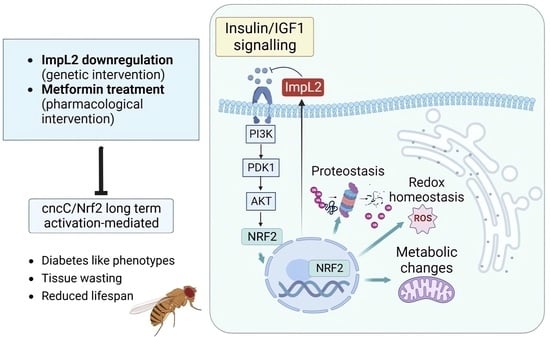
Sustained Nrf2 Overexpression-Induced Metabolic Deregulation Can Be Attenuated by Modulating Insulin/Insulin-like Growth Factor Signaling
Abstract
:1. Introduction
2. Materials and Methods
2.1. Fly Stocks
2.2. Flies’ Culture, Exposure to Compounds, and Longevity Assays
2.3. Genomic DNA Extraction and Conventional PCR Analyses
2.4. RNA Extraction, cDNA Synthesis, and Quantitative Real-Time PCR (Q-PCR) Analysis
2.5. Preparation of Tissue Protein Extracts, Immunoblot Analysis, Measurement of ROS, and Proteasome Enzymatic Activities
2.6. Measurement of GLU/TREH Levels
2.7. Antibodies
2.8. CSLM and Immunofluorescence Staining
2.9. Statistical Analyses
3. Results
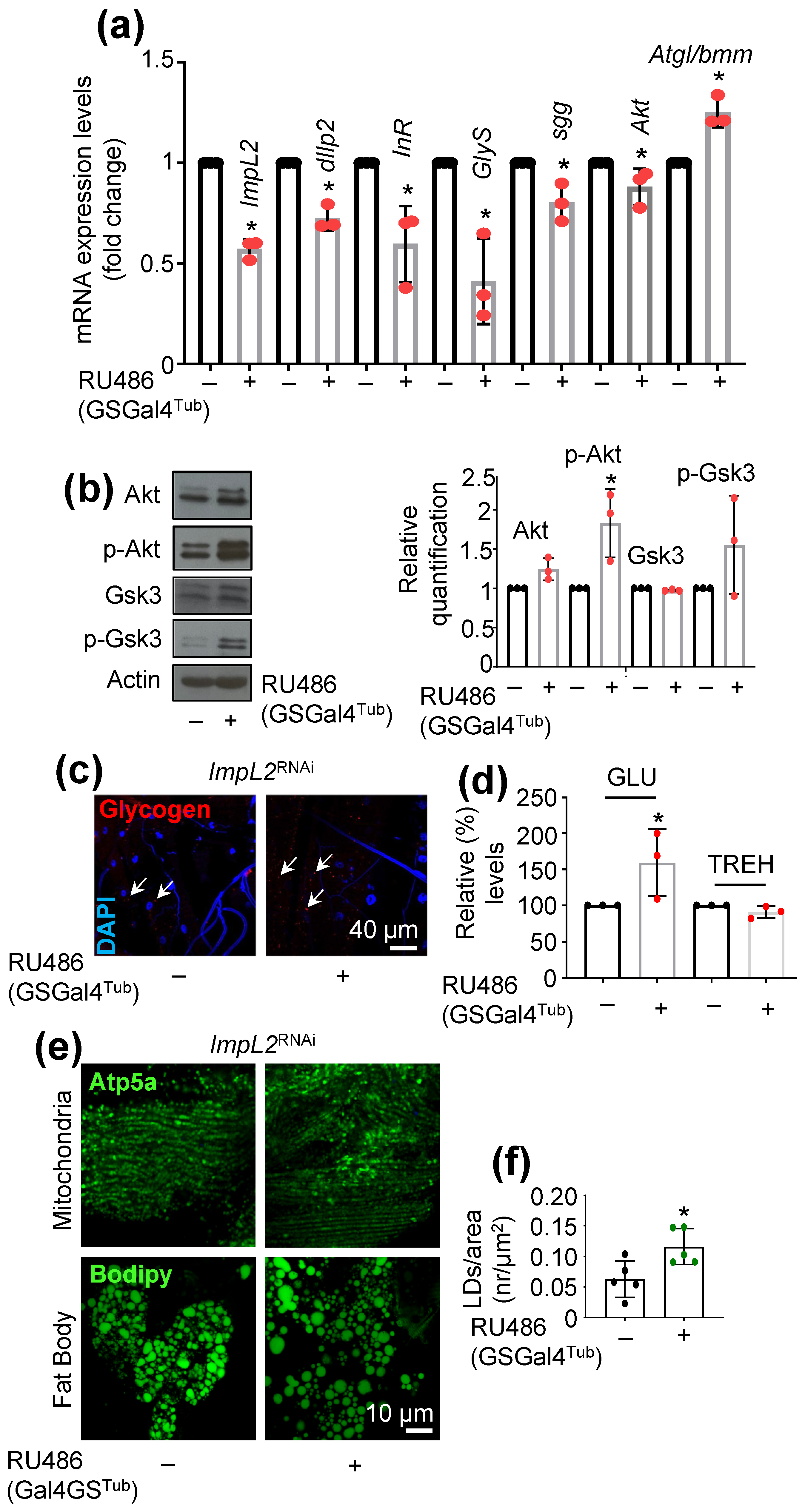
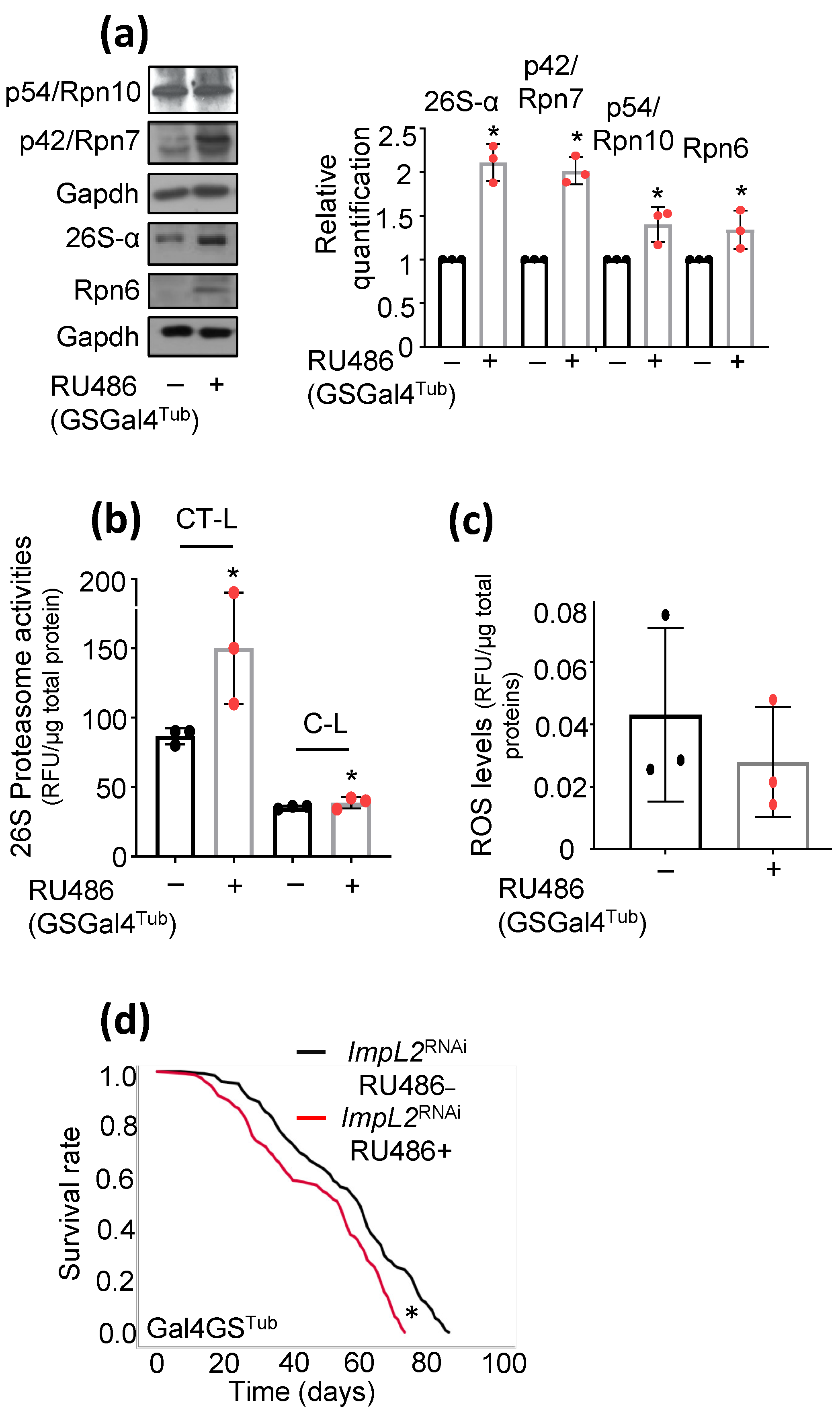
3.1. ImpL2 Knockdown (KD) Activates IIS-, Antioxidant- and Proteostasis-Related Modules
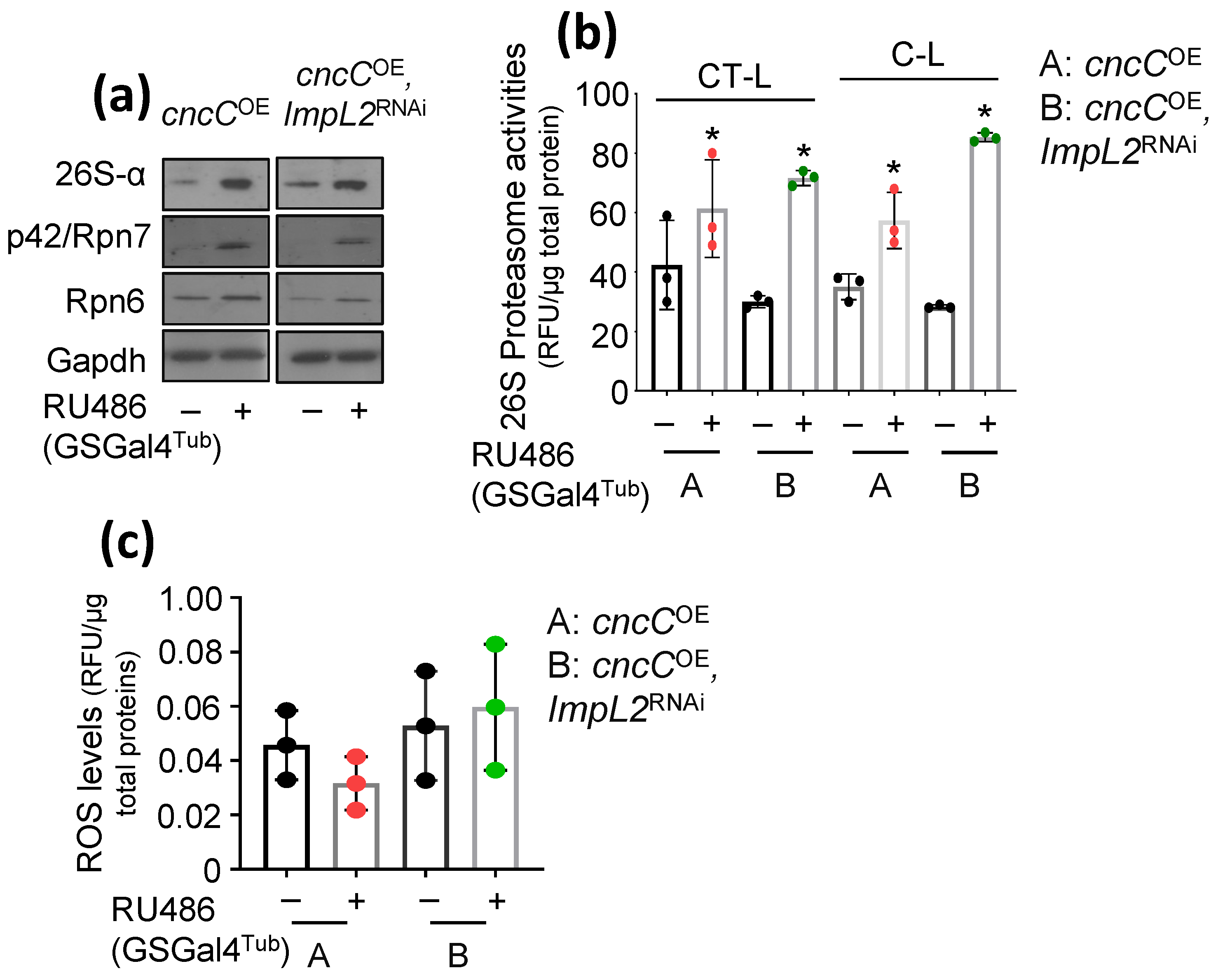
3.2. Proteostatic Responses Remain Upregulated after Concomitant ImpL2 KD in cncCOE Flies
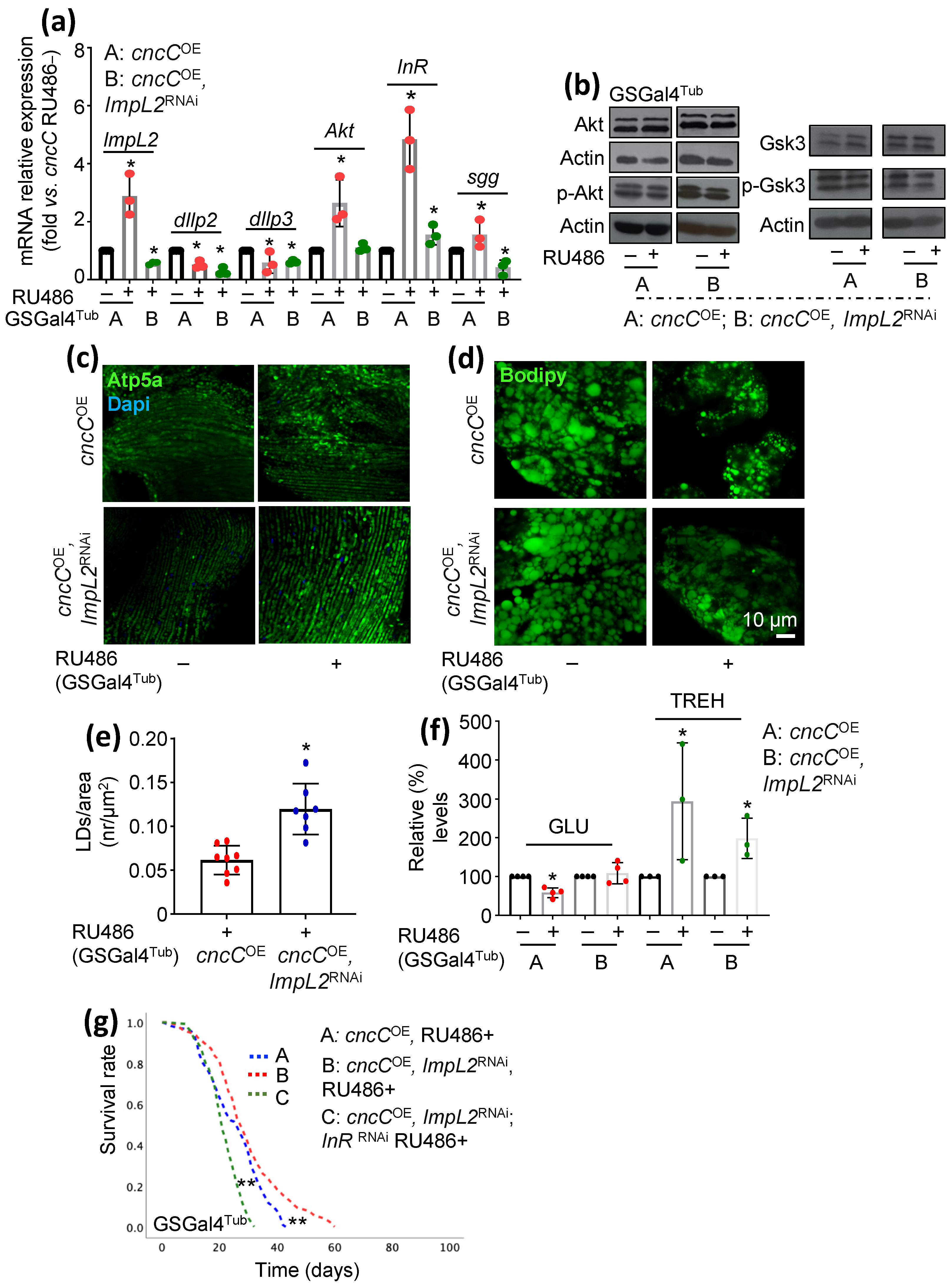
3.3. Concomitant to cncCOE, ImpL2 KD Partially Restores IIS Activity and Attenuates the Sustained cncCOE-Induced Metabolic Deregulation, Leading to Lifespan Extension
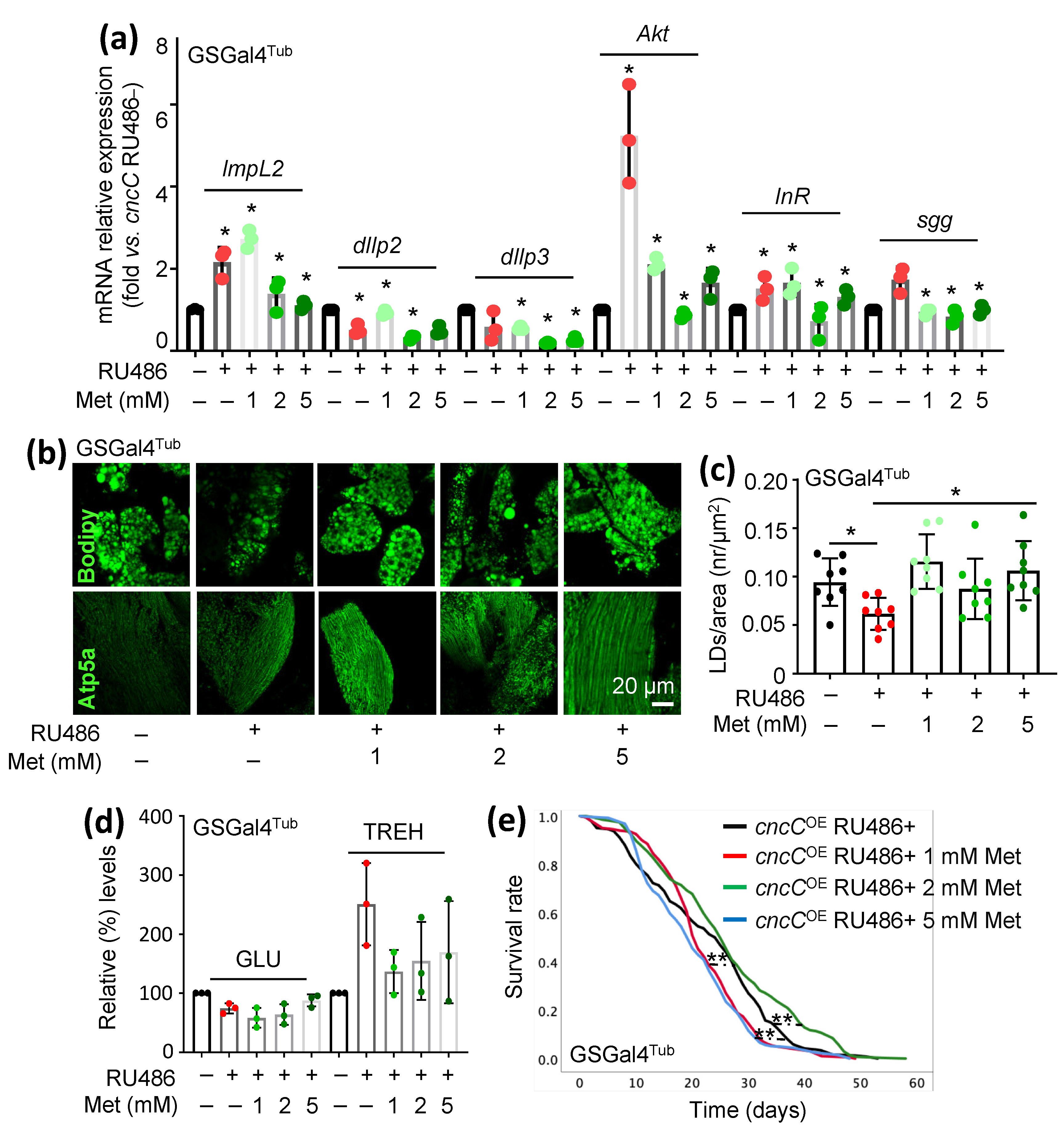
3.4. Treatment of cncCOE Flies with the Anti-Diabetic Drug Metformin (Met) Partially Restores the Functionality of Metabolic/Mitostatic Pathways and Extends Flies’ Lifespan
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fontana, L.; Partridge, L.; Longo, V.D. Extending healthy life span--from yeast to humans. Science 2010, 328, 321–326. [Google Scholar] [CrossRef] [PubMed]
- Boucher, J.; Kleinridders, A.; Kahn, C.R. Insulin receptor signaling in normal and insulin-resistant states. Cold Spring Harb. Perspect. Biol. 2014, 6, a009191. [Google Scholar] [CrossRef] [PubMed]
- Taniguchi, C.M.; Emanuelli, B.; Kahn, C.R. Critical nodes in signalling pathways: Insights into insulin action. Nat. Rev. Mol. Cell Biol. 2006, 7, 85–96. [Google Scholar] [CrossRef] [PubMed]
- Adams, T.E.; Epa, V.C.; Garrett, T.P.; Ward, C.W. Structure and function of the type 1 insulin-like growth factor receptor. Cell Mol. Life Sci. 2000, 57, 1050–1093. [Google Scholar] [CrossRef] [PubMed]
- De Meyts, P. Insulin and its receptor: Structure, function and evolution. BioEssays 2004, 26, 1351–1362. [Google Scholar] [CrossRef]
- Röder, P.V.; Wu, B.; Liu, Y.; Han, W. Pancreatic regulation of glucose homeostasis. Exp. Mol. Med. 2016, 48, e219. [Google Scholar]
- Brogiolo, W.; Stocker, H.; Ikeya, T.; Rintelen, F.; Fernandez, R.; Hafen, E. An evolutionarily conserved function of the Drosophila insulin receptor and insulin-like peptides in growth control. Curr. Biol. 2001, 11, 213–221. [Google Scholar] [CrossRef]
- Rulifson, E.J.; Kim, S.K.; Nusse, R. Ablation of insulin-producing neurons in flies: Growth and diabetic phenotypes. Science 2002, 296, 1118–1120. [Google Scholar] [CrossRef]
- Honegger, B.; Galic, M.; Köhler, K.; Wittwer, F.; Brogiolo, W.; Hafen, E.; Stocker, H. Imp-L2, a putative homolog of vertebrate IGF-binding protein 7, counteracts insulin signaling in Drosophila and is essential for starvation resistance. J. Biol. 2008, 7, 10. [Google Scholar] [CrossRef]
- Alic, N.; Hoddinott, M.P.; Vinti, G.; Partridge, L. Lifespan extension by increased expression of the Drosophila homologue of the IGFBP7 tumour suppressor. Aging Cell 2011, 10, 137–147. [Google Scholar] [CrossRef]
- Sloth Andersen, A.; Hertz Hansen, P.; Schaffer, L.; Kristensen, C. A new secreted insect protein belonging to the immunoglobulin superfamily binds insulin and related peptides and inhibits their activities. J. Biol. Chem. 2000, 275, 16948–16953. [Google Scholar] [PubMed]
- Oh, Y.; Nagalla, S.R.; Yamanaka, Y.; Kim, H.S.; Wilson, E.; Rosenfeld, R.G. Synthesis and characterization of insulin-like growth factor-binding protein (IGFBP)-7. Recombinant human mac25 protein specifically binds IGF-I and -II. J. Biol. Chem. 1996, 29, 271. [Google Scholar]
- Yamanaka, Y.; Wilson, E.M.; Rosenfeld, R.G.; Oh, Y. Inhibition of insulin receptor activation by insulin-like growth factor binding proteins. J. Biol. Chem. 1997, 272, 30729–30734. [Google Scholar] [CrossRef] [PubMed]
- Roed, N.K.; Viola, C.M.; Kristensen, O.; Schluckebier, G.; Norrman, M.; Sajid, W.; Wade, J.D.; Andersen, A.S.; Kristensen, C.; Ganderton, T.R.; et al. Structures of insect Imp-L2 suggest an alternative strategy for regulating the bioavailability of insulin-like hormones. Nat. Commun. 2018, 9, 3860. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, T.; Yamamoto, M. Molecular basis of the Keap1-Nrf2 system. Free Radic. Biol. Med. 2015, 88, 93–100. [Google Scholar]
- Chu, X.Y.; Liu, Y.M.; Zhang, H.Y. Activating or Inhibiting Nrf2? Trends Pharmacol. Sci. 2017, 38, 953–955. [Google Scholar] [CrossRef]
- Itoh, K.; Wakabayashi, N.; Katoh, Y.; Ishii, T.; Igarashi, K.; Engel, J.D.; Yamamoto, M. Keap1 represses nuclear activation of antioxidant responsive elements by Nrf2 through binding to the amino-terminal Neh2 domain. Genes. Dev. 1999, 13, 76–86. [Google Scholar] [CrossRef]
- Cuadrado, A.; Rojo, A.I.; Wells, G.; Hayes, J.D.; Cousin, S.P.; Rumsey, W.L.; Attucks, O.C.; Franklin, S.; Levonen, A.L.; Kensler, T.W.; et al. Therapeutic targeting of the NRF2 and KEAP1 partnership in chronic diseases. Nat. Rev. Drug Discov. 2019, 18, 295–317. [Google Scholar]
- Tsakiri, E.N.; Gumeni, S.; Iliaki, K.K.; Benaki, D.; Vougas, K.; Sykiotis, G.P.; Gorgoulis, V.G.; Mikros, E.; Scorrano, L.; Trougakos, I.P. Hyperactivation of Nrf2 increases stress tolerance at the cost of aging acceleration due to metabolic deregulation. Aging Cell 2019, 18, e12845. [Google Scholar] [CrossRef]
- Sykiotis, G.P.; Bohmann, D. Keap1/Nrf2 signaling regulates oxidative stress tolerance and lifespan in Drosophila. Dev. Cell 2008, 14, 76–85. [Google Scholar] [CrossRef]
- Osterwalder, T.; Yoon, K.S.; White, B.H.; Keshishian, H. A conditional tissue-specific transgene expression system using inducible GAL4. Proc. Natl. Acad. Sci. USA 2001, 98, 12596–12601. [Google Scholar] [CrossRef]
- Roman, G.; Endo, K.; Zong, L.; Davis, R.L. P[Switch], a system for spatial and temporal control of gene expression in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 2001, 98, 12602–12607. [Google Scholar] [CrossRef] [PubMed]
- Tsakiri, E.N.; Sykiotis, G.P.; Papassideri, I.S.; Gorgoulis, V.G.; Bohmann, D.; Trougakos, I.P. Differential regulation of proteasome functionality in reproductive vs. somatic tissues of Drosophila during aging or oxidative stress. FASEB J. 2013, 27, 2407–2420. [Google Scholar] [CrossRef] [PubMed]
- Tsakiri, E.N.; Sykiotis, G.P.; Papassideri, I.S.; Terpos, E.; Dimopoulos, M.A.; Gorgoulis, V.G.; Bohmann, D.; Trougakos, I.P. Proteasome dysfunction in Drosophila signals to an Nrf2-dependent regulatory circuit aiming to restore proteostasis and prevent premature aging. Aging Cell 2013, 12, 802–813. [Google Scholar] [PubMed]
- Manola, M.S.; Gumeni, S.; Trougakos, I.P. Differential Dose- and Tissue-Dependent Effects of foxo on Aging, Metabolic and Proteostatic Pathways. Cells 2021, 10, 3577. [Google Scholar] [CrossRef]
- Géminard, C.; Rulifson, E.J.; Léopold, P. Remote control of insulin secretion by fat cells in Drosophila. Cell Metab. 2009, 10, 199–207. [Google Scholar] [CrossRef]
- Li, Z.; Zhang, Z.; Ren, Y.; Wang, Y.; Fang, J.; Yue, H.; Ma, S.; Guan, F. Aging and age-related diseases: From mechanisms to therapeutic strategies. Biogerontology 2021, 22, 165–187. [Google Scholar]
- López-Otín, C.; Blasco, M.A.; Partridge, L.; Serrano, M.; Kroemer, G. The hallmarks of aging. Cell 2013, 153, 1194–1217. [Google Scholar]
- Foretz, M.; Guigas, B.; Viollet, B. Metformin: Update on mechanisms of action and repurposing potential. Nat. Rev. Endocrinol. 2023, 19, 460–476. [Google Scholar]
- Soukas, A.A.; Hao, H.; Wu, L. Metformin as Anti-Aging Therapy: Is It for Everyone? Trends Endocrinol. Metab. 2019, 30, 745–755. [Google Scholar]
- Nakaso, K.; Yano, H.; Fukuhara, Y.; Takeshima, T.; Wada-Isoe, K.; Nakashima, K. PI3K is a key molecule in the Nrf2-mediated regulation of antioxidative proteins by hemin in human neuroblastoma cells. FEBS Lett. 2003, 546, 181–184. [Google Scholar] [CrossRef] [PubMed]
- Dinkova-Kostova, A.T.; Abramov, A.Y. The emerging role of Nrf2 in mitochondrial function. Free Radic. Biol. Med. 2015, 88, 179–188. [Google Scholar] [PubMed]
- Chatterjee, N.; Perrimon, N. What fuels the fly: Energy metabolism in Drosophila and its application to the study of obesity and diabetes. Sci. Adv. 2021, 7, eabg4336. [Google Scholar] [PubMed]
- Figueroa-Clarevega, A.; Bilder, D. Malignant Drosophila tumors interrupt insulin signaling to induce cachexia-like wasting. Dev. Cell 2015, 33, 47–55. [Google Scholar] [CrossRef]
- Kwon, Y.; Song, W.; Droujinine, I.A.; Hu, Y.; Asara, J.M.; Perrimon, N. Systemic organ wasting induced by localized expression of the secreted insulin/IGF antagonist ImpL2. Dev. Cell 2015, 33, 36–46. [Google Scholar]
- Lee, J.; Ng, K.G.; Dombek, K.M.; Eom, D.S.; Kwon, Y.V. Tumors overcome the action of the wasting factor ImpL2 by locally elevating Wnt/Wingless. Proc. Natl. Acad. Sci. USA 2021, 118, e2020120118. [Google Scholar] [CrossRef] [PubMed]
- Chevalier, S.; Farsijani, S. Cancer cachexia and diabetes: Similarities in metabolic alterations and possible treatment. Appl. Physiol. Nutr. Metab. 2014, 39, 643–653. [Google Scholar] [CrossRef]
- Barzilai, N.; Crandall, J.P.; Kritchevsky, S.B.; Espeland, M.A. Metformin as a Tool to Target Aging. Cell Metab. 2016, 23, 1060–1065. [Google Scholar] [CrossRef]
- Triggle, C.R.; Mohammed, I.; Bshesh, K.; Marei, I.; Ye, K.; Ding, H.; MacDonald, R.; Hollenberg, M.D.; Hill, M.A. Metformin: Is it a drug for all reasons and diseases? Metabolism 2022, 133, 155223. [Google Scholar]
- Owen, M.R.; Doran, E.; Halestrap, A.P. Evidence that metformin exerts its anti-diabetic effects through inhibition of complex 1 of the mitochondrial respiratory chain. Biochem. J. 2000, 348, 607–614. [Google Scholar] [CrossRef]
- Hawley, S.A.; Gadalla, A.E.; Olsen, G.S.; Hardie, D.G. The antidiabetic drug metformin activates the AMP-activated protein kinase cascade via an adenine nucleotide-independent mechanism. Diabetes 2002, 51, 2420–2425. [Google Scholar] [CrossRef] [PubMed]
- Kjøbsted, R.; Kristensen, J.M.; Birk, J.B.; Eskesen, N.O.; Kido, K.; Andersen, N.R.; Larsen, J.K.; Foretz, M.; Viollet, B.; Nielsen, F.; et al. Metformin improves glycemia independently of skeletal muscle AMPK via enhanced intestinal glucose clearance. bioRxiv. 2022. Available online: https://www.biorxiv.org/content/10.1101/2022.05.22.492936v1 (accessed on 21 November 2022).
- Foretz, M.; Hébrard, S.; Leclerc, J.; Zarrinpashneh, E.; Soty, M.; Mithieux, G.; Sakamoto, K.; Andreelli, F.; Viollet, B. Metformin inhibits hepatic gluconeogenesis in mice independently of the LKB1/AMPK pathway via a decrease in hepatic energy state. J. Clin. Investig. 2010, 120, 2355–2369. [Google Scholar] [CrossRef] [PubMed]
- Vial, G.; Detaille, D.; Guigas, B. Role of Mitochondria in the Mechanism(s) of Action of Metformin. Front. Endocrinol. 2019, 10, 294. [Google Scholar] [CrossRef] [PubMed]
- He, L.; Wondisford, F.E. Metformin action: Concentrations matter. Cell Metab. 2015, 21, 159–162. [Google Scholar] [CrossRef] [PubMed]
- Fontaine, E. Metformin-Induced Mitochondrial Complex I Inhibition: Facts, Uncertainties, and Consequences. Front. Endocrinol. 2018, 9, 753. [Google Scholar] [CrossRef] [PubMed]
- Kulkarni, A.S.; Gubbi, S.; Barzilai, N. Benefits of Metformin in Attenuating the Hallmarks of Aging. Cell Metab. 2020, 32, 15–30. [Google Scholar]
- Elbein, A.D.; Pan, Y.T.; Pastuszak, I.; Carroll, D. New insights on trehalose: A multifunctional molecule. Glycobiology 2003, 13, 17R–27R. [Google Scholar] [CrossRef]
- Aatsinki, S.M.; Buler, M.; Salomäki, H.; Koulu, M.; Pavek, P.; Hakkola, J. Metformin induces PGC-1α expression and selectively affects hepatic PGC-1α functions. Br. J. Pharmacol. 2014, 171, 2351–2363. [Google Scholar] [CrossRef]
- Murphy, M.P.; Hartley, R.C. Mitochondria as a therapeutic target for common pathologies. Nat. Rev. Drug Discov. 2018, 17, 865–886. [Google Scholar]
- Slack, C.; Foley, A.; Partridge, L. Activation of AMPK by the putative dietary restriction mimetic metformin is insufficient to extend lifespan in Drosophila. PLoS ONE 2012, 7, e47699. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gumeni, S.; Lamprou, M.; Evangelakou, Z.; Manola, M.S.; Trougakos, I.P. Sustained Nrf2 Overexpression-Induced Metabolic Deregulation Can Be Attenuated by Modulating Insulin/Insulin-like Growth Factor Signaling. Cells 2023, 12, 2650. https://doi.org/10.3390/cells12222650
Gumeni S, Lamprou M, Evangelakou Z, Manola MS, Trougakos IP. Sustained Nrf2 Overexpression-Induced Metabolic Deregulation Can Be Attenuated by Modulating Insulin/Insulin-like Growth Factor Signaling. Cells. 2023; 12(22):2650. https://doi.org/10.3390/cells12222650
Chicago/Turabian StyleGumeni, Sentiljana, Maria Lamprou, Zoi Evangelakou, Maria S. Manola, and Ioannis P. Trougakos. 2023. "Sustained Nrf2 Overexpression-Induced Metabolic Deregulation Can Be Attenuated by Modulating Insulin/Insulin-like Growth Factor Signaling" Cells 12, no. 22: 2650. https://doi.org/10.3390/cells12222650
APA StyleGumeni, S., Lamprou, M., Evangelakou, Z., Manola, M. S., & Trougakos, I. P. (2023). Sustained Nrf2 Overexpression-Induced Metabolic Deregulation Can Be Attenuated by Modulating Insulin/Insulin-like Growth Factor Signaling. Cells, 12(22), 2650. https://doi.org/10.3390/cells12222650