SORT) nanoparticles for tissue-specific mRNA delivery and CRISPR-Cas gene editing. Nat. Nanotechnol. 2020, 15, 313–320. [Google Scholar] [CrossRef]
- Yin, H.; Song, C.-Q.; Suresh, S.; Wu, Q.; Walsh, S.; Rhym, L.H.; Mintzer, E.; Bolukbasi, M.F.; Zhu, L.J.; Kauffman, K.; et al. Structure-guided chemical modification of guide RNA enables potent non-viral in vivo genome editing. Nat. Biotechnol. 2017, 35, 1179–1187. [Google Scholar] [CrossRef]
- Carreras, A.; Pane, L.S.; Nitsch, R.; Madeyski-Bengtson, K.; Porritt, M.; Akcakaya, P.; Taheri-Ghahfarokhi, A.; Ericson, E.; Bjursell, M.; Perez-Alcazar, M.; et al. In vivo genome and base editing of a human PCSK9 knock-in hypercholesterolemic mouse model. BMC Biol. 2019, 17, 4. [Google Scholar] [CrossRef]
- Wang, L.; Breton, C.; Warzecha, C.C.; Bell, P.; Yan, H.; He, Z.; White, J.; Zhu, Y.; Li, M.; Buza, E.L.; et al. Long-term stable reduction of low-density lipoprotein in nonhuman primates following in vivo genome editing of PCSK9. Mol. Ther. 2021, 29, 2019–2029. [Google Scholar] [CrossRef]
- Silva, G.; Poirot, L.; Galetto, R.; Smith, J.; Montoya, G.; Duchateau, P.; Paques, F. Meganucleases and other tools for targeted genome engineering: Perspectives and challenges for gene therapy. Curr. Gene Ther. 2011, 11, 11–27. [Google Scholar] [CrossRef]
- Tröder, S.E.; Zevnik, B. History of genome editing: From meganucleases to CRISPR. Lab. Anim. 2022, 56, 60–68. [Google Scholar] [CrossRef] [PubMed]
- Musunuru, K.; Chadwick, A.C.; Mizoguchi, T.; Garcia, S.P.; DeNizio, J.E.; Reiss, C.W.; Wang, K.; Iyer, S.; Dutta, C.; Clendaniel, V.; et al. In vivo CRISPR base editing of PCSK9 durably lowers cholesterol in primates. Nature 2021, 593, 429–434. [Google Scholar] [CrossRef] [PubMed]
- Rothgangl, T.; Dennis, M.K.; Lin, P.J.; Oka, R.; Witzigmann, D.; Villiger, L.; Qi, W.; Hruzova, M.; Kissling, L.; Lenggenhager, D.; et al. In vivo adenine base editing of PCSK9 in macaques reduces LDL cholesterol levels. Nat. Biotechnol. 2021, 39, 949–957. [Google Scholar] [CrossRef] [PubMed]
- Gaudelli, N.M.; Komor, A.C.; Rees, H.A.; Packer, M.S.; Badran, A.H.; Bryson, D.I.; Liu, D.R. Programmable base editing of A*T to G*C in genomic DNA without DNA cleavage. Nature 2017, 551, 464–471. [Google Scholar] [CrossRef] [PubMed]
- Lee, R.G.; Mazzola, A.M.; Braun, M.C.; Platt, C.; Vafai, S.B.; Kathiresan, S.; Rohde, E.; Bellinger, A.M.; Khera, A.V. Efficacy and Safety of an Investigational Single-Course CRISPR Base-Editing Therapy Targeting PCSK9 in Nonhuman Primate and Mouse Models. Circulation 2023, 147, 242–253. [Google Scholar] [CrossRef] [PubMed]
- Verve takes base editors into humans. Nat. Biotechnol. 2022, 40, 1159.
- Hassan, M. ANGPLT3: A novel modulator of lipid metabolism. Glob. Cardiol. Sci. Pract. 2017, 2017, e201706. [Google Scholar] [CrossRef]
- Su, X.; Peng, D. New insights into ANGPLT3 in controlling lipoprotein metabolism and risk of cardiovascular diseases. Lipids Health Dis. 2018, 17, 12. [Google Scholar] [CrossRef]
- Dewey, F.E.; Gusarova, V.; Dunbar, R.L.; O’dushlaine, C.; Schurmann, C.; Gottesman, O.; McCarthy, S.; Van Hout, C.V.; Bruse, S.; Dansky, H.M.; et al. Genetic and Pharmacologic Inactivation of ANGPTL3 and Cardiovascular Disease. N. Engl. J. Med. 2017, 377, 211–221. [Google Scholar] [CrossRef]
- Stitziel, N.O.; Khera, A.V.; Wang, X.; Bierhals, A.J.; Vourakis, A.C.; Sperry, A.E.; Natarajan, P.; Klarin, D.; Emdin, C.A.; Zekavat, S.M.; et al. ANGPTL3 Deficiency and Protection Against Coronary Artery Disease. J. Am. Coll. Cardiol. 2017, 69, 2054–2063. [Google Scholar] [CrossRef] [PubMed]
- Graham, M.J.; Lee, R.G.; Brandt, T.A.; Tai, L.-J.; Fu, W.; Peralta, R.; Yu, R.; Hurh, E.; Paz, E.; McEvoy, B.W.; et al. Cardiovascular and Metabolic Effects of ANGPTL3 Antisense Oligonucleotides. N. Engl. J. Med. 2017, 377, 222–232. [Google Scholar] [CrossRef] [PubMed]
- Gusarova, V.; Alexa, C.A.; Wang, Y.; Rafique, A.; Kim, J.H.; Buckler, D.; Mintah, I.J.; Shihanian, L.M.; Cohen, J.C.; Hobbs, H.H.; et al. ANGPTL3 blockade with a human monoclonal antibody reduces plasma lipids in dyslipidemic mice and monkeys. J. Lipid Res. 2015, 56, 1308–1317. [Google Scholar] [CrossRef] [PubMed]
- Komor, A.C.; Kim, Y.B.; Packer, M.S.; Zuris, J.A.; Liu, D.R. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 2016, 533, 420–424. [Google Scholar] [CrossRef] [PubMed]
- Chadwick, A.C.; Evitt, N.H.; Lv, W.; Musunuru, K. Reduced Blood Lipid Levels with In Vivo CRISPR-Cas9 Base Editing of ANGPTL3. Circulation 2018, 137, 975–977. [Google Scholar] [CrossRef]
- Glass, Z.; Lee, M.; Li, Y.; Xu, Q. Engineering the Delivery System for CRISPR-Based Genome Editing. Trends Biotechnol. 2018, 36, 173–185. [Google Scholar] [CrossRef]
- Yin, H.; Kanasty, R.L.; Eltoukhy, A.A.; Vegas, A.J.; Dorkin, J.R.; Anderson, D.G. Non-viral vectors for gene-based therapy. Nat. Rev. Genet. 2014, 15, 541–555. [Google Scholar] [CrossRef]
- Qiu, M.; Glass, Z.; Chen, J.; Haas, M.; Jin, X.; Zhao, X.; Rui, X.; Ye, Z.; Li, Y.; Zhang, F.; et al. Lipid nanoparticle-mediated codelivery of Cas9 mRNA and single-guide RNA achieves liver-specific in vivo genome editing of Angptl3. Proc. Natl. Acad. Sci. USA 2021, 118, e2020401118. [Google Scholar] [CrossRef]
- Zuo, Y.; Zhang, C.; Zhou, Y.; Li, H.; Xiao, W.; Herzog, R.W.; Xu, J.; Zhang, J.; Chen, Y.E.; Han, R. Liver-specific in vivo base editing of Angptl3 via AAV delivery efficiently lowers blood lipid levels in mice. Cell Biosci. 2023, 13, 109. [Google Scholar] [CrossRef]
- Davis, J.R.; Wang, X.; Witte, I.P.; Huang, T.P.; Levy, J.M.; Raguram, A.; Banskota, S.; Seidah, N.G.; Musunuru, K.; Liu, D.R. Efficient in vivo base editing via single adeno-associated viruses with size-optimized genomes encoding compact adenine base editors. Nat. Biomed. Eng. 2022, 6, 1272–1283. [Google Scholar] [CrossRef]
- Nair, J.K.; Willoughby, J.L.S.; Chan, A.; Charisse, K.; Alam, R.; Wang, Q.; Hoekstra, M.; Kandasamy, P.; Kel’in, A.V.; Milstein, S.; et al. Multivalent N-acetylgalactosamine-conjugated siRNA localizes in hepatocytes and elicits robust RNAi-mediated gene silencing. J. Am. Chem. Soc. 2014, 136, 16958–16961. [Google Scholar] [CrossRef]
- Paunovska, K.; Loughrey, D.; Dahlman, J.E. Drug delivery systems for RNA therapeutics. Nat. Rev. Genet. 2022, 23, 265–280. [Google Scholar] [CrossRef]
- Kasiewicz, L.N.; Biswas, S.; Beach, A.; Ren, H.; Dutta, C.; Mazzola, A.M.; Rohde, E.; Chadwick, A.; Cheng, C.; Garcia, S.P.; et al. GalNAc-Lipid nanoparticles enable non-LDLR dependent hepatic delivery of a CRISPR base editing therapy. Nat. Commun. 2023, 14, 2776. [Google Scholar] [CrossRef] [PubMed]
- Soutar, A.K.; Naoumova, R.P. Mechanisms of disease: Genetic causes of familial hypercholesterolemia. Nat. Clin. Pract. Cardiovasc. Med. 2007, 4, 214–225. [Google Scholar] [CrossRef] [PubMed]
- Hobbs, H.H.; Brown, M.S.; Goldstein, J.L. Molecular genetics of the LDL receptor gene in familial hypercholesterolemia. Hum. Mutat. 1992, 1, 445–466. [Google Scholar] [CrossRef] [PubMed]
- Tolleshaug, H.; Goldstein, J.L.; Schneider, W.J.; Brown, M.S. Posttranslational processing of the LDL receptor and its genetic disruption in familial hypercholesterolemia. Cell 1982, 30, 715–724. [Google Scholar] [CrossRef] [PubMed]
- Jarrett, K.E.; Lee, C.; De Giorgi, M.; Hurley, A.; Gillard, B.K.; Doerfler, A.M.; Li, A.; Pownall, H.J.; Bao, G.; Lagor, W.R. Somatic Editing of Ldlr with Adeno-Associated Viral-CRISPR Is an Efficient Tool for Atherosclerosis Research. Arterioscler. Thromb. Vasc. Biol. 2018, 38, 1997–2006. [Google Scholar] [CrossRef]
- Sun, X.M.; Patel, D.D.; Webb, J.C.; Knight, B.L.; Fan, L.M.; Cai, H.J.; Soutar, A.K. Familial hypercholesterolemia in China. Identification of mutations in the LDL-receptor gene that result in a receptor-negative phenotype. Arterioscler. Thromb. 1994, 14, 85–94. [Google Scholar] [CrossRef]
- Zhao, H.; Li, Y.; He, L.; Pu, W.; Yu, W.; Li, Y.; Wu, Y.-T.; Xu, C.; Wei, Y.; Ding, Q.; et al. In Vivo AAV-CRISPR/Cas9-Mediated Gene Editing Ameliorates Atherosclerosis in Familial Hypercholesterolemia. Circulation 2020, 141, 67–79. [Google Scholar] [CrossRef]
- Greig, J.A.; Limberis, M.P.; Bell, P.; Chen, S.J.; Calcedo, R.; Rader, D.J.; Wilson, J.M. Nonclinical Pharmacology/Toxicology Study of AAV8.TBG.mLDLR and AAV8.TBG.hLDLR in a Mouse Model of Homozygous Familial Hypercholesterolemia. Hum. Gene Ther. Clin. Dev. 2017, 28, 28–38. [Google Scholar] [CrossRef]
- Kassim, S.H.; Li, H.; Bell, P.; Somanathan, S.; Lagor, W.; Jacobs, F.; Billheimer, J.; Wilson, J.M.; Rader, D.J.; Greig, J.A.; et al. Adeno-associated virus serotype 8 gene therapy leads to significant lowering of plasma cholesterol levels in humanized mouse models of homozygous and heterozygous familial hypercholesterolemia. Hum. Gene Ther. 2013, 24, 19–26. [Google Scholar] [CrossRef]
- Kassim, S.H.; Li, H.; Vandenberghe, L.H.; Hinderer, C.; Bell, P.; Marchadier, D.; Wilson, A.; Cromley, D.; Redon, V.; Yu, H.; et al. Gene therapy in a humanized mouse model of familial hypercholesterolemia leads to marked regression of atherosclerosis. PLoS ONE 2010, 5, e13424. [Google Scholar] [CrossRef]
- Fan, J.; Kitajima, S.; Watanabe, T.; Xu, J.; Zhang, J.; Liu, E.; Chen, Y.E. Rabbit models for the study of human atherosclerosis: From pathophysiological mechanisms to translational medicine. Pharmacol. Ther. 2015, 146, 104–119. [Google Scholar] [CrossRef] [PubMed]
- Gisterå, A.; Ketelhuth, D.F.; Malin, S.G.; Hansson, G.K. Animal Models of Atherosclerosis-Supportive Notes and Tricks of the Trade. Circ. Res. 2022, 130, 1869–1887. [Google Scholar] [CrossRef] [PubMed]
- Lu, R.; Yuan, T.; Wang, Y.; Zhang, T.; Yuan, Y.; Wu, D.; Zhou, M.; He, Z.; Lu, Y.; Chen, Y.; et al. Spontaneous severe hypercholesterolemia and atherosclerosis lesions in rabbits with deficiency of low-density lipoprotein receptor (LDLR) on exon 7. EBioMedicine 2018, 36, 29–38. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Gao, M.; Wang, Y.; Lin, X.; Yang, L.; Cong, N.; An, X.; Wang, F.; Qu, K.; Yu, L.; et al. LDL Receptor Gene-ablated Hamsters: A Rodent Model of Familial Hypercholesterolemia with Dominant Inheritance and Diet-induced Coronary Atherosclerosis. EBioMedicine 2018, 27, 214–224. [Google Scholar] [CrossRef] [PubMed]
- Ishibashi, S.; Brown, M.S.; Goldstein, J.L.; Gerard, R.D.; Hammer, R.E.; Herz, J. Hypercholesterolemia in low density lipoprotein receptor knockout mice and its reversal by adenovirus-mediated gene delivery. J. Clin. Investig. 1993, 92, 883–893. [Google Scholar] [CrossRef] [PubMed]
- Xian, X.; Wang, Y.; Liu, G. Genetically Engineered Hamster Models of Dyslipidemia and Atherosclerosis. Methods Mol. Biol. 2022, 2419, 433–459. [Google Scholar]
- Zhou, Y.; Luo, G. Apolipoproteins, as the carrier proteins for lipids, are involved in the development of breast cancer. Clin. Transl. Oncol. 2020, 22, 1952–1962. [Google Scholar] [CrossRef]
- Liu, T.; Chen, J.M.; Zhang, D.; Zhang, Q.; Peng, B.; Xu, L.; Tang, H. ApoPred: Identification of Apolipoproteins and Their Subfamilies With Multifarious Features. Front. Cell Dev. Biol. 2020, 8, 621144. [Google Scholar] [CrossRef]
- Zhao, G.J.; Yin, K.; Fu, Y.C.; Tang, C.K. The interaction of ApoA-I and ABCA1 triggers signal transduction pathways to mediate efflux of cellular lipids. Mol. Med. 2012, 18, 149–158. [Google Scholar] [CrossRef] [PubMed]
- Mangaraj, M.; Nanda, R.; Panda, S. Apolipoprotein A-I: A Molecule of Diverse Function. Indian J. Clin. Biochem. 2016, 31, 253–259. [Google Scholar] [CrossRef] [PubMed]
- De Giorgi, M.; Li, A.; Hurley, A.; Barzi, M.; Doerfler, A.M.; Cherayil, N.A.; Smith, H.E.; Brown, J.D.; Lin, C.Y.; Bissig, K.-D.; et al. Targeting the Apoa1 locus for liver-directed gene therapy. Mol. Ther. Methods Clin. Dev. 2021, 21, 656–669. [Google Scholar] [CrossRef] [PubMed]
- Bertolini, S.; Pisciotta, L.; Rabacchi, C.; Cefalù, A.B.; Noto, D.; Fasano, T.; Signori, A.; Fresa, R.; Averna, M.; Calandra, S. Spectrum of mutations and phenotypic expression in patients with autosomal dominant hypercholesterolemia identified in Italy. Atherosclerosis 2013, 227, 342–348. [Google Scholar] [CrossRef]
- Young, S.G.; Northey, S.T.; McCarthy, B.J. Low plasma cholesterol levels caused by a short deletion in the apolipoprotein B gene. Science 1988, 241, 591–593. [Google Scholar] [CrossRef]
- Cohn, J.S.; Tremblay, M.; Batal, R.; Jacques, H.; Rodriguez, C.; Steiner, G.; Mamer, O.; Davignon, J. Increased apoC-III production is a characteristic feature of patients with hypertriglyceridemia. Atherosclerosis 2004, 177, 137–145. [Google Scholar] [CrossRef]
- Sacks, F.M.; Alaupovic, P.; Moye, L.A.; Cole, T.G.; Sussex, B.; Stampfer, M.J.; Pfeffer, M.A.; Braunwald, E. VLDL, apolipoproteins B, CIII, and E, and risk of recurrent coronary events in the Cholesterol and Recurrent Events (CARE) trial. Circulation 2000, 102, 1886–1892. [Google Scholar] [CrossRef]
- Li, H.; Han, Y.; Qi, R.; Wang, Y.; Zhang, X.; Yu, M.; Tang, Y.; Wang, M.; Shu, Y.-N.; Huang, W.; et al. Aggravated restenosis and atherogenesis in ApoCIII transgenic mice but lack of protection in ApoCIII knockouts: The effect of authentic triglyceride-rich lipoproteins with and without ApoCIII. Cardiovasc. Res. 2015, 107, 579–589. [Google Scholar] [CrossRef]
- Guo, M.; Xu, Y.; Dong, Z.; Zhou, Z.; Cong, N.X.; Gao, M.; Huang, W.; Wang, Y.; Liu, G.; Xian, X. Inactivation of ApoC3 by CRISPR/Cas9 Protects Against Atherosclerosis in Hamsters. Circ. Res. 2020, 127, 1456–1458. [Google Scholar] [CrossRef]
- Zha, Y.; Lu, Y.; Zhang, T.; Yan, K.; Zhuang, W.; Liang, J.; Cheng, Y.; Wang, Y. CRISPR/Cas9-mediated knockout of APOC3 stabilizes plasma lipids and inhibits atherosclerosis in rabbits. Lipids Health Dis. 2021, 20, 180. [Google Scholar] [CrossRef]
- Modrzejewski, D.; Hartung, F.; Lehnert, H.; Sprink, T.; Kohl, C.; Keilwagen, J.; Wilhelm, R. Which Factors Affect the Occurrence of Off-Target Effects Caused by the Use of CRISPR/Cas: A Systematic Review in Plants. Front. Plant Sci. 2020, 11, 574959. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.; Foden, J.A.; Khayter, C.; Maeder, M.L.; Reyon, D.; Joung, J.K.; Sander, J.D. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat. Biotechnol. 2013, 31, 822–826. [Google Scholar] [CrossRef] [PubMed]
- Alkan, F.; Wenzel, A.; Anthon, C.; Havgaard, J.H.; Gorodkin, J. CRISPR-Cas9 off-targeting assessment with nucleic acid duplex energy parameters. Genome Biol. 2018, 19, 177. [Google Scholar] [CrossRef] [PubMed]
- Hsu, P.D.; Scott, D.A.; Weinstein, J.A.; Ran, F.A.; Konermann, S.; Agarwala, V.; Li, Y.; Fine, E.J.; Wu, X.; Shalem, O.; et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol. 2013, 31, 827–832. [Google Scholar] [CrossRef]
- Zuo, E.; Sun, Y.; Wei, W.; Yuan, T.; Ying, W.; Sun, H.; Yuan, L.; Steinmetz, L.M.; Li, Y.; Yang, H. Cytosine base editor generates substantial off-target single-nucleotide variants in mouse embryos. Science 2019, 364, 289–292. [Google Scholar] [CrossRef]
- Grünewald, J.; Zhou, R.; Garcia, S.P.; Iyer, S.; Lareau, C.A.; Aryee, M.J.; Joung, J.K. Transcriptome-wide off-target RNA editing induced by CRISPR-guided DNA base editors. Nature 2019, 569, 433–437. [Google Scholar] [CrossRef]
- Yin, J.; Liu, M.; Liu, Y.; Wu, J.; Gan, T.; Zhang, W.; Li, Y.; Zhou, Y.; Hu, J. Optimizing genome editing strategy by primer-extension-mediated sequencing. Cell Discov. 2019, 5, 18. [Google Scholar] [CrossRef]
- Ayabe, S.; Nakashima, K.; Yoshiki, A. Off- and on-target effects of genome editing in mouse embryos. J. Reprod. Dev. 2019, 65, 1–5. [Google Scholar] [CrossRef]
- Lee, J.K.; Jeong, E.; Lee, J.; Jung, M.; Shin, E.; Kim, Y.-H.; Lee, K.; Jung, I.; Kim, D.; Kim, S.; et al. Directed evolution of CRISPR-Cas9 to increase its specificity. Nat. Commun. 2018, 9, 3048. [Google Scholar] [CrossRef]
- Chen, J.S.; Dagdas, Y.S.; Kleinstiver, B.P.; Welch, M.M.; Sousa, A.A.; Harrington, L.B.; Sternberg, S.H.; Joung, J.K.; Yildiz, A.; Doudna, J.A. Enhanced proofreading governs CRISPR-Cas9 targeting accuracy. Nature 2017, 550, 407–410. [Google Scholar] [CrossRef]
- Frock, R.L.; Hu, J.; Meyers, R.M.; Ho, Y.-J.; Kii, E.; Alt, F.W. Genome-wide detection of DNA double-stranded breaks induced by engineered nucleases. Nat. Biotechnol. 2015, 33, 179–186. [Google Scholar] [CrossRef]
- Fu, Y.; Sander, J.D.; Reyon, D.; Cascio, V.M.; Joung, J.K. Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nat. Biotechnol. 2014, 32, 279–284. [Google Scholar] [CrossRef]
- Kim, D.; Kim, S.; Kim, S.; Park, J.; Kim, J.-S. Genome-wide target specificities of CRISPR-Cas9 nucleases revealed by multiplex Digenome-seq. Genome Res. 2016, 26, 406–415. [Google Scholar] [CrossRef]
- Cho, S.W.; Kim, S.; Kim, Y.; Kweon, J.; Kim, H.S.; Bae, S.; Kim, J.-S. Analysis of off-target effects of CRISPR/Cas-derived RNA-guided endonucleases and nickases. Genome Res. 2014, 24, 132–141. [Google Scholar] [CrossRef]
- Taha, E.A.; Lee, J.; Hotta, A. Delivery of CRISPR-Cas tools for in vivo genome editing therapy: Trends and challenges. J. Control. Release 2022, 342, 345–361. [Google Scholar] [CrossRef]
- Lu, B.; Javidi-Parsijani, P.; Makani, V.; Mehraein-Ghomi, F.; Sarhan, W.M.; Sun, D.; Yoo, K.W.; Atala, Z.P.; Lyu, P.; Atala, A. Delivering SaCas9 mRNA by lentivirus-like bionanoparticles for transient expression and efficient genome editing. Nucleic Acids Res. 2019, 47, e44. [Google Scholar] [CrossRef]
- Hanlon, K.S.; Kleinstiver, B.P.; Garcia, S.P.; Zaborowski, M.P.; Volak, A.; Spirig, S.E.; Muller, A.; Sousa, A.A.; Tsai, S.Q.; Bengtsson, N.E.; et al. High levels of AAV vector integration into CRISPR-induced DNA breaks. Nat. Commun. 2019, 10, 4439. [Google Scholar] [CrossRef]
- Arjomandnejad, M.; Dasgupta, I.; Flotte, T.R.; Keeler, A.M. Immunogenicity of Recombinant Adeno-Associated Virus (AAV) Vectors for Gene Transfer. BioDrugs 2023, 37, 311–329. [Google Scholar] [CrossRef]
- Ghani, M.W.; Iqbal, A.; Ghani, H.; Bibi, S.; Wang, Z.; Pei, R. Recent advances in nanocomposite-based delivery systems for targeted CRISPR/Cas delivery and therapeutic genetic manipulation. J. Mater. Chem. B 2023, 11, 5251–5271. [Google Scholar] [CrossRef]
- Martin, F.; Sánchez-Hernández, S.; Gutiérrez-Guerrero, A.; Pinedo-Gomez, J.; Benabdellah, K. Biased and Unbiased Methods for the Detection of Off-Target Cleavage by CRISPR/Cas9: An Overview. Int. J. Mol. Sci. 2016, 17, 1507. [Google Scholar] [CrossRef]
- Zhang, X.-H.; Tee, L.Y.; Wang, X.-G.; Huang, Q.-S.; Yang, S.-H. Off-target Effects in CRISPR/Cas9-mediated Genome Engineering. Mol. Ther. Nucleic Acids 2015, 4, e264. [Google Scholar] [CrossRef]
- Chew, W.L.; Tabebordbar, M.; Cheng, J.K.; Mali, P.; Wu, E.Y.; Ng, A.H.; Zhu, K.; Wagers, A.J.; Church, G.M. A multifunctional AAV-CRISPR-Cas9 and its host response. Nat. Methods 2016, 13, 868–874. [Google Scholar] [CrossRef]
- Charlesworth, C.T.; Deshpande, P.S.; Dever, D.P.; Camarena, J.; Lemgart, V.T.; Cromer, M.K.; Vakulskas, C.A.; Collingwood, M.A.; Zhang, L.; Bode, N.M.; et al. Identification of preexisting adaptive immunity to Cas9 proteins in humans. Nat. Med. 2019, 25, 249–254. [Google Scholar] [CrossRef]
- Zhao, Z.; Tuakli-Wosornu, Y.; Lagace, T.A.; Kinch, L.; Grishin, N.V.; Horton, J.D.; Cohen, J.C.; Hobbs, H.H. Molecular characterization of loss-of-function mutations in PCSK9 and identification of a compound heterozygote. Am. J. Hum. Genet. 2006, 79, 514–523. [Google Scholar] [CrossRef]
- Hooper, A.J.; Marais, A.D.; Tanyanyiwa, D.M.; Burnett, J.R. The C679X mutation in PCSK9 is present and lowers blood cholesterol in a Southern African population. Atherosclerosis 2007, 193, 445–448. [Google Scholar] [CrossRef]
- Romeo, S.; Yin, W.; Kozlitina, J.; Pennacchio, L.A.; Boerwinkle, E.; Hobbs, H.H.; Cohen, J.C. Rare loss-of-function mutations in ANGPTL family members contribute to plasma triglyceride levels in humans. J. Clin. Investig. 2009, 119, 70–79. [Google Scholar] [CrossRef]
| Species | Delivery Vehicle | Editing Method | References | ||
|---|---|---|---|---|---|
| PCSK9 | Mouse | Adenovirus | CRISPR | [57] | |
| Mouse | AAV | CRISPR | [58] | ||
| Mouse | AAV | CRISPR | [63] | ||
| Mouse | AAV | CRISPR | [64] | ||
| Mouse | LLN | CRISPR | [65] | ||
| Mouse | LNP | CRISPR | [67] | ||
| Mouse | LNP | CRISPR | [68] | ||
| Mouse | Adenovirus | Base editor | [69] | ||
| NHP | AAV | Meganuclease | [70] | ||
| NHP | LNP | Base editor | [73] | ||
| NHP | LNP | Base editor | [74] | ||
| ANGPTL3 | Mouse | Adenovirus | Base editor | [85] | |
| Mouse | LNP | CRISPR | [88] | ||
| Mouse | AAV | Base editor | [89] | ||
| Mouse | AAV | Base editor | [90] | ||
| NHP | GalNAc-LNP | Base editor | [93] | ||
| LDLR | Mouse | AAV | CRISPR | [97] | |
| Mouse | AAV | CRISPR | [99] | ||
| Rabbit | * | CRISPR | [105] | ||
| Hamster | * | CRISPR | [106] | ||
| APO | A1 | Mouse | AAV | CRISPR | [113] |
| B | Mouse | AAV | CRISPR | [3] | |
| B | Mouse | AAV | CRISPR | [63] | |
| C3 | Hamster | * | CRISPR | [119] | |
| C3 | Rabbit | * | CRISPR | [120] | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Preta, G. Development of New Genome Editing Tools for the Treatment of Hyperlipidemia. Cells 2023, 12, 2466. https://doi.org/10.3390/cells12202466
Preta G. Development of New Genome Editing Tools for the Treatment of Hyperlipidemia. Cells. 2023; 12(20):2466. https://doi.org/10.3390/cells12202466
Chicago/Turabian StylePreta, Giulio. 2023. "Development of New Genome Editing Tools for the Treatment of Hyperlipidemia" Cells 12, no. 20: 2466. https://doi.org/10.3390/cells12202466




