Exploration of the Noncoding Genome for Human-Specific Therapeutic Targets—Recent Insights at Molecular and Cellular Level
Abstract
:1. Introduction
2. Overwhelming Expansion of the Noncoding Genome in Higher Organisms
3. Recent Searches for Evolutionary Relevant Primate/Human-Specific Noncoding Genes
4. Exploration of the Noncoding Genome for Human-Specific Therapeutic Targets—Three Levels of Increasingly Complex Genome–Disease Relationships
5. Genetics of Immune System and Neuro-Immune Interactions Impact upon a Large Spectrum of Human Diseases
6. Noncoding Genomic Regions Impact upon Cardiovascular Pathogenesis in Humans
7. Primate/Human-Specific ncRNAs in Neural/Neuroimmune Cells and Their Impact upon Brain Development and Neuropsychiatric Disorders
8. Multi-Level Functional Integration of Extended Noncoding Regions of the Human Genome—Critical Impact upon Fundamental Cellular Processes Governing Immune Response and Oncogenesis
9. Impact of Human-Specific Genes and ncRNAs upon Pathogenesis via Peculiar Interactions between Human Brain and Mind and the Immune System
10. Current Status of Translational Research into Nucleic Acids-Based Therapeutics
11. State-of-the-Art in Nucleic Acid-Based Therapeutics and Their Molecular and Cell-Biological Foundations
12. Unsolved Challenges and Novel Therapeutic Approaches Guided by Mechanistic Insights at the Molecular and Cell Biological Level
13. Summary and Outlook
14. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lander, E.S.; Linton, L.M.; Birren, B.; Nusbaum, C.; Zody, M.C.; Baldwin, J.; Devon, K.; Dewar, K.; Doyle, M.; FitzHugh, W.; et al. Initial sequencing and analysis of the human genome. Nature 2001, 409, 860–921. [Google Scholar] [PubMed]
- International Human Genome Sequencing Consortium. Finishing the euchromatic sequence of the human genome. Nature 2004, 431, 931–945. [Google Scholar] [CrossRef]
- Encode Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature 2012, 489, 57–74. [Google Scholar] [CrossRef]
- Djebali, S.; Davis, C.A.; Merkel, A.; Dobin, A.; Lassmann, T.; Mortazavi, A.; Tanzer, A.; Lagarde, J.; Lin, W.; Schlesinger, F.; et al. Landscape of transcription in human cells. Nature 2012, 489, 101–108. [Google Scholar] [CrossRef]
- Gerstein, M.B.; Rozowsky, J.; Yan, K.K.; Wang, D.; Cheng, C.; Brown, J.B.; Davis, C.A.; Hillier, L.; Sisu, C.; Li, J.J.; et al. Comparative analysis of the transcriptome across distant species. Nature 2014, 512, 445–448. [Google Scholar] [CrossRef] [PubMed]
- Kapranov, P.; Cheng, J.; Dike, S.; Nix, D.A.; Duttagupta, R.; Willingham, A.T.; Stadler, P.F.; Hertel, J.; Hackermuller, J.; Hofacker, I.L.; et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science 2007, 316, 1484–1488. [Google Scholar] [CrossRef]
- Harrow, J.; Frankish, A.; Gonzalez, J.M.; Tapanari, E.; Diekhans, M.; Kokocinski, F.; Aken, B.L.; Barrell, D.; Zadissa, A.; Searle, S.; et al. GENCODE: The reference human genome annotation for The ENCODE Project. Genome Res. 2012, 22, 1760–1774. [Google Scholar] [CrossRef]
- Poller, W.; Dimmeler, S.; Heymans, S.; Zeller, T.; Haas, J.; Karakas, M.; Leistner, D.M.; Jakob, P.; Nakagawa, S.; Blankenberg, S.; et al. Non-coding RNAs in cardiovascular diseases: Diagnostic and therapeutic perspectives. Eur. Heart J. 2018, 39, 2704–2716. [Google Scholar] [CrossRef]
- Poller, W.; Tank, J.; Skurk, C.; Gast, M. Cardiovascular RNA interference therapy: The broadening tool and target spectrum. Circ. Res. 2013, 113, 588–602. [Google Scholar] [CrossRef]
- Landmesser, U.; Poller, W.; Tsimikas, S.; Most, P.; Paneni, F.; Luscher, T.F. From traditional pharmacological towards nucleic acid-based therapies for cardiovascular diseases. Eur. Heart J. 2020, 41, 3884–3899. [Google Scholar] [CrossRef]
- Poller, W.; Heidecker, B.; Ammirati, E.; Kuss, A.W.; Tzvetkova, A.; Poller, W.C.; Skurk, C.; Haghikia, A. Innate Immunity in Cardiovascular Diseases-Identification of Novel Molecular Players and Targets. J. Clin. Med. 2023, 12, 335. [Google Scholar] [CrossRef] [PubMed]
- Gast, M.; Nageswaran, V.; Kuss, A.W.; Tzvetkova, A.; Wang, X.; Mochmann, L.H.; Rad, P.R.; Weiss, S.; Simm, S.; Zeller, T.; et al. tRNA-like Transcripts from the NEAT1-MALAT1 Genomic Region Critically Influence Human Innate Immunity and Macrophage Functions. Cells 2022, 11, 3970. [Google Scholar] [CrossRef] [PubMed]
- Hahn, M.W.; Wray, G.A. The g-value paradox. Evol. Dev. 2002, 4, 73–75. [Google Scholar] [CrossRef]
- Leypold, N.A.; Speicher, M.R. Evolutionary conservation in noncoding genomic regions. Trends Genet. 2021, 37, 903–918. [Google Scholar] [CrossRef] [PubMed]
- Ramirez-Colmenero, A.; Oktaba, K.; Fernandez-Valverde, S.L. Evolution of Genome-Organizing Long Non-coding RNAs in Metazoans. Front. Genet. 2020, 11, 589697. [Google Scholar] [CrossRef]
- Sandmann, C.L.; Schulz, J.F.; Ruiz-Orera, J.; Kirchner, M.; Ziehm, M.; Adami, E.; Marczenke, M.; Christ, A.; Liebe, N.; Greiner, J.; et al. Evolutionary origins and interactomes of human, young microproteins and small peptides translated from short open reading frames. Mol. Cell 2023, 83, 994–1011.e18. [Google Scholar] [CrossRef]
- Liu, S.J.; Horlbeck, M.A.; Cho, S.W.; Birk, H.S.; Malatesta, M.; He, D.; Attenello, F.J.; Villalta, J.E.; Cho, M.Y.; Chen, Y.; et al. CRISPRi-based genome-scale identification of functional long noncoding RNA loci in human cells. Science 2017, 355, eaah7111. [Google Scholar] [CrossRef]
- RNAcentral Consortium. RNAcentral 2021: Secondary structure integration, improved sequence search and new member databases. Nucleic Acids Res. 2021, 49, D212–D220. [Google Scholar] [CrossRef]
- Seifuddin, F.; Singh, K.; Suresh, A.; Judy, J.T.; Chen, Y.C.; Chaitankar, V.; Tunc, I.; Ruan, X.; Li, P.; Chen, Y.; et al. lncRNAKB, a knowledgebase of tissue-specific functional annotation and trait association of long noncoding RNA. Sci. Data 2020, 7, 326. [Google Scholar] [CrossRef]
- Volders, P.J.; Anckaert, J.; Verheggen, K.; Nuytens, J.; Martens, L.; Mestdagh, P.; Vandesompele, J. LNCipedia 5: Towards a reference set of human long non-coding RNAs. Nucleic Acids Res. 2019, 47, D135–D139. [Google Scholar] [CrossRef]
- Ma, L.; Cao, J.; Liu, L.; Du, Q.; Li, Z.; Zou, D.; Bajic, V.B.; Zhang, Z. LncBook: A curated knowledgebase of human long non-coding RNAs. Nucleic Acids Res. 2019, 47, 2699. [Google Scholar] [CrossRef] [PubMed]
- Fang, S.; Zhang, L.; Guo, J.; Niu, Y.; Wu, Y.; Li, H.; Zhao, L.; Li, X.; Teng, X.; Sun, X.; et al. NONCODEV5: A comprehensive annotation database for long non-coding RNAs. Nucleic Acids Res. 2018, 46, D308–D314. [Google Scholar] [CrossRef] [PubMed]
- Mas-Ponte, D.; Carlevaro-Fita, J.; Palumbo, E.; Hermoso Pulido, T.; Guigo, R.; Johnson, R. LncATLAS database for subcellular localization of long noncoding RNAs. RNA 2017, 23, 1080–1087. [Google Scholar] [CrossRef] [PubMed]
- Derrien, T.; Johnson, R.; Bussotti, G.; Tanzer, A.; Djebali, S.; Tilgner, H.; Guernec, G.; Martin, D.; Merkel, A.; Knowles, D.G.; et al. The GENCODE v7 catalog of human long noncoding RNAs: Analysis of their gene structure, evolution, and expression. Genome Res. 2012, 22, 1775–1789. [Google Scholar] [CrossRef]
- Mattick, J.S.; Amaral, P.P.; Carninci, P.; Carpenter, S.; Chang, H.Y.; Chen, L.L.; Chen, R.; Dean, C.; Dinger, M.E.; Fitzgerald, K.A.; et al. Long non-coding RNAs: Definitions, functions, challenges and recommendations. Nat. Rev. Mol. Cell Biol. 2023, 24, 430–447. [Google Scholar] [CrossRef]
- Ulitsky, I.; Shkumatava, A.; Jan, C.H.; Sive, H.; Bartel, D.P. Conserved function of lincRNAs in vertebrate embryonic development despite rapid sequence evolution. Cell 2011, 147, 1537–1550. [Google Scholar] [CrossRef]
- Ross, C.J.; Ulitsky, I. Discovering functional motifs in long noncoding RNAs. Wiley Interdiscip. Rev. RNA 2022, 13, e1708. [Google Scholar] [CrossRef]
- Ross, C.J.; Rom, A.; Spinrad, A.; Gelbard-Solodkin, D.; Degani, N.; Ulitsky, I. Uncovering deeply conserved motif combinations in rapidly evolving noncoding sequences. Genome Biol. 2021, 22, 29. [Google Scholar] [CrossRef]
- Degani, N.; Lubelsky, Y.; Perry, R.B.; Ainbinder, E.; Ulitsky, I. Highly conserved and cis-acting lncRNAs produced from paralogous regions in the center of HOXA and HOXB clusters in the endoderm lineage. PLoS Genet. 2021, 17, e1009681. [Google Scholar] [CrossRef]
- Paabo, S. The human condition-a molecular approach. Cell 2014, 157, 216–226. [Google Scholar] [CrossRef]
- Essel, E.; Zavala, E.I.; Schulz-Kornas, E.; Kozlikin, M.B.; Fewlass, H.; Vernot, B.; Shunkov, M.V.; Derevianko, A.P.; Douka, K.; Barnes, I.; et al. Ancient human DNA recovered from a Palaeolithic pendant. Nature 2023, 618, 328–332. [Google Scholar] [CrossRef] [PubMed]
- Bornschein, U.; Zeberg, H.; Enard, W.; Hevers, W.; Paabo, S. Functional dissection of two amino acid substitutions unique to the human FOXP2 protein. Sci. Rep. 2023, 13, 3747. [Google Scholar] [CrossRef] [PubMed]
- Skov, L.; Peyregne, S.; Popli, D.; Iasi, L.N.M.; Deviese, T.; Slon, V.; Zavala, E.I.; Hajdinjak, M.; Sumer, A.P.; Grote, S.; et al. Genetic insights into the social organization of Neanderthals. Nature 2022, 610, 519–525. [Google Scholar] [CrossRef]
- Pinson, A.; Xing, L.; Namba, T.; Kalebic, N.; Peters, J.; Oegema, C.E.; Traikov, S.; Reppe, K.; Riesenberg, S.; Maricic, T.; et al. Human TKTL1 implies greater neurogenesis in frontal neocortex of modern humans than Neanderthals. Science 2022, 377, eabl6422. [Google Scholar] [CrossRef]
- Mora-Bermudez, F.; Kanis, P.; Macak, D.; Peters, J.; Naumann, R.; Xing, L.; Sarov, M.; Winkler, S.; Oegema, C.E.; Haffner, C.; et al. Longer metaphase and fewer chromosome segregation errors in modern human than Neanderthal brain development. Sci. Adv. 2022, 8, eabn7702. [Google Scholar] [CrossRef]
- Haeggstrom, S.; Ingelman-Sundberg, M.; Paabo, S.; Zeberg, H. The clinically relevant CYP2C8*3 and CYP2C9*2 haplotype is inherited from Neandertals. Pharmacogenom. J. 2022, 22, 247–249. [Google Scholar] [CrossRef] [PubMed]
- Brown, S.; Massilani, D.; Kozlikin, M.B.; Shunkov, M.V.; Derevianko, A.P.; Stoessel, A.; Jope-Street, B.; Meyer, M.; Kelso, J.; Paabo, S.; et al. The earliest Denisovans and their cultural adaptation. Nat. Ecol. Evol. 2022, 6, 28–35. [Google Scholar] [CrossRef]
- Hajdinjak, M.; Mafessoni, F.; Skov, L.; Vernot, B.; Hubner, A.; Fu, Q.; Essel, E.; Nagel, S.; Nickel, B.; Richter, J.; et al. Initial Upper Palaeolithic humans in Europe had recent Neanderthal ancestry. Nature 2021, 592, 253–257. [Google Scholar] [CrossRef]
- Prabhakar, S.; Noonan, J.P.; Paabo, S.; Rubin, E.M. Accelerated evolution of conserved noncoding sequences in humans. Science 2006, 314, 786. [Google Scholar] [CrossRef]
- Madupe, P.P.; Koenig, C.; Patramanis, I.; Ruther, P.L.; Hlazo, N.; Mackie, M.; Tawane, M.; Krueger, J.; Tarouzzi, A.J.; Troche, G.; et al. Enamel proteins reveal biological sex and genetic variability within southern African Paranthropus. bioRxiv 2023. [Google Scholar] [CrossRef]
- Mortlock, D.P.; Fang, Z.M.; Chandler, K.J.; Hou, Y.; Bickford, L.R.; de Bock, C.E.; Eapen, V.; Clarke, R.A. Transcriptional Interference Regulates the Evolutionary Development of Speech. Genes 2022, 13, 1195. [Google Scholar] [CrossRef] [PubMed]
- Pollard, K.S.; Salama, S.R.; Lambert, N.; Lambot, M.A.; Coppens, S.; Pedersen, J.S.; Katzman, S.; King, B.; Onodera, C.; Siepel, A.; et al. An RNA gene expressed during cortical development evolved rapidly in humans. Nature 2006, 443, 167–172. [Google Scholar] [CrossRef] [PubMed]
- Pembroke, W.G.; Hartl, C.L.; Geschwind, D.H. Evolutionary conservation and divergence of the human brain transcriptome. Genome Biol. 2021, 22, 52. [Google Scholar] [CrossRef] [PubMed]
- Espinos, A.; Fernandez-Ortuno, E.; Negri, E.; Borrell, V. Evolution of genetic mechanisms regulating cortical neurogenesis. Dev. Neurobiol. 2022, 82, 428–453. [Google Scholar] [CrossRef] [PubMed]
- Florio, M.; Heide, M.; Pinson, A.; Brandl, H.; Albert, M.; Winkler, S.; Wimberger, P.; Huttner, W.B.; Hiller, M. Evolution and cell-type specificity of human-specific genes preferentially expressed in progenitors of fetal neocortex. eLife 2018, 7, e32332. [Google Scholar] [CrossRef] [PubMed]
- Hubisz, M.J.; Pollard, K.S. Exploring the genesis and functions of Human Accelerated Regions sheds light on their role in human evolution. Curr. Opin. Genet. Dev. 2014, 29, 15–21. [Google Scholar] [CrossRef]
- Lindblad-Toh, K.; Garber, M.; Zuk, O.; Lin, M.F.; Parker, B.J.; Washietl, S.; Kheradpour, P.; Ernst, J.; Jordan, G.; Mauceli, E.; et al. A high-resolution map of human evolutionary constraint using 29 mammals. Nature 2011, 478, 476–482. [Google Scholar] [CrossRef]
- Uebbing, S.; Gockley, J.; Reilly, S.K.; Kocher, A.A.; Geller, E.; Gandotra, N.; Scharfe, C.; Cotney, J.; Noonan, J.P. Massively parallel discovery of human-specific substitutions that alter enhancer activity. Proc. Natl. Acad. Sci. USA 2021, 118, e2007049118. [Google Scholar] [CrossRef]
- Won, H.; Huang, J.; Opland, C.K.; Hartl, C.L.; Geschwind, D.H. Human evolved regulatory elements modulate genes involved in cortical expansion and neurodevelopmental disease susceptibility. Nat. Commun. 2019, 10, 2396. [Google Scholar] [CrossRef]
- Gast, M.; Rauch, B.H.; Haghikia, A.; Nakagawa, S.; Haas, J.; Stroux, A.; Schmidt, D.; Schumann, P.; Weiss, S.; Jensen, L.; et al. Long noncoding RNA NEAT1 modulates immune cell functions and is suppressed in early onset myocardial infarction patients. Cardiovasc. Res. 2019, 115, 1886–1906. [Google Scholar] [CrossRef]
- Drury, S.; Claussen, G.; Zetterman, A.; Moriyama, H.; Moriyama, E.N.; Zhang, L. Evolution and emergence of primate-specific interferon regulatory factor 9. J. Med. Virol. 2023, 95, e28521. [Google Scholar] [CrossRef] [PubMed]
- Ellwanger, K.; Becker, E.; Kienes, I.; Sowa, A.; Postma, Y.; Cardona Gloria, Y.; Weber, A.N.R.; Kufer, T.A. The NLR family pyrin domain-containing 11 protein contributes to the regulation of inflammatory signaling. J. Biol. Chem. 2018, 293, 2701–2710. [Google Scholar] [CrossRef] [PubMed]
- Van Solingen, C.; Cyr, Y.; Scacalossi, K.R.; de Vries, M.; Barrett, T.J.; de Jong, A.; Gourvest, M.; Zhang, T.; Peled, D.; Kher, R.; et al. Long noncoding RNA CHROMR regulates antiviral immunity in humans. Proc. Natl. Acad. Sci. USA 2022, 119, e2210321119. [Google Scholar] [CrossRef]
- Hennessy, E.J.; van Solingen, C.; Scacalossi, K.R.; Ouimet, M.; Afonso, M.S.; Prins, J.; Koelwyn, G.J.; Sharma, M.; Ramkhelawon, B.; Carpenter, S.; et al. The long noncoding RNA CHROME regulates cholesterol homeostasis in primate. Nat. Metab. 2019, 1, 98–110. [Google Scholar] [CrossRef] [PubMed]
- Salvador, A.F.; de Lima, K.A.; Kipnis, J. Neuromodulation by the immune system: A focus on cytokines. Nat. Rev. Immunol. 2021, 21, 526–541. [Google Scholar] [CrossRef] [PubMed]
- Silverstein, A.M. Promethean Evolution: A Comparison of the Immune and Neural Systems. Perspect. Biol. Med. 2014, 57, 449–469. [Google Scholar] [CrossRef]
- Malekos, E.; Carpenter, S. Short open reading frame genes in innate immunity: From discovery to characterization. Trends Immunol. 2022, 43, 741–756. [Google Scholar] [CrossRef]
- Michieletto, M.F.; Henao-Mejia, J. Ontogeny and heterogeneity of innate lymphoid cells and the noncoding genome. Immunol. Rev. 2021, 300, 152–166. [Google Scholar] [CrossRef]
- Sadeq, S.; Al-Hashimi, S.; Cusack, C.M.; Werner, A. Endogenous Double-Stranded RNA. Noncoding RNA 2021, 7, 15. [Google Scholar] [CrossRef]
- Imamura, K.; Imamachi, N.; Akizuki, G.; Kumakura, M.; Kawaguchi, A.; Nagata, K.; Kato, A.; Kawaguchi, Y.; Sato, H.; Yoneda, M.; et al. Long noncoding RNA NEAT1-dependent SFPQ relocation from promoter region to paraspeckle mediates IL8 expression upon immune stimuli. Mol. Cell 2014, 53, 393–406. [Google Scholar] [CrossRef]
- Gomez, J.A.; Wapinski, O.L.; Yang, Y.W.; Bureau, J.F.; Gopinath, S.; Monack, D.M.; Chang, H.Y.; Brahic, M.; Kirkegaard, K. The NeST long ncRNA controls microbial susceptibility and epigenetic activation of the interferon-gamma locus. Cell 2013, 152, 743–754. [Google Scholar] [CrossRef] [PubMed]
- Collier, S.P.; Henderson, M.A.; Tossberg, J.T.; Aune, T.M. Regulation of the Th1 genomic locus from Ifng through Tmevpg1 by T-bet. J. Immunol. 2014, 193, 3959–3965. [Google Scholar] [CrossRef] [PubMed]
- Ouyang, J.; Zhu, X.; Chen, Y.; Wei, H.; Chen, Q.; Chi, X.; Qi, B.; Zhang, L.; Zhao, Y.; Gao, G.F.; et al. NRAV, a long noncoding RNA, modulates antiviral responses through suppression of interferon-stimulated gene transcription. Cell Host Microbe 2014, 16, 616–626. [Google Scholar] [CrossRef] [PubMed]
- Ouyang, J.; Hu, J.; Chen, J.L. lncRNAs regulate the innate immune response to viral infection. Wiley Interdiscip. Rev. RNA 2016, 7, 129–143. [Google Scholar] [CrossRef]
- Li, Z.; Chao, T.C.; Chang, K.Y.; Lin, N.; Patil, V.S.; Shimizu, C.; Head, S.R.; Burns, J.C.; Rana, T.M. The long noncoding RNA THRIL regulates TNFalpha expression through its interaction with hnRNPL. Proc. Natl. Acad. Sci. USA 2014, 111, 1002–1007. [Google Scholar] [CrossRef]
- Plasek, L.M.; Valadkhan, S. lncRNAs in T lymphocytes: RNA regulation at the heart of the immune response. Am. J. Physiol. Cell Physiol. 2021, 320, C415–C427. [Google Scholar] [CrossRef]
- Kambara, H.; Niazi, F.; Kostadinova, L.; Moonka, D.K.; Siegel, C.T.; Post, A.B.; Carnero, E.; Barriocanal, M.; Fortes, P.; Anthony, D.D.; et al. Negative regulation of the interferon response by an interferon-induced long non-coding RNA. Nucleic Acids Res. 2014, 42, 10668–10680. [Google Scholar] [CrossRef]
- Winterling, C.; Koch, M.; Koeppel, M.; Garcia-Alcalde, F.; Karlas, A.; Meyer, T.F. Evidence for a crucial role of a host non-coding RNA in influenza A virus replication. RNA Biol. 2014, 11, 66–75. [Google Scholar] [CrossRef]
- Spurlock, C.F., 3rd; Tossberg, J.T.; Guo, Y.; Collier, S.P.; Crooke, P.S., 3rd; Aune, T.M. Expression and functions of long noncoding RNAs during human T helper cell differentiation. Nat. Commun. 2015, 6, 6932. [Google Scholar] [CrossRef]
- Aune, T.M.; Spurlock, C.F. 3rd, Long non-coding RNAs in innate and adaptive immunity. Virus Res. 2016, 212, 146–160. [Google Scholar] [CrossRef]
- Koh, B.H.; Hwang, S.S.; Kim, J.Y.; Lee, W.; Kang, M.J.; Lee, C.G.; Park, J.W.; Flavell, R.A.; Lee, G.R. Th2 LCR is essential for regulation of Th2 cytokine genes and for pathogenesis of allergic asthma. Proc. Natl. Acad. Sci. USA 2010, 107, 10614–10619. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Xue, Y.; Han, Y.; Lin, L.; Wu, C.; Xu, S.; Jiang, Z.; Xu, J.; Liu, Q.; Cao, X. The STAT3-binding long noncoding RNA lnc-DC controls human dendritic cell differentiation. Science 2014, 344, 310–313. [Google Scholar] [CrossRef] [PubMed]
- Ranzani, V.; Rossetti, G.; Panzeri, I.; Arrigoni, A.; Bonnal, R.J.; Curti, S.; Gruarin, P.; Provasi, E.; Sugliano, E.; Marconi, M.; et al. The long intergenic noncoding RNA landscape of human lymphocytes highlights the regulation of T cell differentiation by linc-MAF-4. Nat. Immunol. 2015, 16, 318–325. [Google Scholar] [CrossRef] [PubMed]
- Barriocanal, M.; Carnero, E.; Segura, V.; Fortes, P. Long Non-Coding RNA BST2/BISPR is Induced by IFN and Regulates the Expression of the Antiviral Factor Tetherin. Front. Immunol. 2014, 5, 655. [Google Scholar] [CrossRef] [PubMed]
- Rapicavoli, N.A.; Qu, K.; Zhang, J.; Mikhail, M.; Laberge, R.M.; Chang, H.Y. A mammalian pseudogene lncRNA at the interface of inflammation and anti-inflammatory therapeutics. eLife 2013, 2, e00762. [Google Scholar] [CrossRef] [PubMed]
- Xiong, Y.; Yuan, J.; Zhang, C.; Zhu, Y.; Kuang, X.; Lan, L.; Wang, X. The STAT3-regulated long non-coding RNA Lethe promote the HCV replication. Biomed. Pharmacother. 2015, 72, 165–171. [Google Scholar] [CrossRef]
- Hu, G.; Tang, Q.; Sharma, S.; Yu, F.; Escobar, T.M.; Muljo, S.A.; Zhu, J.; Zhao, K. Expression and regulation of intergenic long noncoding RNAs during T cell development and differentiation. Nat. Immunol. 2013, 14, 1190–1198. [Google Scholar] [CrossRef]
- Cui, H.; Xie, N.; Tan, Z.; Banerjee, S.; Thannickal, V.J.; Abraham, E.; Liu, G. The human long noncoding RNA lnc-IL7R regulates the inflammatory response. Eur. J. Immunol. 2014, 44, 2085–2095. [Google Scholar] [CrossRef]
- Dijkstra, J.M.; Alexander, D.B. The “NF-k B interacting long noncoding RNA” (NKILA) transcript is antisense to cancer-associated gene PMEPA1. F1000Research 2015, 4, 96. [Google Scholar] [CrossRef]
- Liu, B.; Sun, L.; Liu, Q.; Gong, C.; Yao, Y.; Lv, X.; Lin, L.; Yao, H.; Su, F.; Li, D.; et al. A cytoplasmic NF-kappaB interacting long noncoding RNA blocks IkappaB phosphorylation and suppresses breast cancer metastasis. Cancer Cell 2015, 27, 370–381. [Google Scholar] [CrossRef]
- Imam, H.; Bano, A.S.; Patel, P.; Holla, P.; Jameel, S. The lncRNA NRON modulates HIV-1 replication in a NFAT-dependent manner and is differentially regulated by early and late viral proteins. Sci. Rep. 2015, 5, 8639. [Google Scholar] [CrossRef] [PubMed]
- Krawczyk, M.; Emerson, B.M. p50-associated COX-2 extragenic RNA (PACER) activates COX-2 gene expression by occluding repressive NF-kappaB complexes. eLife 2014, 3, e01776. [Google Scholar] [CrossRef] [PubMed]
- Khan, S.; Masood, M.; Gaur, H.; Ahmad, S.; Syed, M.A. Long non-coding RNA: An immune cells perspective. Life Sci. 2021, 271, 119152. [Google Scholar] [CrossRef]
- Henzinger, H.; Barth, D.A.; Klec, C.; Pichler, M. Non-Coding RNAs and SARS-Related Coronaviruses. Viruses 2020, 12, 1374. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; Lin, X.; Chen, M. LncRNA NEAT1 correlates with Th17 cells and proinflammatory cytokines, also reflects stenosis degree and cholesterol level in coronary heart disease patients. J. Clin. Lab. Anal. 2022, 36, e23975. [Google Scholar] [CrossRef] [PubMed]
- Vlachogiannis, N.I.; Sachse, M.; Georgiopoulos, G.; Zormpas, E.; Bampatsias, D.; Delialis, D.; Bonini, F.; Galyfos, G.; Sigala, F.; Stamatelopoulos, K.; et al. Adenosine-to-inosine Alu RNA editing controls the stability of the pro-inflammatory long noncoding RNA NEAT1 in atherosclerotic cardiovascular disease. J. Mol. Cell. Cardiol. 2021, 160, 111–120. [Google Scholar] [CrossRef]
- Jing, D.; Zhu, F.; Xu, Z.; Zhang, G.; Zhou, G. The role of long noncoding RNA (lncRNA) nuclear-enriched abundant transcript 1 (NEAT1) in immune diseases. Transpl. Immunol. 2022, 75, 101716. [Google Scholar] [CrossRef]
- Deforzh, E.; Uhlmann, E.J.; Das, E.; Galitsyna, A.; Arora, R.; Saravanan, H.; Rabinovsky, R.; Wirawan, A.D.; Teplyuk, N.M.; El Fatimy, R.; et al. Promoter and enhancer RNAs regulate chromatin reorganization and activation of miR-10b/HOXD locus, and neoplastic transformation in glioma. Mol. Cell 2022, 82, 1894–1908 e5. [Google Scholar] [CrossRef]
- Roy, P.; Orecchioni, M.; Ley, K. How the immune system shapes atherosclerosis: Roles of innate and adaptive immunity. Nat. Rev. Immunol. 2022, 22, 251–265. [Google Scholar] [CrossRef]
- Rodriguez, R.M.; Lopez-Vazquez, A.; Lopez-Larrea, C. Immune systems evolution. Adv. Exp. Med. Biol. 2012, 739, 237–251. [Google Scholar]
- Gourbal, B.; Pinaud, S.; Beckers, G.J.M.; Van Der Meer, J.W.M.; Conrath, U.; Netea, M.G. Innate immune memory: An evolutionary perspective. Immunol. Rev. 2018, 283, 21–40. [Google Scholar] [CrossRef] [PubMed]
- Davis, M.M.; Brodin, P. Rebooting Human Immunology. Annu. Rev. Immunol. 2018, 36, 843–864. [Google Scholar] [CrossRef] [PubMed]
- Suckau, L.; Fechner, H.; Chemaly, E.; Krohn, S.; Hadri, L.; Kockskamper, J.; Westermann, D.; Bisping, E.; Ly, H.; Wang, X.; et al. Long-term cardiac-targeted RNA interference for the treatment of heart failure restores cardiac function and reduces pathological hypertrophy. Circulation 2009, 119, 1241–1252. [Google Scholar] [CrossRef] [PubMed]
- Fechner, H.; Sipo, I.; Westermann, D.; Pinkert, S.; Wang, X.; Suckau, L.; Kurreck, J.; Zeichhardt, H.; Muller, O.; Vetter, R.; et al. Cardiac-targeted RNA interference mediated by an AAV9 vector improves cardiac function in coxsackievirus B3 cardiomyopathy. J. Mol. Med. 2008, 86, 987–997. [Google Scholar] [CrossRef] [PubMed]
- Bonauer, A.; Carmona, G.; Iwasaki, M.; Mione, M.; Koyanagi, M.; Fischer, A.; Burchfield, J.; Fox, H.; Doebele, C.; Ohtani, K.; et al. MicroRNA-92a controls angiogenesis and functional recovery of ischemic tissues in mice. Science 2009, 324, 1710–1713. [Google Scholar] [CrossRef]
- Daniel, J.M.; Penzkofer, D.; Teske, R.; Dutzmann, J.; Koch, A.; Bielenberg, W.; Bonauer, A.; Boon, R.A.; Fischer, A.; Bauersachs, J.; et al. Inhibition of miR-92a improves re-endothelialization and prevents neointima formation following vascular injury. Cardiovasc. Res. 2014, 103, 564–572. [Google Scholar] [CrossRef]
- Thum, T.; Gross, C.; Fiedler, J.; Fischer, T.; Kissler, S.; Bussen, M.; Galuppo, P.; Just, S.; Rottbauer, W.; Frantz, S.; et al. MicroRNA-21 contributes to myocardial disease by stimulating MAP kinase signalling in fibroblasts. Nature 2008, 456, 980–984. [Google Scholar] [CrossRef]
- Sassi, Y.; Avramopoulos, P.; Ramanujam, D.; Gruter, L.; Werfel, S.; Giosele, S.; Brunner, A.D.; Esfandyari, D.; Papadopoulou, A.S.; De Strooper, B.; et al. Cardiac myocyte miR-29 promotes pathological remodeling of the heart by activating Wnt signaling. Nat. Commun. 2017, 8, 1614. [Google Scholar] [CrossRef]
- Ganesan, J.; Ramanujam, D.; Sassi, Y.; Ahles, A.; Jentzsch, C.; Werfel, S.; Leierseder, S.; Loyer, X.; Giacca, M.; Zentilin, L.; et al. MiR-378 controls cardiac hypertrophy by combined repression of mitogen-activated protein kinase pathway factors. Circulation 2013, 127, 2097–2106. [Google Scholar] [CrossRef]
- Janssen, H.L.; Reesink, H.W.; Lawitz, E.J.; Zeuzem, S.; Rodriguez-Torres, M.; Patel, K.; van der Meer, A.J.; Patick, A.K.; Chen, A.; Zhou, Y.; et al. Treatment of HCV infection by targeting microRNA. N. Engl. J. Med. 2013, 368, 1685–1694. [Google Scholar] [CrossRef]
- Corsten, M.F.; Papageorgiou, A.; Verhesen, W.; Carai, P.; Lindow, M.; Obad, S.; Summer, G.; Coort, S.L.; Hazebroek, M.; van Leeuwen, R.; et al. MicroRNA profiling identifies microRNA-155 as an adverse mediator of cardiac injury and dysfunction during acute viral myocarditis. Circ. Res. 2012, 111, 415–425. [Google Scholar] [CrossRef] [PubMed]
- Foinquinos, A.; Batkai, S.; Genschel, C.; Viereck, J.; Rump, S.; Gyongyosi, M.; Traxler, D.; Riesenhuber, M.; Spannbauer, A.; Lukovic, D.; et al. Preclinical development of a miR-132 inhibitor for heart failure treatment. Nat. Commun. 2020, 11, 633. [Google Scholar] [CrossRef]
- Iwamoto, N.; Butler, D.C.D.; Svrzikapa, N.; Mohapatra, S.; Zlatev, I.; Sah, D.W.Y.; Meena; Standley, S.M.; Lu, G.; Apponi, L.H.; et al. Control of phosphorothioate stereochemistry substantially increases the efficacy of antisense oligonucleotides. Nat. Biotechnol. 2017, 35, 845–851. [Google Scholar] [CrossRef] [PubMed]
- Hara, R.I.; Hisada, Y.; Maeda, Y.; Yokota, T.; Wada, T. Artificial cationic oligosaccharides for heteroduplex oligonucleotide-type drugs. Sci. Rep. 2018, 8, 4323. [Google Scholar] [CrossRef] [PubMed]
- Farkas, N.; Scaria, P.V.; Woodle, M.C.; Dagata, J.A. Physical-chemical measurement method development for self-assembled, core-shell nanoparticles. Sci. Rep. 2019, 9, 1655. [Google Scholar] [CrossRef]
- Smith, C.I.E.; Zain, R. Therapeutic Oligonucleotides: State of the Art. Annu. Rev. Pharmacol. Toxicol. 2019, 59, 605–630. [Google Scholar] [CrossRef]
- Yu, A.M.; Choi, Y.H.; Tu, M.J. RNA Drugs and RNA Targets for Small Molecules: Principles, Progress, and Challenges. Pharmacol. Rev. 2020, 72, 862–898. [Google Scholar] [CrossRef]
- Graham, M.J.; Lee, R.G.; Brandt, T.A.; Tai, L.J.; Fu, W.; Peralta, R.; Yu, R.; Hurh, E.; Paz, E.; McEvoy, B.W.; et al. Cardiovascular and Metabolic Effects of ANGPTL3 Antisense Oligonucleotides. N. Engl. J. Med. 2017, 377, 222–232. [Google Scholar] [CrossRef]
- Zlatev, I.; Castoreno, A.; Brown, C.R.; Qin, J.; Waldron, S.; Schlegel, M.K.; Degaonkar, R.; Shulga-Morskaya, S.; Xu, H.; Gupta, S.; et al. Reversal of siRNA-mediated gene silencing in vivo. Nat. Biotechnol. 2018, 36, 509–511. [Google Scholar] [CrossRef]
- Fechner, H.; Suckau, L.; Kurreck, J.; Sipo, I.; Wang, X.; Pinkert, S.; Loschen, S.; Rekittke, J.; Weger, S.; Dekkers, D.; et al. Highly efficient and specific modulation of cardiac calcium homeostasis by adenovector-derived short hairpin RNA targeting phospholamban. Gene Ther. 2007, 14, 211–218. [Google Scholar] [CrossRef]
- Le, B.T.; Paul, S.; Jastrzebska, K.; Langer, H.; Caruthers, M.H.; Veedu, R.N. Thiomorpholino oligonucleotides as a robust class of next generation platforms for alternate mRNA splicing. Proc. Natl. Acad. Sci. USA 2022, 119, e2207956119. [Google Scholar] [CrossRef] [PubMed]
- Hwang, J.; Yokota, T. Recent advancements in exon-skipping therapies using antisense oligonucleotides and genome editing for the treatment of various muscular dystrophies. Expert Rev. Mol. Med. 2019, 21, e5. [Google Scholar] [CrossRef] [PubMed]
- Alexander, V.J.; Xia, S.; Hurh, E.; Hughes, S.G.; O’Dea, L.; Geary, R.S.; Witztum, J.L.; Tsimikas, S. N-acetyl galactosamine-conjugated antisense drug to APOC3 mRNA, triglycerides and atherogenic lipoprotein levels. Eur. Heart J. 2019, 40, 2785–2796. [Google Scholar] [CrossRef] [PubMed]
- Ray, K.K.; Wright, R.S.; Kallend, D.; Koenig, W.; Leiter, L.A.; Raal, F.J.; Bisch, J.A.; Richardson, T.; Jaros, M.; Wijngaard, P.L.J.; et al. Two Phase 3 Trials of Inclisiran in Patients with Elevated LDL Cholesterol. N. Engl. J. Med. 2020, 382, 1507–1519. [Google Scholar] [CrossRef]
- Raal, F.J.; Kallend, D.; Ray, K.K.; Turner, T.; Koenig, W.; Wright, R.S.; Wijngaard, P.L.J.; Curcio, D.; Jaros, M.J.; Leiter, L.A.; et al. Inclisiran for the Treatment of Heterozygous Familial Hypercholesterolemia. N. Engl. J. Med. 2020, 382, 1520–1530. [Google Scholar] [CrossRef]
- Benson, M.D.; Waddington-Cruz, M.; Berk, J.L.; Polydefkis, M.; Dyck, P.J.; Wang, A.K.; Plante-Bordeneuve, V.; Barroso, F.A.; Merlini, G.; Obici, L.; et al. Inotersen Treatment for Patients with Hereditary Transthyretin Amyloidosis. N. Engl. J. Med. 2018, 379, 22–31. [Google Scholar] [CrossRef]
- Gertz, M.A.; Scheinberg, M.; Waddington-Cruz, M.; Heitner, S.B.; Karam, C.; Drachman, B.; Khella, S.; Whelan, C.; Obici, L. Inotersen for the treatment of adults with polyneuropathy caused by hereditary transthyretin-mediated amyloidosis. Expert Rev. Clin. Pharmacol. 2019, 12, 701–711. [Google Scholar] [CrossRef]
- Wright, R.S.; Ray, K.K.; Raal, F.J.; Kallend, D.G.; Jaros, M.; Koenig, W.; Leiter, L.A.; Landmesser, U.; Schwartz, G.G.; Friedman, A.; et al. Pooled Patient-Level Analysis of Inclisiran Trials in Patients with Familial Hypercholesterolemia or Atherosclerosis. J. Am. Coll. Cardiol. 2021, 77, 1182–1193. [Google Scholar] [CrossRef]
- Ray, K.K.; Landmesser, U.; Leiter, L.A.; Kallend, D.; Dufour, R.; Karakas, M.; Hall, T.; Troquay, R.P.; Turner, T.; Visseren, F.L.; et al. Inclisiran in Patients at High Cardiovascular Risk with Elevated LDL Cholesterol. N. Engl. J. Med. 2017, 376, 1430–1440. [Google Scholar] [CrossRef]
- Ray, K.K.; Stoekenbroek, R.M.; Kallend, D.; Leiter, L.A.; Landmesser, U.; Wright, R.S.; Wijngaard, P.; Kastelein, J.J.P. Effect of an siRNA Therapeutic Targeting PCSK9 on Atherogenic Lipoproteins. Circulation 2018, 138, 1304–1316. [Google Scholar] [CrossRef]
- Ray, K.K.; Stoekenbroek, R.M.; Kallend, D.; Nishikido, T.; Leiter, L.A.; Landmesser, U.; Wright, R.S.; Wijngaard, P.L.J.; Kastelein, J.J.P. Effect of 1 or 2 Doses of Inclisiran on Low-Density Lipoprotein Cholesterol Levels: One-Year Follow-up of the ORION-1 Randomized Clinical Trial. JAMA Cardiol. 2019, 4, 1067–1075. [Google Scholar] [CrossRef] [PubMed]
- Witztum, J.L.; Gaudet, D.; Freedman, S.D.; Alexander, V.J.; Digenio, A.; Williams, K.R.; Yang, Q.; Hughes, S.G.; Geary, R.S.; Arca, M.; et al. Volanesorsen and Triglyceride Levels in Familial Chylomicronemia Syndrome. N. Engl. J. Med. 2019, 381, 531–542. [Google Scholar] [CrossRef] [PubMed]
- Ramms, B.; Patel, S.; Nora, C.; Pessentheiner, A.R.; Chang, M.W.; Green, C.R.; Golden, G.J.; Secrest, P.; Krauss, R.M.; Metallo, C.M.; et al. ApoC-III ASO promotes tissue LPL activity in the absence of apoE-mediated TRL clearance. J. Lipid Res. 2019, 60, 1379–1395. [Google Scholar] [CrossRef] [PubMed]
- Tsimikas, S.; Karwatowska-Prokopczuk, E.; Gouni-Berthold, I.; Tardif, J.C.; Baum, S.J.; Steinhagen-Thiessen, E.; Shapiro, M.D.; Stroes, E.S.; Moriarty, P.M.; Nordestgaard, B.G.; et al. Lipoprotein(a) Reduction in Persons with Cardiovascular Disease. N. Engl. J. Med. 2020, 382, 244–255. [Google Scholar] [CrossRef] [PubMed]
- Viney, N.J.; van Capelleveen, J.C.; Geary, R.S.; Xia, S.; Tami, J.A.; Yu, R.Z.; Marcovina, S.M.; Hughes, S.G.; Graham, M.J.; Crooke, R.M.; et al. Antisense oligonucleotides targeting apolipoprotein(a) in people with raised lipoprotein(a): Two randomised, double-blind, placebo-controlled, dose-ranging trials. Lancet 2016, 388, 2239–2253. [Google Scholar] [CrossRef]
- Tsimikas, S.; Viney, N.J.; Hughes, S.G.; Singleton, W.; Graham, M.J.; Baker, B.F.; Burkey, J.L.; Yang, Q.; Marcovina, S.M.; Geary, R.S.; et al. Antisense therapy targeting apolipoprotein(a): A randomised, double-blind, placebo-controlled phase 1 study. Lancet 2015, 386, 1472–1483. [Google Scholar] [CrossRef]
- Alfano, L.N.; Charleston, J.S.; Connolly, A.M.; Cripe, L.; Donoghue, C.; Dracker, R.; Dworzak, J.; Eliopoulos, H.; Frank, D.E.; Lewis, S.; et al. Long-term treatment with eteplirsen in nonambulatory patients with Duchenne muscular dystrophy. Medicine 2019, 98, e15858. [Google Scholar] [CrossRef]
- Minamisawa, M.; Claggett, B.; Adams, D.; Kristen, A.V.; Merlini, G.; Slama, M.S.; Dispenzieri, A.; Shah, A.M.; Falk, R.H.; Karsten, V.; et al. Association of Patisiran, an RNA Interference Therapeutic, With Regional Left Ventricular Myocardial Strain in Hereditary Transthyretin Amyloidosis: The APOLLO Study. JAMA Cardiol. 2019, 4, 466–472. [Google Scholar] [CrossRef]
- Ray, K.K.; Nicholls, S.J.; Buhr, K.A.; Ginsberg, H.N.; Johansson, J.O.; Kalantar-Zadeh, K.; Kulikowski, E.; Toth, P.P.; Wong, N.; Sweeney, M.; et al. Effect of Apabetalone Added to Standard Therapy on Major Adverse Cardiovascular Events in Patients with Recent Acute Coronary Syndrome and Type 2 Diabetes: A Randomized Clinical Trial. JAMA 2020, 323, 1565–1573. [Google Scholar] [CrossRef]
- Elman, L.; Youn, B.; Proud, C.M.; Frey, M.R.; Ajroud-Driss, S.; McCormick, M.E.; Michelson, D.; Cartwright, M.S.; Heiman-Patterson, T.; Choi, J.M.; et al. Real-world Adherence to Nusinersen in Adults with Spinal Muscular Atrophy in the US: A Multi-site Chart Review Study. J. Neuromuscul. Dis. 2022, 9, 655–660. [Google Scholar] [CrossRef]
- Qiu, J.; Wu, L.; Qu, R.; Jiang, T.; Bai, J.; Sheng, L.; Feng, P.; Sun, J. History of development of the life-saving drug “Nusinersen” in spinal muscular atrophy. Front. Cell. Neurosci. 2022, 16, 942976. [Google Scholar] [CrossRef] [PubMed]
- Adams, D.; Gonzalez-Duarte, A.; O’Riordan, W.D.; Yang, C.C.; Ueda, M.; Kristen, A.V.; Tournev, I.; Schmidt, H.H.; Coelho, T.; Berk, J.L.; et al. Patisiran, an RNAi Therapeutic, for Hereditary Transthyretin Amyloidosis. N. Engl. J. Med. 2018, 379, 11–21. [Google Scholar] [CrossRef] [PubMed]
- Solomon, S.D.; Adams, D.; Kristen, A.; Grogan, M.; Gonzalez-Duarte, A.; Maurer, M.S.; Merlini, G.; Damy, T.; Slama, M.S.; Brannagan, T.H., 3rd; et al. Effects of Patisiran, an RNA Interference Therapeutic, on Cardiac Parameters in Patients with Hereditary Transthyretin-Mediated Amyloidosis. Circulation 2019, 139, 431–443. [Google Scholar] [CrossRef] [PubMed]
- Levin, A.A. Treating Disease at the RNA Level with Oligonucleotides. N. Engl. J. Med. 2019, 380, 57–70. [Google Scholar] [CrossRef]
- Taubel, J.; Hauke, W.; Rump, S.; Viereck, J.; Batkai, S.; Poetzsch, J.; Rode, L.; Weigt, H.; Genschel, C.; Lorch, U.; et al. Novel antisense therapy targeting microRNA-132 in patients with heart failure: Results of a first-in-human Phase 1b randomized, double-blind, placebo-controlled study. Eur. Heart J. 2021, 42, 178–188. [Google Scholar] [CrossRef]
- Deogharia, M.; Gurha, P. The “guiding” principles of noncoding RNA function. Wiley Interdiscip. Rev. RNA 2022, 13, e1704. [Google Scholar] [CrossRef]
- Degenhardt, F.; Ellinghaus, D.; Juzenas, S.; Lerga-Jaso, J.; Wendorff, M.; Maya-Miles, D.; Uellendahl-Werth, F.; ElAbd, H.; Ruhlemann, M.C.; Arora, J.; et al. Detailed stratified GWAS analysis for severe COVID-19 in four European populations. Hum. Mol. Genet. 2022, 31, 3945–3966. [Google Scholar] [CrossRef]
- Dannebaum, R.; Suwalski, P.; Asgharian, H.; Du Zhipei, G.; Lin, H.; Weiner, J.; Holtgrewe, M.; Thibeault, C.; Muller, M.; Wang, X.; et al. Highly multiplexed immune repertoire sequencing links multiple lymphocyte classes with severity of response to COVID-19. EClinicalMedicine 2022, 48, 101438. [Google Scholar] [CrossRef]
- Weiner, J.; Suwalski, P.; Holtgrewe, M.; Rakitko, A.; Thibeault, C.; Muller, M.; Patriki, D.; Quedenau, C.; Kruger, U.; Ilinsky, V.; et al. Increased risk of severe clinical course of COVID-19 in carriers of HLA-C*04:01. EClinicalMedicine 2021, 40, 101099. [Google Scholar] [CrossRef]
- Schultheiss, H.P.; Baumeier, C.; Pietsch, H.; Bock, C.T.; Poller, W.; Escher, F. Cardiovascular consequences of viral infections: From COVID to other viral diseases. Cardiovasc. Res. 2021, 117, 2610–2623. [Google Scholar] [CrossRef]
- Prodromidou, K.; Matsas, R. Species-Specific miRNAs in Human Brain Development and Disease. Front. Cell. Neurosci. 2019, 13, 559. [Google Scholar] [CrossRef] [PubMed]
- Prodromidou, K.; Vlachos, I.S.; Gaitanou, M.; Kouroupi, G.; Hatzigeorgiou, A.G.; Matsas, R. MicroRNA-934 is a novel primate-specific small non-coding RNA with neurogenic function during early development. eLife 2020, 9, e50561. [Google Scholar] [CrossRef] [PubMed]
- Choe, S.H.; Park, S.J.; Cho, H.M.; Park, H.R.; Lee, J.R.; Kim, Y.H.; Huh, J.W. A single mutation in the ACTR8 gene associated with lineage-specific expression in primates. BMC Evol. Biol. 2020, 20, 66. [Google Scholar] [CrossRef] [PubMed]
- Vaid, S.; Huttner, W.B. Transcriptional Regulators and Human-Specific/Primate-Specific Genes in Neocortical Neurogenesis. Int. J. Mol. Sci. 2020, 21, 4614. [Google Scholar] [CrossRef] [PubMed]
- Field, A.R.; Jacobs, F.M.J.; Fiddes, I.T.; Phillips, A.P.R.; Reyes-Ortiz, A.M.; LaMontagne, E.; Whitehead, L.; Meng, V.; Rosenkrantz, J.L.; Olsen, M.; et al. Structurally Conserved Primate LncRNAs Are Transiently Expressed during Human Cortical Differentiation and Influence Cell-Type-Specific Genes. Stem Cell Rep. 2019, 12, 245–257. [Google Scholar] [CrossRef]
- Shao, Y.; Chen, C.; Shen, H.; He, B.Z.; Yu, D.; Jiang, S.; Zhao, S.; Gao, Z.; Zhu, Z.; Chen, X.; et al. GenTree, an integrated resource for analyzing the evolution and function of primate-specific coding genes. Genome Res. 2019, 29, 682–696. [Google Scholar] [CrossRef]
- Pruunsild, P.; Bengtson, C.P.; Loss, I.; Lohrer, B.; Bading, H. Expression of the primate-specific LINC00473 RNA in mouse neurons promotes excitability and CREB-regulated transcription. J. Biol. Chem. 2023, 299, 104671. [Google Scholar] [CrossRef]
- Whalen, S.; Pollard, K.S. Enhancer Function and Evolutionary Roles of Human Accelerated Regions. Annu. Rev. Genet. 2022, 56, 423–439. [Google Scholar] [CrossRef]
- Franchini, L.F.; Pollard, K.S. Human evolution: The non-coding revolution. BMC Biol. 2017, 15, 89. [Google Scholar] [CrossRef]
- Waters, E.; Pucci, P.; Hirst, M.; Chapman, S.; Wang, Y.; Crea, F.; Heath, C.J. HAR1: An insight into lncRNA genetic evolution. Epigenomics 2021, 13, 1831–1843. [Google Scholar] [CrossRef]
- Guardiola-Ripoll, M.; Fatjo-Vilas, M. A Systematic Review of the Human Accelerated Regions in Schizophrenia and Related Disorders: Where the Evolutionary and Neurodevelopmental Hypotheses Converge. Int. J. Mol. Sci. 2023, 24, 3597. [Google Scholar] [CrossRef] [PubMed]
- Girskis, K.M.; Stergachis, A.B.; DeGennaro, E.M.; Doan, R.N.; Qian, X.; Johnson, M.B.; Wang, P.P.; Sejourne, G.M.; Nagy, M.A.; Pollina, E.A.; et al. Rewiring of human neurodevelopmental gene regulatory programs by human accelerated regions. Neuron 2021, 109, 3239–3251.e7. [Google Scholar] [CrossRef] [PubMed]
- Fulton, S.L.; Bendl, J.; Gameiro-Ros, I.; Fullard, J.F.; Al-Kachak, A.; Lepack, A.E.; Stewart, A.F.; Singh, S.; Poller, W.C.; Bastle, R.M.; et al. ZBTB7A regulates MDD-specific chromatin signatures and astrocyte-mediated stress vulnerability in orbitofrontal cortex. bioRxiv 2023. [Google Scholar] [CrossRef]
- Luria, V.; Ma, S.; Shibata, M.; Pattabiraman, K.; Sestan, N. Molecular and cellular mechanisms of human cortical connectivity. Curr. Opin. Neurobiol. 2023, 80, 102699. [Google Scholar] [CrossRef] [PubMed]
- Baird, D.A.; Liu, J.Z.; Zheng, J.; Sieberts, S.K.; Perumal, T.; Elsworth, B.; Richardson, T.G.; Chen, C.Y.; Carrasquillo, M.M.; Allen, M.; et al. Identifying drug targets for neurological and psychiatric disease via genetics and the brain transcriptome. PLoS Genet. 2021, 17, e1009224. [Google Scholar] [CrossRef] [PubMed]
- Chu, X.Y.; Quan, Y.; Zhang, H.Y. Human accelerated genome regions with value in medical genetics and drug discovery. Drug Discov. Today 2020, 25, 821–827. [Google Scholar] [CrossRef]
- Xu, K.; Schadt, E.E.; Pollard, K.S.; Roussos, P.; Dudley, J.T. Genomic and network patterns of schizophrenia genetic variation in human evolutionary accelerated regions. Mol. Biol. Evol. 2015, 32, 1148–1160. [Google Scholar] [CrossRef]
- Doan, R.N.; Shin, T.; Walsh, C.A. Evolutionary Changes in Transcriptional Regulation: Insights into Human Behavior and Neurological Conditions. Annu. Rev. Neurosci. 2018, 41, 185–206. [Google Scholar] [CrossRef]
- Singh, D.D.; Verma, R.; Parimoo, P.; Sahu, A.; Kumar, V.; Upadhyay, E.; Yadav, D.K. Potential Therapeutic Relevance of CRISPR/Cas9 Guided Epigenetic Regulations for Neuropsychiatric Disorders. Curr. Top. Med. Chem. 2021, 21, 878–894. [Google Scholar] [CrossRef]
- Preussner, M.; Smith, H.L.; Hughes, D.; Zhang, M.; Emmerichs, A.K.; Scalzitti, S.; Peretti, D.; Swinden, D.; Neumann, A.; Haltenhof, T.; et al. ASO targeting RBM3 temperature-controlled poison exon splicing prevents neurodegeneration in vivo. EMBO Mol. Med. 2023, 15, e17157. [Google Scholar] [CrossRef]
- Nguyen, L.D.; Chau, R.K.; Krichevsky, A.M. Small Molecule Drugs Targeting Non-Coding RNAs as Treatments for Alzheimer’s Disease and Related Dementias. Genes 2021, 12, 2005. [Google Scholar] [CrossRef] [PubMed]
- Srinivas, T.; Mathias, C.; Oliveira-Mateos, C.; Guil, S. Roles of lncRNAs in brain development and pathogenesis: Emerging therapeutic opportunities. Mol. Ther. 2023, 31, 1550–1561. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Duan, Y.; Duan, G.; Wang, Q.; Zhang, K.; Deng, X.; Qian, B.; Gu, J.; Ma, Z.; Zhang, S.; et al. Stress Induces Dynamic, Cytotoxicity-Antagonizing TDP-43 Nuclear Bodies via Paraspeckle LncRNA NEAT1-Mediated Liquid-Liquid Phase Separation. Mol. Cell 2020, 79, 443–458.e7. [Google Scholar] [CrossRef] [PubMed]
- Tollervey, J.R.; Curk, T.; Rogelj, B.; Briese, M.; Cereda, M.; Kayikci, M.; Konig, J.; Hortobagyi, T.; Nishimura, A.L.; Zupunski, V.; et al. Characterizing the RNA targets and position-dependent splicing regulation by TDP-43. Nat. Neurosci. 2011, 14, 452–458. [Google Scholar] [CrossRef]
- Tollervey, J.R.; Wang, Z.; Hortobagyi, T.; Witten, J.T.; Zarnack, K.; Kayikci, M.; Clark, T.A.; Schweitzer, A.C.; Rot, G.; Curk, T.; et al. Analysis of alternative splicing associated with aging and neurodegeneration in the human brain. Genome Res. 2011, 21, 1572–1582. [Google Scholar] [CrossRef]
- Sekar, D.; Tusubira, D.; Ross, K. TDP-43 and NEAT long non-coding RNA: Roles in neurodegenerative disease. Front. Cell. Neurosci. 2022, 16, 954912. [Google Scholar] [CrossRef]
- Kukharsky, M.S.; Ninkina, N.N.; An, H.; Telezhkin, V.; Wei, W.; Meritens, C.R.; Cooper-Knock, J.; Nakagawa, S.; Hirose, T.; Buchman, V.L.; et al. Long non-coding RNA Neat1 regulates adaptive behavioural response to stress in mice. Transl. Psychiatry 2020, 10, 171. [Google Scholar] [CrossRef]
- Oiwa, K.; Watanabe, S.; Onodera, K.; Iguchi, Y.; Kinoshita, Y.; Komine, O.; Sobue, A.; Okada, Y.; Katsuno, M.; Yamanaka, K. Monomerization of TDP-43 is a key determinant for inducing TDP-43 pathology in amyotrophic lateral sclerosis. Sci. Adv. 2023, 9, eadf6895. [Google Scholar] [CrossRef]
- Butler, A.A.; Johnston, D.R.; Kaur, S.; Lubin, F.D. Long noncoding RNA NEAT1 mediates neuronal histone methylation and age-related memory impairment. Sci. Signal. 2019, 12, eaaw9277. [Google Scholar] [CrossRef]
- Huang, Z.; Zhao, J.; Wang, W.; Zhou, J.; Zhang, J. Depletion of LncRNA NEAT1 Rescues Mitochondrial Dysfunction Through NEDD4L-Dependent PINK1 Degradation in Animal Models of Alzheimer’s Disease. Front. Cell. Neurosci. 2020, 14, 28. [Google Scholar] [CrossRef]
- Grinman, E.; Nakahata, Y.; Avchalumov, Y.; Espadas, I.; Swarnkar, S.; Yasuda, R.; Puthanveettil, S.V. Activity-regulated synaptic targeting of lncRNA ADEPTR mediates structural plasticity by localizing Sptn1 and AnkB in dendrites. Sci. Adv. 2021, 7, eabf0605. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, S.; Takashima, A.; Anzai, K. The dendritic translocation of translin protein in the form of BC1 RNA protein particles in developing rat hippocampal neurons in primary culture. Biochem. Biophys. Res. Commun. 1998, 253, 448–453. [Google Scholar] [CrossRef] [PubMed]
- Zalfa, F.; Giorgi, M.; Primerano, B.; Moro, A.; Di Penta, A.; Reis, S.; Oostra, B.; Bagni, C. The fragile X syndrome protein FMRP associates with BC1 RNA and regulates the translation of specific mRNAs at synapses. Cell 2003, 112, 317–327. [Google Scholar] [CrossRef]
- Briz, V.; Restivo, L.; Pasciuto, E.; Juczewski, K.; Mercaldo, V.; Lo, A.C.; Baatsen, P.; Gounko, N.V.; Borreca, A.; Girardi, T.; et al. The non-coding RNA BC1 regulates experience-dependent structural plasticity and learning. Nat. Commun. 2017, 8, 293. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Pang, P.; Fang, Z.; Guo, Y.; Li, H.; Li, X.; Tian, T.; Yang, X.; Chen, W.; Shu, S.; et al. Expression of BC1 Impairs Spatial Learning and Memory in Alzheimer’s Disease Via APP Translation. Mol. Neurobiol. 2018, 55, 6007–6020. [Google Scholar] [CrossRef]
- Kamal, A.; Swellam, M.; Shalaby, N.M.; Darwish, M.K.; El-Nahrery, E.-M. Long non-coding RNAs BACE1-AS and BC200 in multiple sclerosis and their relation to cognitive function: A gene expression analysis. Brain Res. 2023, 1814, 148424. [Google Scholar] [CrossRef]
- Bampatsias, D.; Mavroeidis, I.; Tual-Chalot, S.; Vlachogiannis, N.I.; Bonini, F.; Sachse, M.; Mavraganis, G.; Mareti, A.; Kritsioti, C.; Laina, A.; et al. Beta-Secretase-1 Antisense RNA Is Associated with Vascular Ageing and Atherosclerotic Cardiovascular Disease. Thromb. Haemost. 2022, 122, 1932–1942. [Google Scholar] [CrossRef]
- Sayad, A.; Najafi, S.; Hussen, B.M.; Abdullah, S.T.; Movahedpour, A.; Taheri, M.; Hajiesmaeili, M. The Emerging Roles of the beta-Secretase BACE1 and the Long Non-coding RNA BACE1-AS in Human Diseases: A Focus on Neurodegenerative Diseases and Cancer. Front. Aging Neurosci. 2022, 14, 853180. [Google Scholar] [CrossRef]
- Cui, X.; Zhang, R.; Yang, Y.; Wu, E.; Tang, Y.; Zhao, Z.; Li, C.; Yang, L.; Teng, X.; Ye, Y.; et al. Identification and characterization of long non-coding RNA Carip in modulating spatial learning and memory. Cell Rep. 2022, 38, 110398. [Google Scholar] [CrossRef]
- El Fatimy, R.; Li, S.; Chen, Z.; Mushannen, T.; Gongala, S.; Wei, Z.; Balu, D.T.; Rabinovsky, R.; Cantlon, A.; Elkhal, A.; et al. MicroRNA-132 provides neuroprotection for tauopathies via multiple signaling pathways. Acta Neuropathol. 2018, 136, 537–555. [Google Scholar] [CrossRef]
- Smith, P.Y.; Hernandez-Rapp, J.; Jolivette, F.; Lecours, C.; Bisht, K.; Goupil, C.; Dorval, V.; Parsi, S.; Morin, F.; Planel, E.; et al. miR-132/212 deficiency impairs tau metabolism and promotes pathological aggregation in vivo. Hum. Mol. Genet. 2015, 24, 6721–6735. [Google Scholar] [CrossRef] [PubMed]
- Walgrave, H.; Balusu, S.; Snoeck, S.; Vanden Eynden, E.; Craessaerts, K.; Thrupp, N.; Wolfs, L.; Horre, K.; Fourne, Y.; Ronisz, A.; et al. Restoring miR-132 expression rescues adult hippocampal neurogenesis and memory deficits in Alzheimer’s disease. Cell Stem Cell 2021, 28, 1805–1821.e8. [Google Scholar] [CrossRef] [PubMed]
- Cao, J.; Huang, M.; Guo, L.; Zhu, L.; Hou, J.; Zhang, L.; Pero, A.; Ng, S.; El Gaamouch, F.; Elder, G.; et al. MicroRNA-195 rescues ApoE4-induced cognitive deficits and lysosomal defects in Alzheimer’s disease pathogenesis. Mol. Psychiatry 2021, 26, 4687–4701. [Google Scholar] [CrossRef] [PubMed]
- Kase, Y.; Sonn, I.; Goto, M.; Murakami, R.; Sato, T.; Okano, H. The original strain of SARS-CoV-2, the Delta variant, and the Omicron variant infect microglia efficiently, in contrast to their inability to infect neurons: Analysis using 2D and 3D cultures. Exp. Neurol. 2023, 363, 114379. [Google Scholar] [CrossRef] [PubMed]
- Takemoto, R.; Hirai, Y.; Watanabe, S.; Harada, H.; Suzuki, T.; Hashiguchi, T.; Yanagi, Y.; Shirogane, Y. Interaction of the Hemagglutinin Stalk Region with Cell Adhesion Molecule (CADM) 1 and CADM2 Mediates the Spread between Neurons and Neuropathogenicity of Measles Virus with a Hyperfusogenic Fusion Protein. J. Virol. 2023, 97, e0034023. [Google Scholar] [CrossRef] [PubMed]
- Dietert, K.; Mahesula, S.; Hegde, S.; Verschelde, J.; Reed, P.; Sprague, S.; Kokovay, E.; Sayre, N.L. Loss of LRP1 in Adult Neural Stem Cells Impairs Migration to Ischemic Lesions. Stem Cells 2023, 41, 570–577. [Google Scholar] [CrossRef] [PubMed]
- Malik, J.R.; Acharya, A.; Avedissian, S.N.; Byrareddy, S.N.; Fletcher, C.V.; Podany, A.T.; Dyavar, S.R. ACE-2, TMPRSS2, and Neuropilin-1 Receptor Expression on Human Brain Astrocytes and Pericytes and SARS-CoV-2 Infection Kinetics. Int. J. Mol. Sci. 2023, 24, 8622. [Google Scholar] [CrossRef] [PubMed]
- Bates, E.A.; Lovatt, C.; Plein, A.R.; Davies, J.A.; Siebzehnrubl, F.A.; Parker, A.L. Engineering Adenoviral Vectors with Improved GBM Selectivity. Viruses 2023, 15, 1086. [Google Scholar] [CrossRef]
- Liu, Y.; Huang, R.; Han, L.; Ke, W.; Shao, K.; Ye, L.; Lou, J.; Jiang, C. Brain-targeting gene delivery and cellular internalization mechanisms for modified rabies virus glycoprotein RVG29 nanoparticles. Biomaterials 2009, 30, 4195–4202. [Google Scholar] [CrossRef]
- Malik, A.R.; Lips, J.; Gorniak-Walas, M.; Broekaart, D.W.M.; Asaro, A.; Kuffner, M.T.C.; Hoffmann, C.J.; Kikhia, M.; Dopatka, M.; Boehm-Sturm, P.; et al. SorCS2 facilitates release of endostatin from astrocytes and controls post-stroke angiogenesis. Glia 2020, 68, 1304–1316. [Google Scholar] [CrossRef]
- Rodgers, T.M.; Muzzio, N.; Valero, A.; Ahmad, I.; Ludtke, T.U.; Moya, S.E.; Romero, G. Poly (beta-amino Ester) Nanoparticles Modified with a Rabies Virus-derived peptide for the Delivery of ASCL1 Across a 3D In Vitro Model of the Blood Brain Barrier. ACS Appl. Nano Mater. 2023, 6, 6299–6311. [Google Scholar] [CrossRef] [PubMed]
- Huang, R.; Ke, W.; Han, L.; Liu, Y.; Shao, K.; Ye, L.; Lou, J.; Jiang, C.; Pei, Y. Brain-targeting mechanisms of lactoferrin-modified DNA-loaded nanoparticles. J. Cereb. Blood Flow Metab. 2009, 29, 1914–1923. [Google Scholar] [CrossRef] [PubMed]
- Endo-Takahashi, Y.; Kurokawa, R.; Sato, K.; Takizawa, N.; Katagiri, F.; Hamano, N.; Suzuki, R.; Maruyama, K.; Nomizu, M.; Takagi, N.; et al. Ternary Complexes of pDNA, Neuron-Binding Peptide, and PEGylated Polyethyleneimine for Brain Delivery with Nano-Bubbles and Ultrasound. Pharmaceutics 2021, 13, 1003. [Google Scholar] [CrossRef]
- Gast, M.; Rauch, B.H.; Nakagawa, S.; Haghikia, A.; Jasina, A.; Haas, J.; Nath, N.; Jensen, L.; Stroux, A.; Bohm, A.; et al. Immune system-mediated atherosclerosis caused by deficiency of long non-coding RNA MALAT1 in ApoE−/− mice. Cardiovasc. Res. 2019, 115, 302–314. [Google Scholar] [CrossRef]
- Cremer, S.; Michalik, K.M.; Fischer, A.; Pfisterer, L.; Jae, N.; Winter, C.; Boon, R.A.; Muhly-Reinholz, M.; John, D.; Uchida, S.; et al. Hematopoietic Deficiency of the Long Noncoding RNA MALAT1 Promotes Atherosclerosis and Plaque Inflammation. Circulation 2019, 139, 1320–1334. [Google Scholar] [CrossRef]
- Mamontova, V.; Trifault, B.; Burger, K. Compartment-Specific Proximity Ligation Expands the Toolbox to Assess the Interactome of the Long Non-Coding RNA NEAT1. Int. J. Mol. Sci. 2022, 23, 4432. [Google Scholar] [CrossRef]
- Gu, J.; Zhang, B.; An, R.; Qian, W.; Han, L.; Duan, W.; Wang, Z.; Ma, Q. Molecular Interactions of the Long Noncoding RNA NEAT1 in Cancer. Cancers 2022, 14, 4009. [Google Scholar] [CrossRef]
- Park, M.K.; Zhang, L.; Min, K.W.; Cho, J.H.; Yeh, C.C.; Moon, H.; Hormaechea-Agulla, D.; Mun, H.; Ko, S.; Lee, J.W.; et al. NEAT1 is essential for metabolic changes that promote breast cancer growth and metastasis. Cell Metab. 2021, 33, 2380–2397.e9. [Google Scholar] [CrossRef]
- Naveed, A.; Cooper, J.A.; Li, R.; Hubbard, A.; Chen, J.; Liu, T.; Wilton, S.D.; Fletcher, S.; Fox, A.H. NEAT1 polyA-modulating antisense oligonucleotides reveal opposing functions for both long non-coding RNA isoforms in neuroblastoma. Cell. Mol. Life Sci. 2021, 78, 2213–2230. [Google Scholar] [CrossRef]
- Bhatt, U.; Kretzmann, A.L.; Guedin, A.; Ou, A.; Kobelke, S.; Bond, C.S.; Evans, C.W.; Hurley, L.H.; Mergny, J.L.; Iyer, K.S.; et al. The role of G-Quadruplex DNA in Paraspeckle formation in cancer. Biochimie 2021, 190, 124–131. [Google Scholar] [CrossRef]
- Knutsen, E.; Lellahi, S.M.; Aure, M.R.; Nord, S.; Fismen, S.; Larsen, K.B.; Gabriel, M.T.; Hedberg, A.; Bjorklund, S.S.; Oslo Breast Cancer Research Consortium (OSBREAC); et al. The expression of the long NEAT1_2 isoform is associated with human epidermal growth factor receptor 2-positive breast cancers. Sci. Rep. 2020, 10, 1277. [Google Scholar] [CrossRef] [PubMed]
- Sunwoo, J.S.; Lee, S.T.; Im, W.; Lee, M.; Byun, J.I.; Jung, K.H.; Park, K.I.; Jung, K.Y.; Lee, S.K.; Chu, K.; et al. Altered Expression of the Long Noncoding RNA NEAT1 in Huntington’s Disease. Mol. Neurobiol. 2017, 54, 1577–1586. [Google Scholar] [CrossRef] [PubMed]
- Modic, M.; Grosch, M.; Rot, G.; Schirge, S.; Lepko, T.; Yamazaki, T.; Lee, F.C.Y.; Rusha, E.; Shaposhnikov, D.; Palo, M.; et al. Cross-Regulation between TDP-43 and Paraspeckles Promotes Pluripotency-Differentiation Transition. Mol. Cell 2019, 74, 951–965.e13. [Google Scholar] [CrossRef] [PubMed]
- Boros, F.A.; Vecsei, L.; Klivenyi, P. NEAT1 on the Field of Parkinson’s Disease: Offense, Defense, or a Player on the Bench? J. Park. Dis. 2021, 11, 123–138. [Google Scholar] [CrossRef] [PubMed]
- McCluggage, F.; Fox, A.H. Paraspeckle nuclear condensates: Global sensors of cell stress? Bioessays 2021, 43, e2000245. [Google Scholar] [CrossRef] [PubMed]
- Gast, M.; Schroen, B.; Voigt, A.; Haas, J.; Kuehl, U.; Lassner, D.; Skurk, C.; Escher, F.; Wang, X.; Kratzer, A.; et al. Long noncoding RNA MALAT1-derived mascRNA is involved in cardiovascular innate immunity. J. Mol. Cell Biol. 2016, 8, 178–181. [Google Scholar] [CrossRef]
- Yamazaki, T.; Yamamoto, T.; Hirose, T. Micellization: A new principle in the formation of biomolecular condensates. Front. Mol. Biosci. 2022, 9, 974772. [Google Scholar] [CrossRef]
- Deforzh, E.; Kharel, P.; Karelin, A.; Ivanov, P.; Krichevsky, A.M. HOXDeRNA activates a cancerous transcription program and super-enhancers genome-wide. bioRxiv 2023. [Google Scholar] [CrossRef]
- Zheng, C.; Wei, Y.; Zhang, Q.; Sun, M.; Wang, Y.; Hou, J.; Zhang, P.; Lv, X.; Su, D.; Jiang, Y.; et al. Multiomics analyses reveal DARS1-AS1/YBX1-controlled posttranscriptional circuits promoting glioblastoma tumorigenesis/radioresistance. Sci. Adv. 2023, 9, eadf3984. [Google Scholar] [CrossRef]
- Gabriely, G.; Yi, M.; Narayan, R.S.; Niers, J.M.; Wurdinger, T.; Imitola, J.; Ligon, K.L.; Kesari, S.; Esau, C.; Stephens, R.M.; et al. Human glioma growth is controlled by microRNA-10b. Cancer Res. 2011, 71, 3563–3572. [Google Scholar] [CrossRef]
- Lang, M.F.; Yang, S.; Zhao, C.; Sun, G.; Murai, K.; Wu, X.; Wang, J.; Gao, H.; Brown, C.E.; Liu, X.; et al. Genome-wide profiling identified a set of miRNAs that are differentially expressed in glioblastoma stem cells and normal neural stem cells. PLoS ONE 2012, 7, e36248. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.; Teo, S.; Lam, D.H.; Jeyaseelan, K.; Wang, S. MicroRNA-10b pleiotropically regulates invasion, angiogenicity and apoptosis of tumor cells resembling mesenchymal subtype of glioblastoma multiforme. Cell Death Dis. 2012, 3, e398. [Google Scholar] [CrossRef] [PubMed]
- El Fatimy, R.; Zhang, Y.; Deforzh, E.; Ramadas, M.; Saravanan, H.; Wei, Z.; Rabinovsky, R.; Teplyuk, N.M.; Uhlmann, E.J.; Krichevsky, A.M. A nuclear function for an oncogenic microRNA as a modulator of snRNA and splicing. Mol. Cancer 2022, 21, 17. [Google Scholar] [CrossRef] [PubMed]
- Teplyuk, N.M.; Uhlmann, E.J.; Gabriely, G.; Volfovsky, N.; Wang, Y.; Teng, J.; Karmali, P.; Marcusson, E.; Peter, M.; Mohan, A.; et al. Therapeutic potential of targeting microRNA-10b in established intracranial glioblastoma: First steps toward the clinic. EMBO Mol. Med. 2016, 8, 268–287. [Google Scholar] [CrossRef] [PubMed]
- El Fatimy, R.; Subramanian, S.; Uhlmann, E.J.; Krichevsky, A.M. Genome Editing Reveals Glioblastoma Addiction to MicroRNA-10b. Mol. Ther. 2017, 25, 368–378. [Google Scholar] [CrossRef]
- Zhang, Y.; Rabinovsky, R.; Wei, Z.; El Fatimy, R.; Deforzh, E.; Luan, B.; Peshkin, L.; Uhlmann, E.J.; Krichevsky, A.M. Secreted PGK1 and IGFBP2 contribute to the bystander effect of miR-10b gene editing in glioma. Mol. Ther. Nucleic Acids 2023, 31, 265–275. [Google Scholar] [CrossRef]
- Wang, Y.; Malik, S.; Suh, H.W.; Xiao, Y.; Deng, Y.; Fan, R.; Huttner, A.; Bindra, R.S.; Singh, V.; Saltzman, W.M.; et al. Anti-seed PNAs targeting multiple oncomiRs for brain tumor therapy. Sci. Adv. 2023, 9, eabq7459. [Google Scholar] [CrossRef]
- Douka, K.; Birds, I.; Wang, D.; Kosteletos, A.; Clayton, S.; Byford, A.; Vasconcelos, E.J.R.; O’Connell, M.J.; Deuchars, J.; Whitehouse, A.; et al. Cytoplasmic long noncoding RNAs are differentially regulated and translated during human neuronal differentiation. RNA 2021, 27, 1082–1101. [Google Scholar] [CrossRef]
- Xu, Y.; Yu, X.; Zhang, M.; Zheng, Q.; Sun, Z.; He, Y.; Guo, W. Promising Advances in LINC01116 Related to Cancer. Front. Cell Dev. Biol. 2021, 9, 736927. [Google Scholar] [CrossRef]
- Viswanathan, K.; Dhabhar, F.S. Stress-induced enhancement of leukocyte trafficking into sites of surgery or immune activation. Proc. Natl. Acad. Sci. USA 2005, 102, 5808–5813. [Google Scholar] [CrossRef]
- Dhabhar, F.S.; Malarkey, W.B.; Neri, E.; McEwen, B.S. Stress-induced redistribution of immune cells--from barracks to boulevards to battlefields: A tale of three hormones—Curt Richter Award winner. Psychoneuroendocrinology 2012, 37, 1345–1368. [Google Scholar] [CrossRef] [PubMed]
- Glaser, R.; Kiecolt-Glaser, J.K. Stress-induced immune dysfunction: Implications for health. Nat. Rev. Immunol. 2005, 5, 243–251. [Google Scholar] [CrossRef] [PubMed]
- Jensterle, M.; Herman, R.; Janez, A.; Mahmeed, W.A.; Al-Rasadi, K.; Al-Alawi, K.; Banach, M.; Banerjee, Y.; Ceriello, A.; Cesur, M.; et al. The Relationship between COVID-19 and Hypothalamic-Pituitary-Adrenal Axis: A Large Spectrum from Glucocorticoid Insufficiency to Excess-The CAPISCO International Expert Panel. Int. J. Mol. Sci. 2022, 23, 7326. [Google Scholar] [CrossRef] [PubMed]
- Schiller, M.; Ben-Shaanan, T.L.; Rolls, A. Neuronal regulation of immunity: Why, how and where? Nat. Rev. Immunol. 2021, 21, 20–36. [Google Scholar] [CrossRef]
- Poller, W.C.; Downey, J.; Mooslechner, A.A.; Khan, N.; Li, L.; Chan, C.T.; McAlpine, C.S.; Xu, C.; Kahles, F.; He, S.; et al. Brain motor and fear circuits regulate leukocytes during acute stress. Nature 2022, 607, 578–584. [Google Scholar] [CrossRef]
- McAlpine, C.S.; Kiss, M.G.; Rattik, S.; He, S.; Vassalli, A.; Valet, C.; Anzai, A.; Chan, C.T.; Mindur, J.E.; Kahles, F.; et al. Sleep modulates haematopoiesis and protects against atherosclerosis. Nature 2019, 566, 383–387. [Google Scholar] [CrossRef]
- Poller, W.C.; Nahrendorf, M.; Swirski, F.K. Hematopoiesis and Cardiovascular Disease. Circ. Res. 2020, 126, 1061–1085. [Google Scholar] [CrossRef]
- McAlpine, C.S.; Kiss, M.G.; Zuraikat, F.M.; Cheek, D.; Schiroli, G.; Amatullah, H.; Huynh, P.; Bhatti, M.Z.; Wong, L.P.; Yates, A.G.; et al. Sleep exerts lasting effects on hematopoietic stem cell function and diversity. J. Exp. Med. 2022, 219, e20220081. [Google Scholar] [CrossRef]
- Janssen, H.; Kahles, F.; Liu, D.; Downey, J.; Koekkoek, L.L.; Roudko, V.; D’Souza, D.; McAlpine, C.S.; Halle, L.; Poller, W.C.; et al. Monocytes re-enter the bone marrow during fasting and alter the host response to infection. Immunity 2023, 56, 783–796.e7. [Google Scholar] [CrossRef]
- Vita, G.; Vita, G.L.; Stancanelli, C.; Gentile, L.; Russo, M.; Mazzeo, A. Genetic neuromuscular disorders: Living the era of a therapeutic revolution. Part 1: Peripheral neuropathies. Neurol. Sci. 2019, 40, 661–669. [Google Scholar] [CrossRef]
- Mathew, V.; Wang, A.K. Inotersen: New promise for the treatment of hereditary transthyretin amyloidosis. Drug Des. Dev. Ther. 2019, 13, 1515–1525. [Google Scholar] [CrossRef]
- Kristen, A.V.; Ajroud-Driss, S.; Conceicao, I.; Gorevic, P.; Kyriakides, T.; Obici, L. Patisiran, an RNAi therapeutic for the treatment of hereditary transthyretin-mediated amyloidosis. Neurodegener. Dis. Manag. 2019, 9, 5–23. [Google Scholar] [CrossRef] [PubMed]
- Krichevsky, A.; Nguyen, L.; Wei, Z.; Silva, M.; Barberan-Soler, S.; Rabinovsky, R.; Muratore, C.; Stricker, J.; Hortman, C.; Young-Pearse, T.; et al. Small Molecule Regulators of microRNAs Identified by High-Throughput Screen Coupled with High-Throughput Sequencing. Res. Sq. 2023. [Google Scholar] [CrossRef]
- Umuhire Juru, A.; Hargrove, A.E. Frameworks for targeting RNA with small molecules. J. Biol. Chem. 2021, 296, 100191. [Google Scholar] [CrossRef] [PubMed]
- Childs-Disney, J.L.; Yang, X.; Gibaut, Q.M.R.; Tong, Y.; Batey, R.T.; Disney, M.D. Targeting RNA structures with small molecules. Nat. Rev. Drug Discov. 2022, 21, 736–762. [Google Scholar] [CrossRef]
- Pilunni, D.; Montrasio, L.; Navarra, P. When innovation goes fast. The case of hemophilia. Curr. Opin. Pharmacol. 2019, 45, 95–101. [Google Scholar] [CrossRef]
- Majowicz, A.; Nijmeijer, B.; Lampen, M.H.; Spronck, L.; de Haan, M.; Petry, H.; van Deventer, S.J.; Meyer, C.; Tangelder, M.; Ferreira, V. Therapeutic hFIX Activity Achieved after Single AAV5-hFIX Treatment in Hemophilia B Patients and NHPs with Pre-existing Anti-AAV5 NABs. Mol. Ther. Methods Clin. Dev. 2019, 14, 27–36. [Google Scholar] [CrossRef]
- Peyvandi, F.; Garagiola, I. Clinical advances in gene therapy updates on clinical trials of gene therapy in haemophilia. Haemophilia 2019, 25, 738–746. [Google Scholar] [CrossRef]
- Samelson-Jones, B.J.; Finn, J.D.; George, L.A.; Camire, R.M.; Arruda, V.R. Hyperactivity of factor IX Padua (R338L) depends on factor VIIIa cofactor activity. JCI Insight 2019, 4, e128683. [Google Scholar] [CrossRef]
- Hunt, B.J.; Corral, J.; Simioni, P. Should we consider gene therapy in severe inherited thrombophilia? Communication from the ISTH SSC subcommittee on Physiological Anticoagulants and Thrombophilia. J. Thromb. Haemost. 2023, 21, 2620–2622. [Google Scholar] [CrossRef]
- Valentino, L.A.; Kaczmarek, R.; Pierce, G.F.; Noone, D.; O’Mahony, B.; Page, D.; Rotellini, D.; Skinner, M.W. Hemophilia Gene Therapy: First, Do No Harm. J. Thromb. Haemost. 2023, 21, 2354–2361. [Google Scholar] [CrossRef] [PubMed]
- Valentino, L.A.; Ozelo, M.C.; Herzog, R.W.; Key, N.S.; Pishko, A.M.; Ragni, M.V.; Samelson-Jones, B.J.; Lillicrap, D. A Review of the Rationale for Gene Therapy for Hemophilia A with Inhibitors: One-Shot Tolerance and Treatment? J. Thromb. Haemost. 2023, 21, 3033–3044. [Google Scholar] [CrossRef]
- Hajighasemi, S.; Mahdavi Gorabi, A.; Bianconi, V.; Pirro, M.; Banach, M.; Ahmadi Tafti, H.; Reiner, Z.; Sahebkar, A. A review of gene- and cell-based therapies for familial hypercholesterolemia. Pharmacol. Res. 2019, 143, 119–132. [Google Scholar] [CrossRef] [PubMed]
- Khorkova, O.; Stahl, J.; Joji, A.; Volmar, C.H.; Wahlestedt, C. Amplifying gene expression with RNA-targeted therapeutics. Nat. Rev. Drug Discov. 2023, 22, 539–561. [Google Scholar] [CrossRef] [PubMed]
- Ray, R.M.; Hansen, A.H.; Slott, S.; Taskova, M.; Astakhova, K.; Morris, K.V. Control of LDL Uptake in Human Cells by Targeting the LDLR Regulatory Long Non-coding RNA BM450697. Mol. Ther. Nucleic Acids 2019, 17, 264–276. [Google Scholar] [CrossRef] [PubMed]
- Costales, M.G.; Aikawa, H.; Li, Y.; Childs-Disney, J.L.; Abegg, D.; Hoch, D.G.; Pradeep Velagapudi, S.; Nakai, Y.; Khan, T.; Wang, K.W.; et al. Small-molecule targeted recruitment of a nuclease to cleave an oncogenic RNA in a mouse model of metastatic cancer. Proc. Natl. Acad. Sci. USA 2020, 117, 2406–2411. [Google Scholar] [CrossRef]
- Benhamou, R.I.; Suresh, B.M.; Tong, Y.; Cochrane, W.G.; Cavett, V.; Vezina-Dawod, S.; Abegg, D.; Childs-Disney, J.L.; Adibekian, A.; Paegel, B.M.; et al. DNA-encoded library versus RNA-encoded library selection enables design of an oncogenic noncoding RNA inhibitor. Proc. Natl. Acad. Sci. USA 2022, 119, e2114971119. [Google Scholar] [CrossRef]
- Campos, L.J.; Arokiaraj, C.M.; Chuapoco, M.R.; Chen, X.; Goeden, N.; Gradinaru, V.; Fox, A.S. Advances in AAV technology for delivering genetically encoded cargo to the nonhuman primate nervous system. Curr. Res. Neurobiol. 2023, 4, 100086. [Google Scholar] [CrossRef]
- Williams, J.A.; Paez, P.A. Improving cell and gene therapy safety and performance using next-generation Nanoplasmid vectors. Mol. Ther. Nucleic Acids 2023, 32, 494–503. [Google Scholar] [CrossRef]
- Ail, D.; Malki, H.; Zin, E.A.; Dalkara, D. Adeno-Associated Virus (AAV)-Based Gene Therapies for Retinal Diseases: Where are We? Appl. Clin. Genet. 2023, 16, 111–130. [Google Scholar] [CrossRef]
- Varshney, S.; Alam, A.; Kaur, A.; Dhoundiyal, S. Niosomes: A Smart Drug Delivery System for Brain Targeting. Pharm. Nanotechnol. 2023. [Google Scholar] [CrossRef] [PubMed]
- Narasipura, E.A.; VanKeulen-Miller, R.; Ma, Y.; Fenton, O.S. Ongoing Clinical Trials of Nonviral siRNA Therapeutics. Bioconjug. Chem. 2023, 34, 1177–1197. [Google Scholar] [CrossRef] [PubMed]
- Alterman, J.F.; Godinho, B.; Hassler, M.R.; Ferguson, C.M.; Echeverria, D.; Sapp, E.; Haraszti, R.A.; Coles, A.H.; Conroy, F.; Miller, R.; et al. A divalent siRNA chemical scaffold for potent and sustained modulation of gene expression throughout the central nervous system. Nat. Biotechnol. 2019, 37, 884–894. [Google Scholar] [CrossRef] [PubMed]
- Conroy, F.; Miller, R.; Alterman, J.F.; Hassler, M.R.; Echeverria, D.; Godinho, B.; Knox, E.G.; Sapp, E.; Sousa, J.; Yamada, K.; et al. Chemical engineering of therapeutic siRNAs for allele-specific gene silencing in Huntington’s disease models. Nat. Commun. 2022, 13, 5802. [Google Scholar] [CrossRef] [PubMed]
- Fechner, H.; Haack, A.; Wang, H.; Wang, X.; Eizema, K.; Pauschinger, M.; Schoemaker, R.; vanVeghel, R.; Houtsmuller, A.; Schultheiss, H.-P.; et al. Expression of Coxsackie-adenovirus-receptor and av-integrin does not correlate with adenovector targeting in vivo indicating anatomical vector barriers. Gene Ther. 1999, 6, 1520–1535. [Google Scholar] [CrossRef]
- Noutsias, M.; Fechner, H.; de Jonge, H.; Wang, X.; Dekkers, D.; Houtsmuller, A.; Pauschinger, M.; Bergelson, J.; Warraich, R.; Yacoub, M.; et al. Human Coxsackie-Adenovirus-Receptor is Co-Localized with Integrins avb3 and avb5 on the Cardiomyocyte Sarcolemma and Upregulated in Dilated Cardiomyopathy—Implications for Cardiotropic Viral Infections. Circulation 2001, 104, 275–280. [Google Scholar] [CrossRef]
- Fechner, H.; Noutsias, M.; Tschoepe, C.; Hinze, K.; Wang, X.; Escher, F.; Pauschinger, M.; Dekkers, D.; Vetter, R.; Paul, M.; et al. Induction of coxsackievirus-adenovirus-receptor expression during myocardial tissue formation and remodeling: Identification of a cell-to-cell contact-dependent regulatory mechanism. Circulation 2003, 107, 876–882. [Google Scholar] [CrossRef]
- Ishikawa, K.; Weber, T.; Hajjar, R.J. Human Cardiac Gene Therapy. Circ. Res. 2018, 123, 601–613. [Google Scholar] [CrossRef]
- O’Reilly, M.; Shipp, A.; Rosenthal, E.; Jambou, R.; Shih, T.; Montgomery, M.; Gargiulo, L.; Patterson, A.; Corrigan-Curay, J. NIH oversight of human gene transfer research involving retroviral, lentiviral, and adeno-associated virus vectors and the role of the NIH recombinant DNA advisory committee. Methods Enzymol. 2012, 507, 313–335. [Google Scholar]
- Consiglieri, G.; Bernardo, M.E.; Brunetti-Pierri, N.; Aiuti, A. Ex Vivo and In Vivo Gene Therapy for Mucopolysaccharidoses: State of the Art. Hematol. Oncol. Clin. North Am. 2022, 36, 865–878. [Google Scholar] [CrossRef]
- Papaioannou, I.; Owen, J.S.; Yanez-Munoz, R.J. Clinical applications of gene therapy for rare diseases: A review. Int. J. Exp. Pathol. 2023, 104, 154–176. [Google Scholar] [CrossRef] [PubMed]
- Grossen, P.; Skaripa Koukelli, I.; van Haasteren, J.; Machado, A.H.E.; Duerr, C. The Ice Age—A Review on Formulation of Adeno-Associated Virus Therapeutics. Eur. J. Pharm. Biopharm. 2023, 190, 1–23. [Google Scholar] [CrossRef] [PubMed]
- Ke, W.; Shao, K.; Huang, R.; Han, L.; Liu, Y.; Li, J.; Kuang, Y.; Ye, L.; Lou, J.; Jiang, C. Gene delivery targeted to the brain using an Angiopep-conjugated polyethyleneglycol-modified polyamidoamine dendrimer. Biomaterials 2009, 30, 6976–6985. [Google Scholar] [CrossRef] [PubMed]
- Arora, S.; Singh, J. In vitro and in vivo optimization of liposomal nanoparticles based brain targeted vgf gene therapy. Int. J. Pharm. 2021, 608, 121095. [Google Scholar] [CrossRef] [PubMed]
- Mendonca, M.C.P.; Cronin, M.F.; Cryan, J.F.; O’Driscoll, C.M. Modified cyclodextrin-based nanoparticles mediated delivery of siRNA for huntingtin gene silencing across an in vitro BBB model. Eur. J. Pharm. Biopharm. 2021, 169, 309–318. [Google Scholar] [CrossRef]
- Zhang, L.; Wu, T.; Shan, Y.; Li, G.; Ni, X.; Chen, X.; Hu, X.; Lin, L.; Li, Y.; Guan, Y.; et al. Therapeutic reversal of Huntington’s disease by in vivo self-assembled siRNAs. Brain 2021, 144, 3421–3435. [Google Scholar] [CrossRef]
- Shalaby, K.E.; Aouida, M.; Gupta, V.; Abdesselem, H.; El-Agnaf, O.M.A. Development of non-viral vectors for neuronal-targeted delivery of CRISPR-Cas9 RNA-proteins as a therapeutic strategy for neurological disorders. Biomater. Sci. 2022, 10, 4959–4977. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, X.; Xie, R.; Burger, J.C.; Tong, Y.; Gong, S. Overcoming the Blood-Brain Barrier for Gene Therapy via Systemic Administration of GSH-Responsive Silica Nanocapsules. Adv. Mater. 2023, 35, e2208018. [Google Scholar] [CrossRef]
- Wohlfart, S.; Gelperina, S.; Kreuter, J. Transport of drugs across the blood-brain barrier by nanoparticles. J. Control. Release 2012, 161, 264–273. [Google Scholar] [CrossRef]
- Andrieux, K.; Couvreur, P. Polyalkylcyanoacrylate nanoparticles for delivery of drugs across the blood-brain barrier. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 2009, 1, 463–474. [Google Scholar] [CrossRef]
- Huang, S.; Li, J.; Han, L.; Liu, S.; Ma, H.; Huang, R.; Jiang, C. Dual targeting effect of Angiopep-2-modified, DNA-loaded nanoparticles for glioma. Biomaterials 2011, 32, 6832–6838. [Google Scholar] [CrossRef] [PubMed]
- Tang, Z.; Hu, S.; Wu, Z.; He, M. Therapeutic effects of engineered exosome-based miR-25 and miR-181a treatment in spinocerebellar ataxia type 3 mice by silencing ATXN3. Mol. Med. 2023, 29, 96. [Google Scholar] [CrossRef]
- Pinheiro, R.G.R.; Granja, A.; Loureiro, J.A.; Pereira, M.C.; Pinheiro, M.; Neves, A.R.; Reis, S. RVG29-Functionalized Lipid Nanoparticles for Quercetin Brain Delivery and Alzheimer’s Disease. Pharm. Res. 2020, 37, 139. [Google Scholar] [CrossRef] [PubMed]
- Sumser, A.; Joesch, M.; Jonas, P.; Ben-Simon, Y. Fast, high-throughput production of improved rabies viral vectors for specific, efficient and versatile transsynaptic retrograde labeling. eLife 2022, 11, e79848. [Google Scholar] [CrossRef] [PubMed]
- Ghanem, A.; Conzelmann, K.K. G gene-deficient single-round rabies viruses for neuronal circuit analysis. Virus Res. 2016, 216, 41–54. [Google Scholar] [CrossRef]
- Salin, P.; Blondel, D.; Kerkerian-Le Goff, L.; Coulon, P. Golgi staining-like retrograde labeling of brain circuits using rabies virus: Focus onto the striatonigral neurons. J. Neurosci. Methods 2020, 344, 108872. [Google Scholar] [CrossRef]
- Sun, L.; Tang, Y.; Yan, K.; Yu, J.; Zou, Y.; Xu, W.; Xiao, K.; Zhang, Z.; Li, W.; Wu, B.; et al. Differences in neurotropism and neurotoxicity among retrograde viral tracers. Mol. Neurodegener. 2019, 14, 8. [Google Scholar] [CrossRef]
- Zhu, X.; Lin, K.; Liu, Q.; Yue, X.; Mi, H.; Huang, X.; He, X.; Wu, R.; Zheng, D.; Wei, D.; et al. Rabies Virus Pseudotyped with CVS-N2C Glycoprotein as a Powerful Tool for Retrograde Neuronal Network Tracing. Neurosci. Bull. 2020, 36, 202–216. [Google Scholar] [CrossRef]
- Lin, K.Z.; Li, L.; Ma, W.Y.; Yang, X.; Han, Z.P.; Luo, N.S.; Wang, J.; Xu, F.Q. A rabies virus-based toolkit for efficient retrograde labeling and monosynaptic tracing. Neural Regen. Res. 2023, 18, 1827–1833. [Google Scholar] [CrossRef]
- Zhong, C.; Chen, Z.; Xia, Y.; Wu, J.; Zhang, F.; Cheng, C.; Wu, X.; Zhuang, Y.; Xiao, X. Treatment of experimental autoimmune encephalomyelitis using AAV gene therapy by blocking T cell costimulatory pathways. Mol. Ther. Methods Clin. Dev. 2022, 25, 461–475. [Google Scholar] [CrossRef]
- Sandin, E.S.; Folberth, J.; Muller-Fielitz, H.; Pietrzik, C.U.; Herold, E.; Willnow, T.E.; Pfluger, P.T.; Nogueiras, R.; Prevot, V.; Krey, T.; et al. Is LRP2 Involved in Leptin Transport over the Blood-Brain Barrier and Development of Obesity? Int. J. Mol. Sci. 2021, 22, 4998. [Google Scholar] [CrossRef] [PubMed]
- Alnaqbi, N.; Mohammad, M.G.; Hamoudi, R.; Mabondzo, A.; Harati, R. Molecular Heterogeneity of the Brain Endothelium. Curr. Issues Mol. Biol. 2023, 45, 3462–3478. [Google Scholar] [CrossRef] [PubMed]
- Mollgard, K.; Beinlich, F.R.M.; Kusk, P.; Miyakoshi, L.M.; Delle, C.; Pla, V.; Hauglund, N.L.; Esmail, T.; Rasmussen, M.K.; Gomolka, R.S.; et al. A mesothelium divides the subarachnoid space into functional compartments. Science 2023, 379, 84–88. [Google Scholar] [CrossRef] [PubMed]
- Hartmann, K.; Neyazi, B.; Stein, K.P.; Haghikia, A.; Sandalcioglu, I.E. Is the central nervous system enclosed by a mesothel? Ther. Adv. Neurol. Disord. 2023, 16, 17562864231180335. [Google Scholar] [CrossRef] [PubMed]
- Hartmann, K.; Stein, K.P.; Neyazi, B.; Sandalcioglu, I.E. First in vivo visualization of the human subarachnoid space and brain cortex via optical coherence tomography. Ther. Adv. Neurol. Disord. 2019, 12, 1756286419843040. [Google Scholar] [CrossRef]
- Andreone, B.J.; Chow, B.W.; Tata, A.; Lacoste, B.; Ben-Zvi, A.; Bullock, K.; Deik, A.A.; Ginty, D.D.; Clish, C.B.; Gu, C. Blood-Brain Barrier Permeability Is Regulated by Lipid Transport-Dependent Suppression of Caveolae-Mediated Transcytosis. Neuron 2017, 94, 581–594.e5. [Google Scholar] [CrossRef]
- Taghian, T.; Batista, A.R.; Kamper, S.; Caldwell, M.; Lilley, L.; Li, H.; Rodriguez, P.; Mesa, K.; Zheng, S.; King, R.M.; et al. Real-time MR tracking of AAV gene therapy with betagal-responsive MR probe in a murine model of GM1-gangliosidosis. Mol. Ther. Methods Clin. Dev. 2021, 23, 128–134. [Google Scholar] [CrossRef]
- Gray-Edwards, H.L.; Maguire, A.S.; Salibi, N.; Ellis, L.E.; Voss, T.L.; Diffie, E.B.; Koehler, J.; Randle, A.N.; Taylor, A.R.; Brunson, B.L.; et al. 7T MRI Predicts Amelioration of Neurodegeneration in the Brain after AAV Gene Therapy. Mol. Ther. Methods Clin. Dev. 2020, 17, 258–270. [Google Scholar] [CrossRef]
- Gray-Edwards, H.L.; Jiang, X.; Randle, A.N.; Taylor, A.R.; Voss, T.L.; Johnson, A.K.; McCurdy, V.J.; Sena-Esteves, M.; Ory, D.S.; Martin, D.R. Lipidomic Evaluation of Feline Neurologic Disease after AAV Gene Therapy. Mol. Ther. Methods Clin. Dev. 2017, 6, 135–142. [Google Scholar] [CrossRef]
- Hudry, E.; Vandenberghe, L.H. Therapeutic AAV Gene Transfer to the Nervous System: A Clinical Reality. Neuron 2019, 101, 839–862. [Google Scholar] [CrossRef]
- Saraiva, J.; Nobre, R.J.; Pereira de Almeida, L. Gene therapy for the CNS using AAVs: The impact of systemic delivery by AAV9. J. Control. Release 2016, 241, 94–109. [Google Scholar] [CrossRef] [PubMed]
- Rosenberg, J.B.; Kaplitt, M.G.; De, B.P.; Chen, A.; Flagiello, T.; Salami, C.; Pey, E.; Zhao, L.; Ricart Arbona, R.J.; Monette, S.; et al. AAVrh.10-Mediated APOE2 Central Nervous System Gene Therapy for APOE4-Associated Alzheimer’s Disease. Hum. Gene Ther. Clin. Dev. 2018, 29, 24–47. [Google Scholar] [CrossRef] [PubMed]
- Hammoudi, N.; Ishikawa, K.; Hajjar, R.J. Adeno-associated virus-mediated gene therapy in cardiovascular disease. Curr. Opin. Cardiol. 2015, 30, 228–234. [Google Scholar] [CrossRef]
- Li, X.; La Salvia, S.; Liang, Y.; Adamiak, M.; Kohlbrenner, E.; Jeong, D.; Chepurko, E.; Ceholski, D.; Lopez-Gordo, E.; Yoon, S.; et al. Extracellular Vesicle-Encapsulated Adeno-Associated Viruses for Therapeutic Gene Delivery to the Heart. Circulation 2023, 148, 405–425. [Google Scholar] [CrossRef]
- Aronson, S.J.; Veron, P.; Collaud, F.; Hubert, A.; Delahais, V.; Honnet, G.; de Knegt, R.J.; Junge, N.; Baumann, U.; Di Giorgio, A.; et al. Prevalence and Relevance of Pre-Existing Anti-Adeno-Associated Virus Immunity in the Context of Gene Therapy for Crigler-Najjar Syndrome. Hum. Gene Ther. 2019, 30, 1297–1305. [Google Scholar] [CrossRef] [PubMed]
- Xiao, W.; Gao, G.; Ling, C.; Herzog, R.W.; Xiao, X.; Samulski, R.J. Impact of neutralizing antibodies against AAV is a key consideration in gene transfer to nonhuman primates. Nat. Med. 2018, 24, 699. [Google Scholar] [CrossRef] [PubMed]
- Di Pasquale, G.; Chiorini, J.A. AAV transcytosis through barrier epithelia and endothelium. Mol. Ther. 2006, 13, 506–516. [Google Scholar] [CrossRef]
- Dennys, C.N.; Sierra-Delgado, J.A.; Ray, S.S.; Hartlaub, A.M.; Roussel, F.S.; Rodriguez, Y.; Meyer, K. In vitro Modeling for Neurological Diseases using Direct Conversion from Fibroblasts to Neuronal Progenitor Cells and Differentiation into Astrocytes. J. Vis. Exp. 2021, 172, e62016. [Google Scholar]
- Farfara, D.; Sooliman, M.; Avrahami, L.; Royal, T.G.; Amram, S.; Rozenstein-Tsalkovich, L.; Trudler, D.; Blanga-Kanfi, S.; Eldar-Finkelman, H.; Pahnke, J.; et al. Physiological expression of mutated TAU impaired astrocyte activity and exacerbates beta-amyloid pathology in 5xFAD mice. J. Neuroinflamm. 2023, 20, 174. [Google Scholar] [CrossRef]
- Zhang, L.; Rossi, A.; Lange, L.; Meumann, N.; Koitzsch, U.; Christie, K.; Nesbit, M.A.; Moore, C.B.T.; Hacker, U.T.; Morgan, M.; et al. Capsid Engineering Overcomes Barriers Toward Adeno-Associated Virus Vector-Mediated Transduction of Endothelial Cells. Hum. Gene Ther. 2019, 30, 1284–1296. [Google Scholar] [CrossRef]
- Greig, J.A.; Limberis, M.P.; Bell, P.; Chen, S.J.; Calcedo, R.; Rader, D.J.; Wilson, J.M. Non-Clinical Study Examining AAV8.TBG.hLDLR Vector-Associated Toxicity in Chow-Fed Wild-Type and LDLR+/− Rhesus Macaques. Hum. Gene Ther. Clin. Dev. 2017, 28, 39–50. [Google Scholar] [CrossRef] [PubMed]
- Meyer, N.L.; Chapman, M.S. Adeno-associated virus (AAV) cell entry: Structural insights. Trends Microbiol. 2022, 30, 432–451. [Google Scholar] [PubMed]
- Peters, C.W.; Maguire, C.A.; Hanlon, K.S. Delivering AAV to the Central Nervous and Sensory Systems. Trends Pharmacol. Sci. 2021, 42, 461–474. [Google Scholar] [CrossRef]
- Pillay, S.; Meyer, N.L.; Puschnik, A.S.; Davulcu, O.; Diep, J.; Ishikawa, Y.; Jae, L.T.; Wosen, J.E.; Nagamine, C.M.; Chapman, M.S.; et al. An essential receptor for adeno-associated virus infection. Nature 2016, 530, 108–112. [Google Scholar] [CrossRef]
- Dudek, A.M.; Zabaleta, N.; Zinn, E.; Pillay, S.; Zengel, J.; Porter, C.; Franceschini, J.S.; Estelien, R.; Carette, J.E.; Zhou, G.L.; et al. GPR108 Is a Highly Conserved AAV Entry Factor. Mol. Ther. 2020, 28, 367–381. [Google Scholar] [CrossRef] [PubMed]
- Jang, S.; Shen, H.K.; Ding, X.; Miles, T.F.; Gradinaru, V. Structural basis of receptor usage by the engineered capsid AAV-PHP.eB. Mol. Ther. Methods Clin. Dev. 2022, 26, 343–354. [Google Scholar] [CrossRef]
- Mathiesen, S.N.; Lock, J.L.; Schoderboeck, L.; Abraham, W.C.; Hughes, S.M. CNS Transduction Benefits of AAV-PHP.eB over AAV9 Are Dependent on Administration Route and Mouse Strain. Mol. Ther. Methods Clin. Dev. 2020, 19, 447–458. [Google Scholar] [CrossRef]
- Kanaan, N.M.; Sellnow, R.C.; Boye, S.L.; Coberly, B.; Bennett, A.; Agbandje-McKenna, M.; Sortwell, C.E.; Hauswirth, W.W.; Boye, S.E.; Manfredsson, F.P. Rationally Engineered AAV Capsids Improve Transduction and Volumetric Spread in the CNS. Mol. Ther. Nucleic Acids 2017, 8, 184–197. [Google Scholar] [CrossRef]
- Chamberlain, K.; Riyad, J.M.; Garnett, T.; Kohlbrenner, E.; Mookerjee, A.; Elmastour, F.; Benard, L.; Chen, J.; VandenDriessche, T.; Chuah, M.K.; et al. A Calsequestrin Cis-Regulatory Motif Coupled to a Cardiac Troponin T Promoter Improves Cardiac Adeno-Associated Virus Serotype 9 Transduction Specificity. Hum. Gene Ther. 2018, 29, 927–937. [Google Scholar] [CrossRef]
- Katz, M.G.; Fargnoli, A.S.; Kendle, A.P.; Hajjar, R.J.; Bridges, C.R. Gene Therapy in Cardiac Surgery: Clinical Trials, Challenges, and Perspectives. Ann. Thorac. Surg. 2016, 101, 2407–2416. [Google Scholar] [CrossRef]
- Hulot, J.S.; Ishikawa, K.; Hajjar, R.J. Gene therapy for the treatment of heart failure: Promise postponed. Eur. Heart J. 2016, 37, 1651–1658. [Google Scholar]
- Hartmann, J.; Thalheimer, F.B.; Hopfner, F.; Kerzel, T.; Khodosevich, K.; Garcia-Gonzalez, D.; Monyer, H.; Diester, I.; Buning, H.; Carette, J.E.; et al. GluA4-Targeted AAV Vectors Deliver Genes Selectively to Interneurons while Relying on the AAV Receptor for Entry. Mol. Ther. Methods Clin. Dev. 2019, 14, 252–260. [Google Scholar]
- Eichhoff, A.M.; Borner, K.; Albrecht, B.; Schafer, W.; Baum, N.; Haag, F.; Korbelin, J.; Trepel, M.; Braren, I.; Grimm, D.; et al. Nanobody-Enhanced Targeting of AAV Gene Therapy Vectors. Mol. Ther. Methods Clin. Dev. 2019, 15, 211–220. [Google Scholar]
- Maturana, C.J.; Verpeut, J.L.; Pisano, T.J.; Dhanerawala, Z.M.; Esteves, A.; Enquist, L.W.; Engel, E.A. Small Alphaherpesvirus Latency-Associated Promoters Drive Efficient and Long-Term Transgene Expression in the CNS. Mol. Ther. Methods Clin. Dev. 2020, 17, 843–857. [Google Scholar]
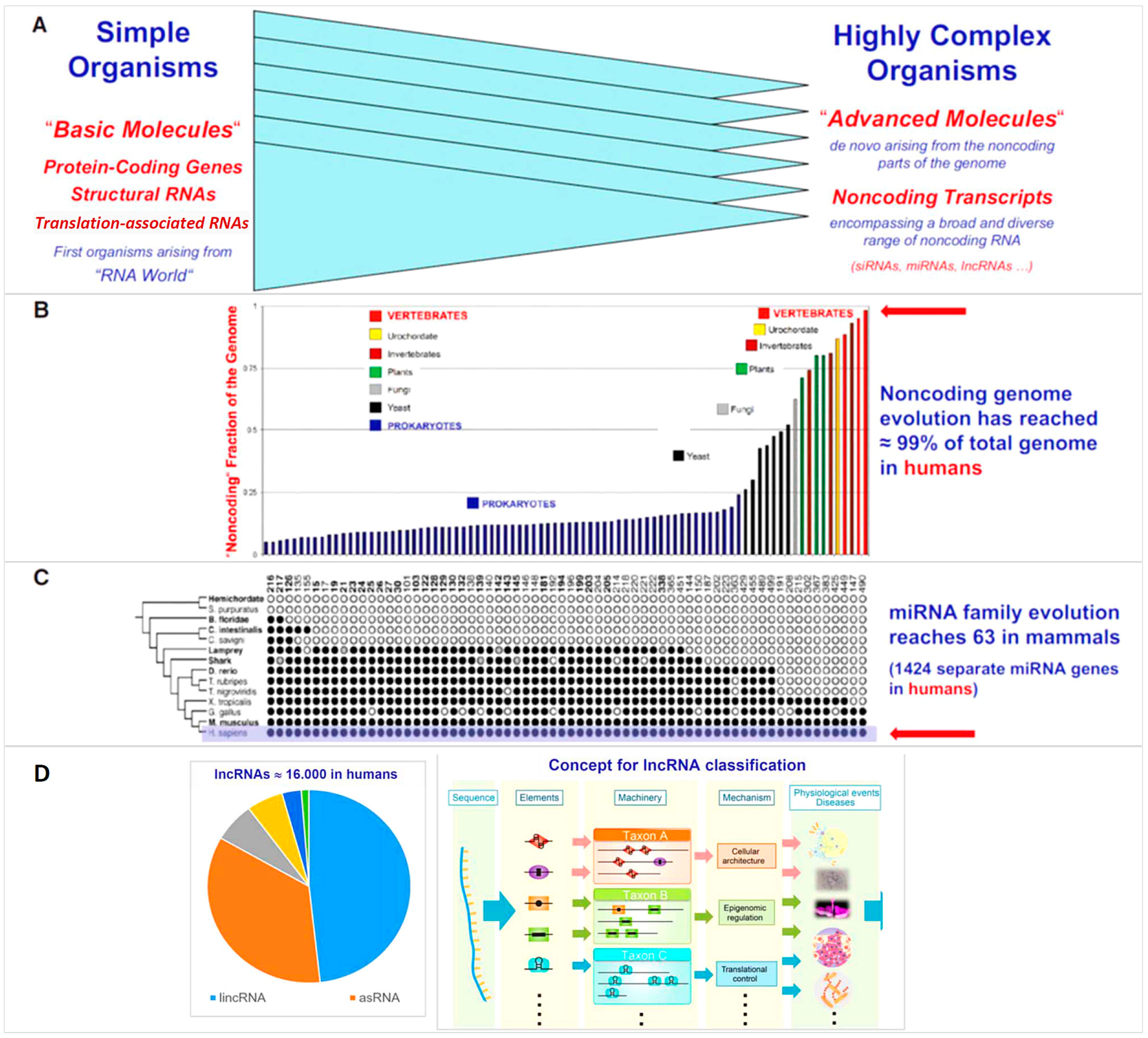






Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Poller, W.; Sahoo, S.; Hajjar, R.; Landmesser, U.; Krichevsky, A.M. Exploration of the Noncoding Genome for Human-Specific Therapeutic Targets—Recent Insights at Molecular and Cellular Level. Cells 2023, 12, 2660. https://doi.org/10.3390/cells12222660
Poller W, Sahoo S, Hajjar R, Landmesser U, Krichevsky AM. Exploration of the Noncoding Genome for Human-Specific Therapeutic Targets—Recent Insights at Molecular and Cellular Level. Cells. 2023; 12(22):2660. https://doi.org/10.3390/cells12222660
Chicago/Turabian StylePoller, Wolfgang, Susmita Sahoo, Roger Hajjar, Ulf Landmesser, and Anna M. Krichevsky. 2023. "Exploration of the Noncoding Genome for Human-Specific Therapeutic Targets—Recent Insights at Molecular and Cellular Level" Cells 12, no. 22: 2660. https://doi.org/10.3390/cells12222660
APA StylePoller, W., Sahoo, S., Hajjar, R., Landmesser, U., & Krichevsky, A. M. (2023). Exploration of the Noncoding Genome for Human-Specific Therapeutic Targets—Recent Insights at Molecular and Cellular Level. Cells, 12(22), 2660. https://doi.org/10.3390/cells12222660






