CAR NK Cell Therapy for the Treatment of Metastatic Melanoma: Potential & Prospects
Abstract
:1. Introduction
2. Biology and Function of NK Cells in the Context of Melanoma
2.1. NK Cell Activating Receptors
2.2. NK Cell Inhibitory Receptors
2.3. NK Cell Cytotoxicity and Significance to Tumor Cell Clearance
3. NK Cell Immunosuppression within the Tumor Microenvironment
4. Car NK Cell Therapies to Treat Melanoma
4.1. Design & Production of CAR-NK Cells
4.2. Sources of NK Cells
4.3. Pre-Clinical CAR-NK Studies with Meaningful Implications for Melanoma
4.4. Potential Targets for CAR-NK Cell Therapies in Melanoma
5. Perspectives and Future Prospects for CAR-NK Cell Therapies to Treat Melanoma
5.1. Structural Optimization of the CAR to Enhance CAR-NK Cell Activation
5.2. Integration of TCR Structural Elements to Expand the Range of Targetable Antigens
5.3. Targeting Melanoma Cell Signaling Pathways
5.4. Targeting Cell Motility to Enhance Immune Infiltration and Impair Malignant Invasion
5.5. Targeting Non-Malignant Cell Types in the TME
5.6. Incoporation with Deletion of Inhibitory Receptor Using Innovated Methods
5.7. Clinical Consideration of Pharmaceutical Impacts on CAR-NK Cell Efficacy
6. Conclusions
Funding
Acknowledgments
Conflicts of Interest
References
- Conforti, C.; Zalaudek, I. Epidemiology and Risk Factors of Melanoma: A Review. Dermatol. Pract. Concept. 2021, 11, 2021161S. [Google Scholar] [CrossRef]
- Strashilov, S.; Yordanov, A. Aetiology and Pathogenesis of Cutaneous Melanoma: Current Concepts and Advances. Int. J. Mol. Sci. 2021, 22, 6395. [Google Scholar] [CrossRef]
- Sundararajan, S.; Thida, A.M.; Yadlapati, S.; Koya, S. Metastatic Melanoma; StatPearls Publishing: Treasure Island, FL, USA, 2022. [Google Scholar]
- National Cancer Institute. SEER Cancer Stat Facts: Melanoma of the Skin. National Cancer Institute. In National Cancer Institute; 2022. Available online: https://seer.cancer.gov/statfacts/html/melan.html (accessed on 26 December 2022).
- Saginala, K.; Barsouk, A.; Aluru, J.S.; Rawla, P.; Barsouk, A. Epidemiology of Melanoma. Med. Sci. 2021, 9, 63. [Google Scholar] [CrossRef]
- Davis, E.J.; Johnson, D.B.; Sosman, J.A.; Chandra, S. Melanoma: What do all the mutations mean? Cancer 2018, 124, 3490–3499. [Google Scholar] [CrossRef] [PubMed]
- Chapman, P.B.; Hauschild, A.; Robert, C.; Haanen, J.B.; Ascierto, P.; Larkin, J.; Dummer, R.; Garbe, C.; Testori, A.; Maio, M.; et al. Improved Survival with Vemurafenib in Melanoma with BRAF V600E Mutation. N. Engl. J. Med. 2011, 364, 2507–2516. [Google Scholar] [CrossRef] [PubMed]
- Chapman, P.B.; Einhorn, L.H.; Meyers, M.L.; Saxman, S.; Destro, A.N.; Panageas, K.S.; Begg, C.B.; Agarwala, S.S.; Schuchter, L.M.; Ernstoff, M.S.; et al. Phase III Multicenter Randomized Trial of the Dartmouth Regimen versus Dacarbazine in Patients with Metastatic Melanoma. J. Clin. Oncol. 1999, 17, 2745–2751. [Google Scholar] [CrossRef] [PubMed]
- Sandru, A.; Voinea, S.; Panaitescu, E.; Blidaru, A. Survival rates of patients with metastatic malignant melanoma. J. Med. Life 2014, 7, 572–576. [Google Scholar] [PubMed]
- Wessely, A.; Steeb, T.; Berking, C.; Heppt, M.V. How Neural Crest Transcription Factors Contribute to Melanoma Heterogeneity, Cellular Plasticity, and Treatment Resistance. Int. J. Mol. Sci. 2021, 22, 5761. [Google Scholar] [CrossRef]
- Priantti, J.N.; Vilbert, M.; Madeira, T.; Moraes, F.C.A.; Hein, E.C.K.; Saeed, A.; Cavalcante, L. Efficacy and Safety of Rechallenge with BRAF/MEK Inhibitors in Advanced Melanoma Patients: A Systematic Review and Meta-Analysis. Cancers 2023, 15, 3754. [Google Scholar] [CrossRef]
- Davis, L.E.; Shalin, S.C.; Tackett, A.J. Current state of melanoma diagnosis and treatment. Cancer Biol. Ther. 2019, 20, 1366–1379. [Google Scholar] [CrossRef]
- Kalal, B.S.; Upadhya, D.; Pai, V.R. Chemotherapy resistance mechanisms in advanced skin cancer. Oncol. Rev. 2017, 11, 19–25. [Google Scholar] [CrossRef] [PubMed]
- June, C.H.; O’Connor, R.S.; Kawalekar, O.U.; Ghassemi, S.; Milone, M.C. CAR T cell immunotherapy for human cancer. Science 2018, 359, 1361–1365. [Google Scholar] [CrossRef] [PubMed]
- Pfefferle, A.; Huntington, N.D. You Have Got a Fast CAR: Chimeric Antigen Receptor NK Cells in Cancer Therapy. Cancers 2020, 12, 706. [Google Scholar] [CrossRef]
- Lee, D.W.; Santomasso, B.D.; Locke, F.L.; Ghobadi, A.; Turtle, C.J.; Brudno, J.N.; Maus, M.V.; Park, J.H.; Mead, E.; Pavletic, S.; et al. ASTCT Consensus Grading for Cytokine Release Syndrome and Neurologic Toxicity Associated with Immune Effector Cells. Biol. Blood Marrow Transplant. 2019, 25, 625–638. [Google Scholar] [CrossRef] [PubMed]
- Schuster, S.J.; Bishop, M.R.; Tam, C.S.; Waller, E.K.; Borchmann, P.; McGuirk, J.P.; Jäger, U.; Jaglowski, S.; Andreadis, C.; Westin, J.R.; et al. Tisagenlecleucel in Adult Relapsed or Refractory Diffuse Large B-Cell Lymphoma. N. Engl. J. Med. 2019, 380, 45–56. [Google Scholar] [CrossRef] [PubMed]
- Neelapu, S.S.; Locke, F.L.; Bartlett, N.L.; Lekakis, L.J.; Miklos, D.B.; Jacobson, C.A.; Braunschweig, I.; Oluwole, O.O.; Siddiqi, T.; Lin, Y.; et al. Axicabtagene Ciloleucel CAR T-Cell Therapy in Refractory Large B-Cell Lymphoma. N. Engl. J. Med. 2017, 377, 2531–2544. [Google Scholar] [CrossRef]
- Liu, P.; Liu, M.; Lyu, C.; Lu, W.; Cui, R.; Wang, J.; Li, Q.; Mou, N.; Deng, Q.; Yang, D. Acute Graft-Versus-Host Disease after Humanized Anti-CD19-CAR T Therapy in Relapsed B-ALL Patients after Allogeneic Hematopoietic Stem Cell Transplant. Front. Oncol. 2020, 10, 799206. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Li, Y.; Cao, W.; Wang, F.; Xie, X.; Li, Y.; Wang, X.; Guo, R.; Jiang, Z.; Guo, R. Single-Cell Analysis of Target Antigens of CAR-T Reveals a Potential Landscape of “On-Target, Off-Tumor Toxicity”. Front. Immunol. 2021, 12, 799206. [Google Scholar] [CrossRef]
- Morgan, R.A.; Yang, J.C.; Kitano, M.; E Dudley, M.; Laurencot, C.M.; Rosenberg, S.A. Case Report of a Serious Adverse Event Following the Administration of T Cells Transduced with a Chimeric Antigen Receptor Recognizing ERBB2. Mol. Ther. 2010, 18, 843–851. [Google Scholar] [CrossRef]
- Castellarin, M.; Sands, C.; Da, T.; Scholler, J.; Graham, K.; Buza, E.; Fraietta, J.A.; Zhao, Y.; June, C.H. A rational mouse model to detect on-target, off-tumor CAR T cell toxicity. JCI Insight 2020, 5, e136012. [Google Scholar] [CrossRef]
- Laskowski, T.J.; Biederstädt, A.; Rezvani, K. Natural killer cells in antitumour adoptive cell immunotherapy. Nat. Rev. Cancer 2022, 22, 557–575. [Google Scholar] [CrossRef]
- Dagogo-Jack, I.; Shaw, A.T. Tumour heterogeneity and resistance to cancer therapies. Nat. Rev. Clin. Oncol. 2018, 15, 81–94. [Google Scholar] [CrossRef] [PubMed]
- Yu, T.; Gao, X.; Zheng, Z.; Zhao, X.; Zhang, S.; Li, C.; Liu, G. Intratumor Heterogeneity as a Prognostic Factor in Solid Tumors: A Systematic Review and Meta-Analysis. Front. Oncol. 2021, 11, 744064. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.X.; Yu, Q. Intra-tumor heterogeneity of cancer cells and its implications for cancer treatment. Acta Pharmacol. Sin. 2015, 36, 1219–1227. [Google Scholar] [CrossRef] [PubMed]
- Grzywa, T.M.; Paskal, W.; Włodarski, P.K. Intratumor and Intertumor Heterogeneity in Melanoma. Transl. Oncol. 2017, 10, 956–975. [Google Scholar] [CrossRef] [PubMed]
- O’rourke, D.M.; Nasrallah, M.P.; Desai, A.; Melenhorst, J.J.; Mansfield, K.; Morrissette, J.J.D.; Martinez-Lage, M.; Brem, S.; Maloney, E.; Shen, A.; et al. A single dose of peripherally infused EGFRvIII-directed CAR T cells mediates antigen loss and induces adaptive resistance in patients with recurrent glioblastoma. Sci. Transl. Med. 2017, 9, eaaa0984. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, N.; Brawley, V.S.; Hegde, M.; Robertson, C.; Ghazi, A.; Gerken, C.; Liu, E.; Dakhova, O.; Ashoori, A.; Corder, A.; et al. Human Epidermal Growth Factor Receptor 2 (HER2)—Specific Chimeric Antigen Receptor—Modified T Cells for the Immunotherapy of HER2-Positive Sarcoma. J. Clin. Oncol. 2015, 33, 1688–1696. [Google Scholar] [CrossRef]
- Pitcovski, J.; Shahar, E.; Aizenshtein, E.; Gorodetsky, R. Melanoma antigens and related immunological markers. Crit. Rev. Oncol. Hematol. 2017, 115, 36–49. [Google Scholar] [CrossRef]
- Shi, J.; Tricot, G.; Szmania, S.; Rosen, N.; Garg, T.K.; Malaviarachchi, P.A.; Moreno, A.; DuPont, B.; Hsu, K.C.; Baxter-Lowe, L.A.; et al. Infusion of haplo-identical killer immunoglobulin-like receptor ligand mismatched NK cells for relapsed myeloma in the setting of autologous stem cell transplantation. Br. J. Haematol. 2008, 143, 641–653. [Google Scholar] [CrossRef]
- Yu, Y. The Function of NK Cells in Tumor Metastasis and NK Cell-Based Immunotherapy. Cancers 2023, 15, 2323. [Google Scholar] [CrossRef]
- Arai, S.; Meagher, R.; Swearingen, M.; Myint, H.; Rich, E.; Martinson, J.; Klingemann, H. Infusion of the allogeneic cell line NK-92 in patients with advanced renal cell cancer or melanoma: A phase I trial. Cytotherapy 2008, 10, 625–632. [Google Scholar] [CrossRef] [PubMed]
- Li, H.K.; Hsiao, C.W.; Yang, S.H.; Yang, H.P.; Wu, T.S.; Lee, C.Y.; Lin, Y.L.; Pan, J.; Cheng, Z.F.; Lai, Y.D.; et al. A Novel off-the-Shelf Trastuzumab-Armed NK Cell Therapy (ACE1702) Using Antibody-Cell-Conjugation Technology. Cancers 2021, 13, 2724. [Google Scholar] [CrossRef] [PubMed]
- Williams, B.A.; Law, A.D.; Routy, B.; Denhollander, N.; Gupta, V.; Wang, X.-H.; Chaboureau, A.; Viswanathan, S.; Keating, A. A phase I trial of NK-92 cells for refractory hematological malignancies relapsing after autologous hematopoietic cell transplantation shows safety and evidence of efficacy. Oncotarget 2017, 8, 89256–89268. [Google Scholar] [CrossRef] [PubMed]
- Rujkijyanont, P.; Chan, W.K.; Eldridge, P.W.; Lockey, T.; Holladay, M.; Rooney, B.; Davidoff, A.M.; Leung, W.; Vong, Q. Ex Vivo Activation of CD56+ Immune Cells That Eradicate Neuroblastoma. Cancer Res. 2013, 73, 2608–2618. [Google Scholar] [CrossRef] [PubMed]
- Otegbeye, F.; Cooper, B.; Caimi, P.; Zamborsky, K.; Reese-Koc, J.; Hillian, A.; Hernandez-Collazo, Y.; Lee, G.; Boughan, K.; Tomlinson, B.; et al. A Phase I Study to Determine the Maximum Tolerated Dose of Ex Vivo Expanded Natural Killer Cells Derived from Unrelated, HLA-Disparate Adult Donors. Transplant. Cell Ther. 2022, 28, 250.e1–250.e8. [Google Scholar] [CrossRef] [PubMed]
- Mansour, A.G.; Teng, K.-Y.; Li, Z.; Zhu, Z.; Chen, H.; Tian, L.; Ali, A.; Zhang, J.; Lu, T.; Ma, S.; et al. Off-the-shelf CAR–engineered natural killer cells targeting FLT3 enhance killing of acute myeloid leukemia. Blood Adv. 2023, 7, 6225–6239. [Google Scholar] [CrossRef] [PubMed]
- Caruso, S.; De Angelis, B.; Del Bufalo, F.; Ciccone, R.; Donsante, S.; Volpe, G.; Manni, S.; Guercio, M.; Pezzella, M.; Iaffaldano, L.; et al. Safe and effective off-the-shelf immunotherapy based on CAR.CD123-NK cells for the treatment of acute myeloid leukaemia. J. Hematol. Oncol. 2022, 15, 163. [Google Scholar] [CrossRef]
- Portillo, A.L.; Hogg, R.; Poznanski, S.M.; Rojas, E.A.; Cashell, N.J.; Hammill, J.A.; Chew, M.V.; Shenouda, M.M.; Ritchie, T.M.; Cao, Q.T.; et al. Expanded human NK cells armed with CAR uncouple potent anti-tumor activity from off-tumor toxicity against solid tumors. iScience 2021, 24, 102619. [Google Scholar] [CrossRef]
- Ruggeri, L.; Capanni, M.; Urbani, E.; Perruccio, K.; Shlomchik, W.D.; Tosti, A.; Posati, S.; Rogaia, D.; Frassoni, F.; Aversa, F.; et al. Effectiveness of Donor Natural Killer Cell Alloreactivity in Mismatched Hematopoietic Transplants. Science 2002, 295, 2097–2100. [Google Scholar] [CrossRef]
- Daher, M.; Garcia, L.M.; Li, Y.; Rezvani, K. CAR-NK cells: The next wave of cellular therapy for cancer. Clin. Transl. Immunol. 2021, 10, e1274. [Google Scholar] [CrossRef]
- Sojka, D.K.; Plougastel-Douglas, B.; Yang, L.; A Pak-Wittel, M.; Artyomov, M.N.; Ivanova, Y.; Zhong, C.; Chase, J.M.; Rothman, P.B.; Yu, J.; et al. Tissue-resident natural killer (NK) cells are cell lineages distinct from thymic and conventional splenic NK cells. Elife 2014, 3, e01659. [Google Scholar] [CrossRef] [PubMed]
- Marofi, F.; Rahman, H.S.; Thangavelu, L.; Dorofeev, A.; Bayas-Morejón, F.; Shirafkan, N.; Shomali, N.; Chartrand, M.S.; Jarahian, M.; Vahedi, G.; et al. Renaissance of armored immune effector cells, CAR-NK cells, brings the higher hope for successful cancer therapy. Stem Cell Res. Ther. 2021, 12, 200. [Google Scholar] [CrossRef] [PubMed]
- Morice, W.G. The Immunophenotypic Attributes of NK Cells and NK-Cell Lineage Lymphoproliferative Disorders. Am. J. Clin. Pathol. 2007, 127, 881–886. [Google Scholar] [CrossRef] [PubMed]
- Xie, G.; Dong, H.; Liang, Y.; Ham, J.D.; Rizwan, R.; Chen, J. CAR-NK cells: A promising cellular immunotherapy for cancer. EBioMedicine 2020, 59, 102975. [Google Scholar] [CrossRef] [PubMed]
- Lorenzo-Herrero, S.; López-Soto, A.; Sordo-Bahamonde, C.; Gonzalez-Rodriguez, A.P.; Vitale, M.; Gonzalez, S. NK Cell-Based Immunotherapy in Cancer Metastasis. Cancers 2018, 11, 29. [Google Scholar] [CrossRef] [PubMed]
- Poli, A.; Michel, T.; Thérésine, M.; Andrès, E.; Hentges, F.; Zimmer, J. CD56bright natural killer (NK) cells: An important NK cell subset. Immunology 2009, 126, 458–465. [Google Scholar] [CrossRef] [PubMed]
- Radaev, S.; Sun, P.D. Structure and Function of Natural Killer Cell Surface Receptors. Annu. Rev. Biophys. Biomol. Struct. 2003, 32, 93–114. [Google Scholar] [CrossRef]
- Rangarajan, S.; Mariuzza, R.A. Natural Killer Cell Receptors. In Structural Biology in Immunology; Elsevier: Amsterdam, The Netherlands, 2018; pp. 101–125. [Google Scholar] [CrossRef]
- Kruse, P.H.; Matta, J.; Ugolini, S.; Vivier, E. Natural cytotoxicity receptors and their ligands. Immunol. Cell Biol. 2014, 92, 221–229. [Google Scholar] [CrossRef]
- Lee, H.; Da Silva, I.P.; Palendira, U.; Scolyer, R.A.; Long, G.V.; Wilmott, J.S. Targeting NK Cells to Enhance Melanoma Response to Immunotherapies. Cancers 2021, 13, 1363. [Google Scholar] [CrossRef]
- Paul, S.; Lal, G. The Molecular Mechanism of Natural Killer Cells Function and Its Importance in Cancer Immunotherapy. Front. Immunol. 2017, 8, 1124. [Google Scholar] [CrossRef]
- Brandt, C.S.; Baratin, M.; Yi, E.C.; Kennedy, J.; Gao, Z.; Fox, B.; Haldeman, B.; Ostrander, C.D.; Kaifu, T.; Chabannon, C.; et al. The B7 family member B7-H6 is a tumor cell ligand for the activating natural killer cell receptor NKp30 in humans. J. Exp. Med. 2009, 206, 1495–1503. [Google Scholar] [CrossRef]
- Duan, S.; Guo, W.; Xu, Z.; He, Y.; Liang, C.; Mo, Y.; Wang, Y.; Xiong, F.; Guo, C.; Li, Y.; et al. Natural killer group 2D receptor and its ligands in cancer immune escape. Mol. Cancer 2019, 18, 29. [Google Scholar] [CrossRef] [PubMed]
- Cluxton, C.D.; Spillane, C.; O’Toole, S.A.; Sheils, O.; Gardiner, C.M.; O’Leary, J.J. Suppression of Natural Killer cell NKG2D and CD226 anti-tumour cascades by platelet cloaked cancer cells: Implications for the metastatic cascade. PLoS ONE 2019, 14, e0211538. [Google Scholar] [CrossRef] [PubMed]
- Tarazona, R.; Duran, E.; Solana, R. Natural Killer Cell Recognition of Melanoma: New Clues for a More Effective Immunotherapy. Front. Immunol. 2016, 6, 649. [Google Scholar] [CrossRef]
- Okumura, G.; Iguchi-Manaka, A.; Murata, R.; Yamashita-Kanemaru, Y.; Shibuya, A.; Shibuya, K. Tumor-derived soluble CD155 inhibits DNAM-1–mediated antitumor activity of natural killer cells. J. Exp. Med. 2020, 217, e20191290. [Google Scholar] [CrossRef]
- Bevelacqua, V.; Bevelacqua, Y.; Candido, S.; Skarmoutsou, E.; Amoroso, A.; Guarneri, C.; Strazzanti, A.; Gangemi, P.; Mazzarino, M.C.; D’amico, F.; et al. Nectin like-5 overexpression correlates with the malignant phenotype in cutaneous melanoma. Oncotarget 2012, 3, 882–892. [Google Scholar] [CrossRef]
- Lepletier, A.; Madore, J.; O’Donnell, J.S.; Johnston, R.L.; Li, X.Y.; McDonald, E.; Ahern, E.; Kuchel, A.; Eastgate, M.; Pearson, S.A.; et al. Tumor CD155 Expression Is Associated with Resistance to Anti-PD1 Immunotherapy in Metastatic Melanoma. Clin. Cancer Res. 2020, 26, 3671–3681. [Google Scholar] [CrossRef]
- Ivarsson, M.A.; Michaã«Lsson, J.; Fauriat, C. Activating killer cell Ig-like receptors in health and disease. Front. Immunol. 2014, 5, 184. [Google Scholar] [CrossRef]
- Chen, Y.; Lu, D.; Churov, A.; Fu, R. Research Progress on NK Cell Receptors and Their Signaling Pathways. Mediat. Inflamm. 2020, 2020, 6437057. [Google Scholar] [CrossRef]
- Tsukerman, P.; Stern-Ginossar, N.; Yamin, R.; Ophir, Y.; Stanietsky, A.M.N.; Mandelboim, O. Expansion of CD16 positive and negative human NK cells in response to tumor stimulation. Eur. J. Immunol. 2014, 44, 1517–1525. [Google Scholar] [CrossRef]
- Lee, H.R.; Son, C.H.; Koh, E.K.; Bae, J.H.; Kang, C.D.; Yang, K.; Park, Y.S. Expansion of cytotoxic natural killer cells using irradiated autologous peripheral blood mononuclear cells and anti-CD16 antibody. Sci. Rep. 2017, 7, 11075. [Google Scholar] [CrossRef] [PubMed]
- Romee, R.; Foley, B.; Lenvik, T.; Wang, Y.; Zhang, B.; Ankarlo, D.; Luo, X.; Cooley, S.; Verneris, M.; Walcheck, B.; et al. NK cell CD16 surface expression and function is regulated by a disintegrin and metalloprotease-17 (ADAM17). Blood 2013, 121, 3599–3608. [Google Scholar] [CrossRef] [PubMed]
- Rajagopalan, S.; Fu, J.; Long, E.O. Cutting Edge: Induction of IFN-γ Production but Not Cytotoxicity by the Killer Cell Ig-Like Receptor KIR2DL4 (CD158d) in Resting NK Cells. J. Immunol. 2001, 167, 1877–1881. [Google Scholar] [CrossRef] [PubMed]
- Kandilarova, S.M.; Paschen, A.; Mihaylova, A.; Ivanova, M.; Schadendorf, D.; Naumova, E. The Influence of HLA and KIR Genes on Malignant Melanoma Development and Progression. Arch. Immunol. Ther. Exp. 2016, 64 (Suppl. S1), 73–81. [Google Scholar] [CrossRef] [PubMed]
- Campillo, J.A.; Legaz, I.; López-Álvarez, M.R.; Bolarín, J.M.; Heras, B.L.; Muro, M.; Minguela, A.; Moya-Quiles, M.R.; Blanco-García, R.; Martínez-Banaclocha, H.; et al. KIR gene variability in cutaneous malignant melanoma: Influence of KIR2D/HLA-C pairings on disease susceptibility and prognosis. Immunogenetics 2013, 65, 333–343. [Google Scholar] [CrossRef] [PubMed]
- Moradi, S.; Stankovic, S.; O’connor, G.M.; Pymm, P.; MacLachlan, B.J.; Faoro, C.; Retière, C.; Sullivan, L.C.; Saunders, P.M.; Widjaja, J.; et al. Structural plasticity of KIR2DL2 and KIR2DL3 enables altered docking geometries atop HLA-C. Nat. Commun. 2021, 12, 2173. [Google Scholar] [CrossRef] [PubMed]
- Frazier, W.R.; Steiner, N.; Hou, L.; Dakshanamurthy, S.; Hurley, C.K. Allelic Variation in KIR2DL3 Generates a KIR2DL2-like Receptor with Increased Binding to its HLA-C Ligand. J. Immunol. 2013, 190, 6198–6208. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, S.; Beziat, V.; Dhedin, N.; Kuentz, M.; Vernant, J.P.; Debre, P.; Vieillard, V. HLA-E upregulation on IFN-γ-activated AML blasts impairs CD94/NKG2A-dependent NK cytolysis after haplo-mismatched hematopoietic SCT. Bone Marrow Transplant. 2009, 43, 693–699. [Google Scholar] [CrossRef]
- André, P.; Denis, C.; Soulas, C.; Bourbon-Caillet, C.; Lopez, J.; Arnoux, T.; Bléry, M.; Bonnafous, C.; Gauthier, L.; Morel, A.; et al. Anti-NKG2A mAb Is a Checkpoint Inhibitor that Promotes Anti-Tumor Immunity by Unleashing Both T and NK Cells. Cell 2018, 175, 1731–1743.e13. [Google Scholar] [CrossRef]
- Kamiya, T.; Seow, S.V.; Wong, D.; Robinson, M.; Campana, D. Blocking expression of inhibitory receptor NKG2A overcomes tumor resistance to NK cells. J. Clin. Investig. 2019, 129, 2094–2106. [Google Scholar] [CrossRef]
- Chauvin, J.M.; Pagliano, O.; Fourcade, J.; Sun, Z.; Wang, H.; Sander, C.; Kirkwood, J.M.; Chen, T.H.T.; Maurer, M.; Korman, A.J.; et al. TIGIT and PD-1 impair tumor antigen–specific CD8+ T cells in melanoma patients. J. Clin. Investig. 2015, 125, 2046–2058. [Google Scholar] [CrossRef] [PubMed]
- Inozume, T.; Yaguchi, T.; Furuta, J.; Harada, K.; Kawakami, Y.; Shimada, S. Melanoma Cells Control Antimelanoma CTL Responses via Interaction between TIGIT and CD155 in the Effector Phase. J. Investig. Dermatol. 2016, 136, 255–263. [Google Scholar] [CrossRef]
- Prager, I.; Watzl, C. Mechanisms of natural killer cell-mediated cellular cytotoxicity. J. Leukoc. Biol. 2019, 105, 1319–1329. [Google Scholar] [CrossRef] [PubMed]
- Elahi, R.; Heidary, A.H.; Hadiloo, K.; Esmaeilzadeh, A. Chimeric Antigen Receptor-Engineered Natural Killer (CAR NK) Cells in Cancer Treatment; Recent Advances and Future Prospects. Stem Cell Rev. Rep. 2021, 17, 2081–2106. [Google Scholar] [CrossRef]
- Chan, C.J.; Smyth, M.J.; Martinet, L. Molecular mechanisms of natural killer cell activation in response to cellular stress. Cell Death Differ. 2014, 21, 5–14. [Google Scholar] [CrossRef] [PubMed]
- Du, N.; Guo, F.; Wang, Y.; Cui, J. NK Cell Therapy: A Rising Star in Cancer Treatment. Cancers 2021, 13, 4129. [Google Scholar] [CrossRef]
- Cursons, J.; Souza-Fonseca-Guimaraes, F.; Foroutan, M.; Anderson, A.; Hollande, F.; Hediyeh-Zadeh, S.; Behren, A.; Huntington, N.D.; Davis, M.J. A Gene Signature Predicting Natural Killer Cell Infiltration and Improved Survival in Melanoma Patients. Cancer Immunol. Res. 2019, 7, 1162–1174. [Google Scholar] [CrossRef]
- Ali, T.H.; Pisanti, S.; Ciaglia, E.; Mortarini, R.; Anichini, A.; Garofalo, C.; Tallerico, R.; Santinami, M.; Gulletta, E.; Ietto, C.; et al. Enrichment of CD56dimKIR+CD57+ highly cytotoxic NK cells in tumour-infiltrated lymph nodes of melanoma patients. Nat. Commun. 2014, 5, 5639. [Google Scholar] [CrossRef]
- de Jonge, K.; Ebering, A.; Nassiri, S.; Hajjami, H.M.-E.; Ouertatani-Sakouhi, H.; Baumgaertner, P.; Speiser, D.E. Circulating CD56bright NK cells inversely correlate with survival of melanoma patients. Sci. Rep. 2019, 9, 4487. [Google Scholar] [CrossRef]
- Attrill, G.H.; Ferguson, P.M.; Palendira, U.; Long, G.V.; Wilmott, J.S.; Scolyer, R.A. The tumour immune landscape and its implications in cutaneous melanoma. Pigment. Cell Melanoma Res. 2021, 34, 529–549. [Google Scholar] [CrossRef]
- Vitale, M.; Cantoni, C.; Pietra, G.; Mingari, M.C.; Moretta, L. Effect of tumor cells and tumor microenvironment on NK-cell function. Eur. J. Immunol. 2014, 44, 1582–1592. [Google Scholar] [CrossRef] [PubMed]
- Falcone, I.; Conciatori, F.; Bazzichetto, C.; Ferretti, G.; Cognetti, F.; Ciuffreda, L.; Milella, M. Tumor Microenvironment: Implications in Melanoma Resistance to Targeted Therapy and Immunotherapy. Cancers 2020, 12, 2870. [Google Scholar] [CrossRef] [PubMed]
- Amalinei, C.; Grigoraș, A.; Lozneanu, L.; Căruntu, I.D.; Giușcă, S.E.; Balan, R.A. The Interplay between Tumour Microenvironment Components in Malignant Melanoma. Medicina 2022, 58, 365. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.-H.; Roszik, J.; Cho, S.-N.; Ogata, D.; Milton, D.R.; Peng, W.; Menter, D.G.; Ekmekcioglu, S.; Grimm, E.A. The COX2 Effector Microsomal PGE2 Synthase 1 is a Regulator of Immunosuppression in Cutaneous Melanoma. Clin. Cancer Res. 2019, 25, 1650–1663. [Google Scholar] [CrossRef] [PubMed]
- Holt, D.; Ma, X.; Kundu, N.; Fulton, A. Prostaglandin E2 (PGE2) suppresses natural killer cell function primarily through the PGE2 receptor EP4. Cancer Immunol. Immunother. 2011, 60, 1577–1586. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, M.; Krykbaeva, I.; Damsky, W.; Kluger, H.M.; Bosenberg, M. Evaluating the role of the COX2/PGE2 pathway in anti-melanoma immunity. J. Clin. Oncol. 2019, 37 (Suppl. S14), e14114. [Google Scholar] [CrossRef]
- Simiczyjew, A.; Dratkiewicz, E.; Mazurkiewicz, J.; Ziętek, M.; Matkowski, R.; Nowak, D. The Influence of Tumor Microenvironment on Immune Escape of Melanoma. Int. J. Mol. Sci. 2020, 21, 8359. [Google Scholar] [CrossRef] [PubMed]
- Hornyák, L.; Dobos, N.; Koncz, G.; Karányi, Z.; Páll, D.; Szabó, Z.; Halmos, G.; Székvölgyi, L. The Role of Indoleamine-2,3-Dioxygenase in Cancer Development, Diagnostics, and Therapy. Front. Immunol. 2018, 9, 151. [Google Scholar] [CrossRef]
- Zhang, J.; Han, X.; Hu, X.; Jin, F.; Gao, Z.; Yin, L.; Qin, J.; Yin, F.; Li, C.; Wang, Y. IDO1 impairs NK cell cytotoxicity by decreasing NKG2D/NKG2DLs via promoting miR-18a. Mol. Immunol. 2018, 103, 144–155. [Google Scholar] [CrossRef]
- Pietra, G.; Manzini, C.; Rivara, S.; Vitale, M.; Cantoni, C.; Petretto, A.; Balsamo, M.; Conte, R.; Benelli, R.; Minghelli, S.; et al. Melanoma Cells Inhibit Natural Killer Cell Function by Modulating the Expression of Activating Receptors and Cytolytic Activity. Cancer Res. 2012, 72, 1407–1415. [Google Scholar] [CrossRef]
- López-Cobo, S.; Pieper, N.; Campos-Silva, C.; García-Cuesta, E.M.; Reyburn, H.T.; Paschen, A.; Valés-Gómez, M. Impaired NK cell recognition of vemurafenib-treated melanoma cells is overcome by simultaneous application of histone deacetylase inhibitors. Oncoimmunology 2018, 7, e1392426. [Google Scholar] [CrossRef] [PubMed]
- Peruzzi, G.; Femnou, L.; Gil-Krzewska, A.; Borrego, F.; Weck, J.; Krzewski, K.; Coligan, J.E. Membrane-Type 6 Matrix Metalloproteinase Regulates the Activation-Induced Downmodulation of CD16 in Human Primary NK Cells. J. Immunol. 2013, 191, 1883–1894. [Google Scholar] [CrossRef]
- Liu, G.; Atteridge, C.L.; Wang, X.; Lundgren, A.D.; Wu, J.D. Cutting Edge: The Membrane Type Matrix Metalloproteinase MMP14 Mediates Constitutive Shedding of MHC Class I Chain-Related Molecule a Independent of a Disintegrin and Metalloproteinases. J. Immunol. 2010, 184, 3346–3350. [Google Scholar] [CrossRef] [PubMed]
- Chitadze, G.; Lettau, M.; Bhat, J.; Wesch, D.; Steinle, A.; Fürst, D.; Mytilineos, J.; Kalthoff, H.; Janssen, O.; Oberg, H.; et al. Shedding of endogenous MHC class I-related chain molecules A and B from different human tumor entities: Heterogeneous involvement of the “a disintegrin and metalloproteases” 10 and 17. Int. J. Cancer 2013, 133, 1557–1566. [Google Scholar] [CrossRef]
- Schlecker, E.; Fiegler, N.; Arnold, A.; Altevogt, P.; Rose-John, S.; Moldenhauer, G.; Sucker, A.; Paschen, A.; von Strandmann, E.P.; Textor, S.; et al. Metalloprotease-Mediated Tumor Cell Shedding of B7-H6, the Ligand of the Natural Killer Cell–Activating Receptor NKp30. Cancer Res. 2014, 74, 3429–3440. [Google Scholar] [CrossRef] [PubMed]
- Deng, W.; Gowen, B.G.; Zhang, L.; Wang, L.; Lau, S.; Iannello, A.; Xu, J.; Rovis, T.L.; Xiong, N.; Raulet, D.H. A shed NKG2D ligand that promotes natural killer cell activation and tumor rejection. Science 2015, 348, 136–139. [Google Scholar] [CrossRef]
- Zhang, J.; Basher, F.; Wu, J.D. NKG2D Ligands in Tumor Immunity: Two Sides of a Coin. Front. Immunol. 2015, 6, 97. [Google Scholar] [CrossRef]
- Ichise, H.; Tsukamoto, S.; Hirashima, T.; Konishi, Y.; Oki, C.; Tsukiji, S.; Iwano, S.; Miyawaki, A.; Sumiyama, K.; Terai, K.; et al. Functional visualization of NK cell-mediated killing of metastatic single tumor cells. Elife 2022, 11, e76269. [Google Scholar] [CrossRef]
- de Andrade, L.F.; Kumar, S.; Luoma, A.M.; Ito, Y.; da Silva, P.H.A.; Pan, D.; Pyrdol, J.W.; Yoon, C.H.; Wucherpfennig, K.W. Inhibition of MICA and MICB Shedding Elicits NK-Cell–Mediated Immunity against Tumors Resistant to Cytotoxic T Cells. Cancer Immunol. Res. 2020, 8, 769–780. [Google Scholar] [CrossRef]
- de Andrade, L.F.; Tay, R.E.; Pan, D.; Luoma, A.M.; Ito, Y.; Badrinath, S.; Tsoucas, D.; Franz, B.; May, K.F., Jr.; Harvey, C.J.; et al. Antibody-mediated inhibition of MICA and MICB shedding promotes NK cell–driven tumor immunity. Science 2018, 359, 1537–1542. [Google Scholar] [CrossRef]
- Wang, X.; Xing, Z.; Chen, H.; Yang, H.; Wang, Q.; Xing, T. High expression of nectin-1 indicates a poor prognosis and promotes metastasis in hepatocellular carcinoma. Front. Oncol. 2022, 12, 953529. [Google Scholar] [CrossRef] [PubMed]
- Deuss, F.A.; Gully, B.S.; Rossjohn, J.; Berry, R. Recognition of nectin-2 by the natural killer cell receptor T cell immunoglobulin and ITIM domain (TIGIT). J. Biol. Chem. 2017, 292, 11413–11422. [Google Scholar] [CrossRef] [PubMed]
- Stanietsky, N.; Simic, H.; Arapovic, J.; Toporik, A.; Levy, O.; Novik, A.; Levine, Z.; Beiman, M.; Dassa, L.; Achdout, H.; et al. The interaction of TIGIT with PVR and PVRL2 inhibits human NK cell cytotoxicity. Proc. Natl. Acad. Sci. USA 2009, 106, 17858–17863. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Xia, P.; Du, Y.; Liu, S.; Huang, G.; Chen, J.; Zhang, H.; Hou, N.; Cheng, X.; Zhou, L.; et al. T-cell Immunoglobulin and ITIM Domain (TIGIT) Receptor/Poliovirus Receptor (PVR) Ligand Engagement Suppresses Interferon-γ Production of Natural Killer Cells via β-Arrestin 2-mediated Negative Signaling. J. Biol. Chem. 2014, 289, 17647–17657. [Google Scholar] [CrossRef] [PubMed]
- Siva, A.; Xin, H.; Qin, F.; Oltean, D.; Bowdish, K.S.; Kretz-Rommel, A. Immune modulation by melanoma and ovarian tumor cells through expression of the immunosuppressive molecule CD200. Cancer Immunol. Immunother. 2008, 57, 987–996. [Google Scholar] [CrossRef] [PubMed]
- Saputro, R.D.; Rinonce, H.T.; Iramawasita, Y.; Ridho, M.R.; Pudjohartono, M.F.; Anwar, S.L.; Setiaji, K.; Aryandono, T. Potential prognostic value of PD-L1 and NKG2A expression in Indonesian patients with skin nodular melanoma. BMC Res. Notes 2021, 14, 206. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Shklovskaya, E.; Lim, S.Y.; Carlino, M.S.; Menzies, A.M.; Stewart, A.; Pedersen, B.; Irvine, M.; Alavi, S.; Yang, J.Y.H.; et al. Transcriptional downregulation of MHC class I and melanoma de-differentiation in resistance to PD-1 inhibition. Nat. Commun. 2020, 11, 1897. [Google Scholar] [CrossRef]
- Soulas, C.; Remark, R.; Brezar, V.; Lopez, J.; Bonnet, E.; Caraguel, F.; Lalanne, A.; Hoffmann, C.; Denis, C.; Arnoux, T.; et al. Abstract 2714: Combination of monalizumab and durvalumab as a potent immunotherapy treatment for solid human cancers. Cancer Res. 2018, 78 (Suppl. S13). [Google Scholar] [CrossRef]
- Carosella, E.D.; Rouas-Freiss, N.; Roux, D.T.L.; Moreau, P.; LeMaoult, J. HLA-G: An Immune Checkpoint Molecule. Adv. Immunol. 2015, 127, 33–144. [Google Scholar] [CrossRef]
- Lin, A.; Yan, W.H. Heterogeneity of HLA-G Expression in Cancers: Facing the Challenges. Front. Immunol. 2018, 9, 2164. [Google Scholar] [CrossRef]
- Fisher, J.G.; Doyle, A.D.P.; Graham, L.V.; Khakoo, S.I.; Blunt, M.D. Disruption of the NKG2A:HLA-E Immune Checkpoint Axis to Enhance NK Cell Activation against Cancer. Vaccines 2022, 10, 1993. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Song, J.; Zhang, H.; Liu, X.; Zuo, F.; Zhao, Y.; Zhao, Y.; Yin, X.; Guo, X.; Wu, X.; et al. Immune checkpoint HLA-E:CD94-NKG2A mediates evasion of circulating tumor cells from NK cell surveillance. Cancer Cell 2023, 41, 272–287.e9. [Google Scholar] [CrossRef] [PubMed]
- Fourcade, J.; Sun, Z.; Benallaoua, M.; Guillaume, P.; Luescher, I.F.; Sander, C.; Kirkwood, J.M.; Kuchroo, V.; Zarour, H.M. Upregulation of Tim-3 and PD-1 expression is associated with tumor antigen–specific CD8+ T cell dysfunction in melanoma patients. J. Exp. Med. 2010, 207, 2175–2186. [Google Scholar] [CrossRef]
- da Silva, I.P.; Gallois, A.; Jimenez-Baranda, S.; Khan, S.; Anderson, A.C.; Kuchroo, V.K.; Osman, I.; Bhardwaj, N. Reversal of NK-Cell Exhaustion in Advanced Melanoma by Tim-3 Blockade. Cancer Immunol. Res. 2014, 2, 410–422. [Google Scholar] [CrossRef] [PubMed]
- Whiteside, T.L. Exosomes and tumor-mediated immune suppression. J. Clin. Investig. 2016, 126, 1216–1223. [Google Scholar] [CrossRef]
- Sharma, P.; Diergaarde, B.; Ferrone, S.; Kirkwood, J.M.; Whiteside, T.L. Melanoma cell-derived exosomes in plasma of melanoma patients suppress functions of immune effector cells. Sci. Rep. 2020, 10, 92. [Google Scholar] [CrossRef] [PubMed]
- Papaccio, F.; Kovacs, D.; Bellei, B.; Caputo, S.; Migliano, E.; Cota, C.; Picardo, M. Profiling Cancer-Associated Fibroblasts in Melanoma. Int. J. Mol. Sci. 2021, 22, 7255. [Google Scholar] [CrossRef]
- Mao, X.; Xu, J.; Wang, W.; Liang, C.; Hua, J.; Liu, J.; Zhang, B.; Meng, Q.; Yu, X.; Shi, S. Crosstalk between cancer-associated fibroblasts and immune cells in the tumor microenvironment: New findings and future perspectives. Mol. Cancer 2021, 20, 131. [Google Scholar] [CrossRef]
- Harper, J.; Sainson, R.C.A. Regulation of the anti-tumour immune response by cancer-associated fibroblasts. Semin. Cancer Biol. 2014, 25, 69–77. [Google Scholar] [CrossRef]
- Ziani, L.; Ben Safta-Saadoun, T.; Gourbeix, J.; Cavalcanti, A.; Robert, C.; Favre, G.; Chouaib, S.; Thiery, J. Melanoma-associated fibroblasts decrease tumor cell susceptibility to NK cell-mediated killing through matrix-metalloproteinases secretion. Oncotarget 2017, 8, 19780–19794. [Google Scholar] [CrossRef]
- Du, Y.; Fang, Q.; Zheng, S.G. Regulatory T Cells: Concept, Classification, Phenotype, and Biological Characteristics. In T Regulatory Cells in Human Health and Diseases; Springer: Berlin/Heidelberg, Germany, 2021; pp. 1–31. [Google Scholar] [CrossRef]
- Allard, B.; Longhi, M.S.; Robson, S.C.; Stagg, J. The ectonucleotidases CD39 and CD73: Novel checkpoint inhibitor targets. Immunol. Rev. 2017, 276, 121–144. [Google Scholar] [CrossRef] [PubMed]
- Cekic, C.; Day, Y.-J.; Sag, D.; Linden, J. Myeloid Expression of Adenosine A2A Receptor Suppresses T and NK Cell Responses in the Solid Tumor Microenvironment. Cancer Res. 2014, 74, 7250–7259. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Yang, L.; Wang, D.; Zhang, Q.; Zhang, L. Pro-tumor activities of macrophages in the progression of melanoma. Hum. Vaccin. Immunother. 2017, 13, 1556–1562. [Google Scholar] [CrossRef] [PubMed]
- Bohn, T.; Rapp, S.; Luther, N.; Klein, M.; Bruehl, T.J.; Kojima, N.; Lopez, P.A.; Hahlbrock, J.; Muth, S.; Endo, S.; et al. Tumor immunoevasion via acidosis-dependent induction of regulatory tumor-associated macrophages. Nat. Immunol. 2018, 19, 1319–1329. [Google Scholar] [CrossRef] [PubMed]
- Chanmee, T.; Ontong, P.; Konno, K.; Itano, N. Tumor-Associated Macrophages as Major Players in the Tumor Microenvironment. Cancers 2014, 6, 1670–1690. [Google Scholar] [CrossRef]
- Rébé, C.; Ghiringhelli, F. Interleukin-1β and Cancer. Cancers 2020, 12, 1791. [Google Scholar] [CrossRef]
- Smith, M.P.; Sanchez-Laorden, B.; O’Brien, K.; Brunton, H.; Ferguson, J.; Young, H.; Dhomen, N.; Flaherty, K.T.; Frederick, D.T.; Cooper, Z.A.; et al. The Immune Microenvironment Confers Resistance to MAPK Pathway Inhibitors through Macrophage-Derived TNFα. Cancer Discov. 2014, 4, 1214–1229. [Google Scholar] [CrossRef]
- Érsek, B.; Silló, P.; Cakir, U.; Molnár, V.; Bencsik, A.; Mayer, B.; Mezey, E.; Kárpáti, S.; Pós, Z.; Németh, K. Melanoma-associated fibroblasts impair CD8+ T cell function and modify expression of immune checkpoint regulators via increased arginase activity. Cell. Mol. Life Sci. 2021, 78, 661–673. [Google Scholar] [CrossRef]
- Brownlie, D.; Doughty-Shenton, D.; Soong, D.Y.; Nixon, C.; O Carragher, N.; Carlin, L.M.; Kitamura, T. Metastasis-associated macrophages constrain antitumor capability of natural killer cells in the metastatic site at least partially by membrane bound transforming growth factor β. J. Immunother. Cancer 2021, 9, e001740. [Google Scholar] [CrossRef]
- D’aguanno, S.; Mallone, F.; Marenco, M.; Del Bufalo, D.; Moramarco, A. Hypoxia-dependent drivers of melanoma progression. J. Exp. Clin. Cancer Res. 2021, 40, 159. [Google Scholar] [CrossRef]
- Böhme, I.; Bosserhoff, A.K. Acidic tumor microenvironment in human melanoma. Pigment. Cell Melanoma Res. 2016, 29, 508–523. [Google Scholar] [CrossRef] [PubMed]
- Muz, B.; de la Puente, P.; Azab, F.; Azab, A.K. The role of hypoxia in cancer progression, angiogenesis, metastasis, and resistance to therapy. Hypoxia 2015, 3, 83–92. [Google Scholar] [CrossRef] [PubMed]
- Terrén, I.; Orrantia, A.; Vitallé, J.; Zenarruzabeitia, O.; Borrego, F. NK Cell Metabolism and Tumor Microenvironment. Front. Immunol. 2019, 10, 2278. [Google Scholar] [CrossRef] [PubMed]
- Garcés-Lázaro, I.; Kotzur, R.; Cerwenka, A.; Mandelboim, O. NK Cells Under Hypoxia: The Two Faces of Vascularization in Tumor and Pregnancy. Front. Immunol. 2022, 13, 924775. [Google Scholar] [CrossRef]
- Lu, H.; Zhao, X.; Li, Z.; Hu, Y.; Wang, H. From CAR-T Cells to CAR-NK Cells: A Developing Immunotherapy Method for Hematological Malignancies. Front. Oncol. 2021, 11, 720501. [Google Scholar] [CrossRef] [PubMed]
- Guedan, S.; Calderon, H.; Posey, A.D., Jr.; Maus, M.V. Engineering and Design of Chimeric Antigen Receptors. Mol. Ther. Methods Clin. Dev. 2018, 12, 145–156. [Google Scholar] [CrossRef] [PubMed]
- Guedan, S.; Posey, A.D., Jr.; Shaw, C.; Wing, A.; Da, T.; Patel, P.R.; McGettigan, S.E.; Casado-Medrano, V.; Kawalekar, O.U.; Uribe-Herranz, M.; et al. Enhancing CAR T cell persistence through ICOS and 4-1BB costimulation. JCI Insight 2018, 3, e96976. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.Y.; Roybal, K.T.; Puchner, E.M.; Onuffer, J.; Lim, W.A. Remote control of therapeutic T cells through a small molecule–gated chimeric receptor. Science 2015, 350, aab4077. [Google Scholar] [CrossRef]
- Christodoulou, I.; Ho, W.J.; Marple, A.; Ravich, J.W.; Tam, A.; Rahnama, R.; Fearnow, A.; Rietberg, C.; Yanik, S.; E Solomou, E.; et al. Engineering CAR-NK cells to secrete IL-15 sustains their anti-AML functionality but is associated with systemic toxicities. J. Immunother. Cancer 2021, 9, e003894. [Google Scholar] [CrossRef]
- Wrona, E.; Borowiec, M.; Potemski, P. CAR-NK Cells in the Treatment of Solid Tumors. Int. J. Mol. Sci. 2021, 22, 5899. [Google Scholar] [CrossRef]
- Zhang, Q.; Tian, K.; Xu, J.; Zhang, H.; Li, L.; Fu, Q.; Chai, D.; Li, H.; Zheng, J. Synergistic Effects of Cabozantinib and EGFR-Specific CAR-NK-92 Cells in Renal Cell Carcinoma. J. Immunol. Res. 2017, 2017, 6915912. [Google Scholar] [CrossRef]
- Gossel, L.D.H.; Heim, C.; Pfeffermann, L.-M.; Moser, L.M.; Bönig, H.B.; Klingebiel, T.E.; Bader, P.; Wels, W.S.; Merker, M.; Rettinger, E. Retargeting of NK-92 Cells against High-Risk Rhabdomyosarcomas by Means of an ERBB2 (HER2/Neu)-Specific Chimeric Antigen Receptor. Cancers 2021, 13, 1443. [Google Scholar] [CrossRef] [PubMed]
- Navarrete-Galvan, L.; Guglielmo, M.; Amaya, J.C.; Smith-Gagen, J.; Lombardi, V.C.; Merica, R.; Hudig, D. Optimizing NK-92 serial killers: Gamma irradiation, CD95/Fas-ligation, and NK or LAK attack limit cytotoxic efficacy. J. Transl. Med. 2022, 20, 151. [Google Scholar] [CrossRef] [PubMed]
- Klingemann, H.; Boissel, L.; Toneguzzo, F. Natural Killer Cells for Immunotherapy—Advantages of the NK-92 Cell Line over Blood NK Cells. Front. Immunol. 2016, 7, 91. [Google Scholar] [CrossRef]
- Maddineni, S.; Silberstein, J.L.; Sunwoo, J.B. Emerging NK cell therapies for cancer and the promise of next generation engineering of iPSC-derived NK cells. J. Immunother. Cancer 2022, 10, e004693. [Google Scholar] [CrossRef] [PubMed]
- Guercio, M.; Manni, S.; Boffa, I.; Caruso, S.; Di Cecca, S.; Sinibaldi, M.; Abbaszadeh, Z.; Camera, A.; Ciccone, R.; Polito, V.A.; et al. Inclusion of the Inducible Caspase 9 Suicide Gene in CAR Construct Increases Safety of CAR.CD19 T Cell Therapy in B-Cell Malignancies. Front. Immunol. 2021, 12, 755639. [Google Scholar] [CrossRef] [PubMed]
- Mehta, R.S.; Shpall, E.J.; Rezvani, K. Cord Blood as a Source of Natural Killer Cells. Front. Med. 2016, 2, 93. [Google Scholar] [CrossRef]
- Shaim, H.; Yvon, E. Cord blood: A promising source of allogeneic natural killer cells for immunotherapy. Cytotherapy 2015, 17, 1–2. [Google Scholar] [CrossRef]
- Liu, E.; Tong, Y.; Dotti, G.; Shaim, H.; Savoldo, B.; Mukherjee, M.; Orange, J.; Wan, X.; Lu, X.; Reynolds, A.; et al. Cord blood NK cells engineered to express IL-15 and a CD19-targeted CAR show long-term persistence and potent antitumor activity. Leukemia 2018, 32, 520–531. [Google Scholar] [CrossRef]
- Li, Y.; Yin, J.; Li, T.; Huang, S.; Yan, H.; Leavenworth, J.; Wang, X. NK cell-based cancer immunotherapy: From basic biology to clinical application. Sci. China Life Sci. 2015, 58, 1233–1245. [Google Scholar] [CrossRef]
- Xiao, L.; Cen, D.; Gan, H.; Sun, Y.; Huang, N.; Xiong, H.; Jin, Q.; Su, L.; Liu, X.; Wang, K.; et al. Adoptive Transfer of NKG2D CAR mRNA-Engineered Natural Killer Cells in Colorectal Cancer Patients. Mol. Ther. 2019, 27, 1114–1125. [Google Scholar] [CrossRef] [PubMed]
- Focaccetti, C.; Benvenuto, M.; Pighi, C.; Vitelli, A.; Napolitano, F.; Cotugno, N.; Fruci, D.; Palma, P.; Rossi, P.; Bei, R.; et al. DNAM-1-chimeric receptor-engineered NK cells, combined with Nutlin-3a, more effectively fight neuroblastoma cells in vitro: A proof-of-concept study. Front. Immunol. 2022, 13, 886319. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Zeng, J.; Liu, T.; Xu, Q.; Song, X.; Zeng, J. DNAM1 and 2B4 Costimulatory Domains Enhance the Cytotoxicity of Anti-GPC3 Chimeric Antigen Receptor-Modified Natural Killer Cells Against Hepatocellular Cancer Cells in vitro. Cancer Manag. Res. 2020, 12, 3247–3255. [Google Scholar] [CrossRef] [PubMed]
- Cao, B.; Liu, M.; Huang, J.; Zhou, J.; Li, J.; Lian, H.; Huang, W.; Guo, Y.; Yang, S.; Lin, L.; et al. Development of mesothelin-specific CAR NK-92 cells for the treatment of gastric cancer. Int. J. Biol. Sci. 2021, 17, 3850–3861. [Google Scholar] [CrossRef]
- Liu, M.; Huang, W.; Guo, Y.; Zhou, Y.; Zhi, C.; Chen, J.; Li, J.; He, J.; Lian, H.; Zhou, J.; et al. CAR NK-92 cells targeting DLL3 kill effectively small cell lung cancer cells in vitro and in vivo. J. Leukoc. Biol. 2022, 112, 901–911. [Google Scholar] [CrossRef] [PubMed]
- Peng, Y.; Zhang, W.; Chen, Y.; Zhang, L.; Shen, H.; Wang, Z.; Tian, S.; Yang, X.; Cui, D.; He, Y.; et al. Engineering c-Met-CAR NK-92 cells as a promising therapeutic candidate for lung adenocarcinoma. Pharmacol. Res. 2023, 188, 106656. [Google Scholar] [CrossRef]
- Yang, S.; Cao, B.; Zhou, G.; Zhu, L.; Wang, L.; Zhang, L.; Kwok, H.F.; Zhang, Z.; Zhao, Q. Targeting B7-H3 Immune Checkpoint with Chimeric Antigen Receptor-Engineered Natural Killer Cells Exhibits Potent Cytotoxicity against Non-Small Cell Lung Cancer. Front. Pharmacol. 2020, 11, 1089. [Google Scholar] [CrossRef]
- Pekar, L.; Klausz, K.; Busch, M.; Valldorf, B.; Kolmar, H.; Wesch, D.; Oberg, H.-H.; Krohn, S.; Boje, A.S.; Gehlert, C.L.; et al. Affinity Maturation of B7-H6 Translates into Enhanced NK Cell–Mediated Tumor Cell Lysis and Improved Proinflammatory Cytokine Release of Bispecific Immunoligands via NKp30 Engagement. J. Immunol. 2021, 206, 225–236. [Google Scholar] [CrossRef]
- Klapdor, R.; Wang, S.; Morgan, M.; Dörk, T.; Hacker, U.; Hillemanns, P.; Büning, H.; Schambach, A. Characterization of a Novel Third-Generation Anti-CD24-CAR against Ovarian Cancer. Int. J. Mol. Sci. 2019, 20, 660. [Google Scholar] [CrossRef]
- Grote, S.; Ureña-Bailén, G.; Chan, K.C.-H.; Baden, C.; Mezger, M.; Handgretinger, R.; Schleicher, S. In Vitro Evaluation of CD276-CAR NK-92 Functionality, Migration and Invasion Potential in the Presence of Immune Inhibitory Factors of the Tumor Microenvironment. Cells 2021, 10, 1020. [Google Scholar] [CrossRef]
- Klapdor, R.; Wang, S.; Morgan, M.A.; Zimmermann, K.; Hachenberg, J.; Büning, H.; Dörk, T.; Hillemanns, P.; Schambach, A. NK Cell-Mediated Eradication of Ovarian Cancer Cells with a Novel Chimeric Antigen Receptor Directed against CD44. Biomedicines 2021, 9, 1339. [Google Scholar] [CrossRef]
- Zhang, Z.; He, B. CLDN6-CAR-NK Cell Therapy for Advanced Solid Tumors. Available online: https://classic.clinicaltrials.gov/ct2/show/NCT05410717 (accessed on 9 October 2023).
- Murakami, T.; Nakazawa, T.; Natsume, A.; Nishimura, F.; Nakamura, M.; Matsuda, R.; Omoto, K.; Tanaka, Y.; Shida, Y.; Park, Y.-S.; et al. Novel Human NK Cell Line Carrying CAR Targeting EGFRvIII Induces Antitumor Effects in Glioblastoma Cells. Anticancer Res. 2018, 38, 5049–5056. [Google Scholar] [CrossRef] [PubMed]
- Müller, N.; Michen, S.; Tietze, S.; Töpfer, K.; Schulte, A.; Lamszus, K.; Schmitz, M.; Schackert, G.; Pastan, I.; Temme, A. Engineering NK Cells Modified with an EGFRvIII-specific Chimeric Antigen Receptor to Overexpress CXCR4 Improves Immunotherapy of CXCL12/SDF-1α-secreting Glioblastoma. J. Immunother. 2015, 38, 197–210. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Zhou, Y.; Huang, K.; Fang, X.; Li, Y.; Wang, F.; An, L.; Chen, Q.; Zhang, Y.; Shi, A.; et al. Targeting epidermal growth factor-overexpressing triple-negative breast cancer by natural killer cells expressing a specific chimeric antigen receptor. Cell Prolif. 2020, 53, e12858. [Google Scholar] [CrossRef] [PubMed]
- Han, J.; Chu, J.; Chan, W.K.; Zhang, J.; Wang, Y.; Cohen, J.B.; Victor, A.; Meisen, W.H.; Kim, S.-H.; Grandi, P.; et al. CAR-Engineered NK Cells Targeting Wild-Type EGFR and EGFRvIII Enhance Killing of Glioblastoma and Patient-Derived Glioblastoma Stem Cells. Sci. Rep. 2015, 5, 11483. [Google Scholar] [CrossRef]
- Sahm, C.; Schönfeld, K.; Wels, W.S. Expression of IL-15 in NK cells results in rapid enrichment and selective cytotoxicity of gene-modified effectors that carry a tumor-specific antigen receptor. Cancer Immunol. Immunother. 2012, 61, 1451–1461. [Google Scholar] [CrossRef] [PubMed]
- Mitwasi, N.; Feldmann, A.; Arndt, C.; Koristka, S.; Berndt, N.; Jureczek, J.; Loureiro, L.R.; Bergmann, R.; Máthé, D.; Hegedüs, N.; et al. “UniCAR”-modified off-the-shelf NK-92 cells for targeting of GD2-expressing tumour cells. Sci. Rep. 2020, 10, 2141. [Google Scholar] [CrossRef] [PubMed]
- Rudek, L.S.; Zimmermann, K.; Galla, M.; Meyer, J.; Kuehle, J.; Stamopoulou, A.; Brand, D.; Sandalcioglu, I.E.; Neyazi, B.; Moritz, T.; et al. Generation of an NFκB-Driven Alpharetroviral “All-in-One” Vector Construct as a Potent Tool for CAR NK Cell Therapy. Front. Immunol. 2021, 12, 751138. [Google Scholar] [CrossRef]
- Kailayangiri, S.; Altvater, B.; Spurny, C.; Jamitzky, S.; Schelhaas, S.; Jacobs, A.H.; Wiek, C.; Roellecke, K.; Hanenberg, H.; Hartmann, W.; et al. Targeting Ewing sarcoma with activated and GD2-specific chimeric antigen receptor-engineered human NK cells induces upregulation of immune-inhibitory HLA-G. Oncoimmunology 2017, 6, e1250050. [Google Scholar] [CrossRef]
- Zhang, G.; Liu, R.; Zhu, X.; Wang, L.; Ma, J.; Han, H.; Wang, X.; Zhang, G.; He, W.; Wang, W.; et al. Retargeting NK-92 for anti-melanoma activity by a TCR-like single-domain antibody. Immunol. Cell Biol. 2013, 91, 615–624. [Google Scholar] [CrossRef]
- Zhang, C.; Burger, M.C.; Jennewein, L.; Genßler, S.; Schönfeld, K.; Zeiner, P.; Hattingen, E.; Harter, P.N.; Mittelbronn, M.; Tonn, T.; et al. ErbB2/HER2-Specific NK Cells for Targeted Therapy of Glioblastoma. J. Natl. Cancer Inst. 2016, 108, djv375. [Google Scholar] [CrossRef] [PubMed]
- Schönfeld, K.; Sahm, C.; Zhang, C.; Naundorf, S.; Brendel, C.; Odendahl, M.; Nowakowska, P.; Bönig, H.; Köhl, U.; Kloess, S.; et al. Selective Inhibition of Tumor Growth by Clonal NK Cells Expressing an ErbB2/HER2-Specific Chimeric Antigen Receptor. Mol. Ther. 2015, 23, 330–338. [Google Scholar] [CrossRef] [PubMed]
- Merker, M.; Wagner, J.; Kreyenberg, H.; Heim, C.; Moser, L.M.; Wels, W.S.; Bonig, H.; Ivics, Z.; Ullrich, E.; Klingebiel, T.; et al. ERBB2-CAR-Engineered Cytokine-Induced Killer Cells Exhibit Both CAR-Mediated and Innate Immunity Against High-Risk Rhabdomyosarcoma. Front. Immunol. 2020, 11, 581468. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Wang, Y.; Lin, M.; Xia, L.; Bao, Y.; Sun, X.; Yang, L. Abstract A014: Phase I clinical trial with PD-1/MUC1 CAR-pNK92 immunotherapy. Cancer Immunol. Res. 2019, 7 (Suppl. S2), A014. [Google Scholar] [CrossRef]
- Kulemzin, S.V.; Gorchakov, A.A.; Chikaev, A.N.; Kuznetsova, V.V.; Volkova, O.Y.; Matvienko, D.A.; Petukhov, A.V.; Zaritskey, A.Y.; Taranin, A.V. VEGFR2-specific FnCAR effectively redirects the cytotoxic activity of T cells and YT NK cells. Oncotarget 2018, 9, 9021–9029. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Chen, K.; Liu, H.; Jing, C.; Zhang, X.; Qu, C.; Yu, S. PMEL as a Prognostic Biomarker and Negatively Associated With Immune Infiltration in Skin Cutaneous Melanoma (SKCM). J. Immunother. 2021, 44, 214–223. [Google Scholar] [CrossRef]
- Strobel, S.B.; Machiraju, D.; Hülsmeyer, I.; Becker, J.C.; Paschen, A.; Jäger, D.; Wels, W.S.; Bachmann, M.; Hassel, J.C. Expression of Potential Targets for Cell-Based Therapies on Melanoma Cells. Life 2021, 11, 269. [Google Scholar] [CrossRef]
- Golinelli, G.; Grisendi, G.; Prapa, M.; Bestagno, M.; Spano, C.; Rossignoli, F.; Bambi, F.; Sardi, I.; Cellini, M.; Horwitz, E.M.; et al. Targeting GD2-positive glioblastoma by chimeric antigen receptor empowered mesenchymal progenitors. Cancer Gene Ther. 2020, 27, 558–570. [Google Scholar] [CrossRef]
- Kailayangiri, S.; Altvater, B.; Meltzer, J.; Pscherer, S.; Luecke, A.; Dierkes, C.; Titze, U.; Leuchte, K.; Landmeier, S.; Hotfilder, M.; et al. The ganglioside antigen GD2 is surface-expressed in Ewing sarcoma and allows for MHC-independent immune targeting. Br. J. Cancer 2012, 106, 1123–1133. [Google Scholar] [CrossRef]
- Reppel, L.; Tsahouridis, O.; Akulian, J.; Davis, I.J.; Lee, H.; Fucà, G.; Weiss, J.; Dotti, G.; Pecot, C.V.; Savoldo, B. Targeting disialoganglioside GD2 with chimeric antigen receptor-redirected T cells in lung cancer. J. Immunother. Cancer 2022, 10, e003897. [Google Scholar] [CrossRef]
- Yu, J.; Wu, X.; Yan, J.; Yu, H.; Xu, L.; Chi, Z.; Sheng, X.; Si, L.; Cui, C.; Dai, J.; et al. Anti-GD2/4-1BB chimeric antigen receptor T cell therapy for the treatment of Chinese melanoma patients. J. Hematol. Oncol. 2018, 11, 1. [Google Scholar] [CrossRef] [PubMed]
- Doronin, I.I.; Vishnyakova, P.A.; Kholodenko, I.V.; Ponomarev, E.D.; Ryazantsev, D.Y.; Molotkovskaya, I.M.; Kholodenko, R.V. Ganglioside GD2 in reception and transduction of cell death signal in tumor cells. BMC Cancer 2014, 14, 295. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, M.; Cheung, N.K.V. Disialoganglioside GD2 as a therapeutic target for human diseases. Expert Opin. Ther. Targets 2015, 19, 349–362. [Google Scholar] [CrossRef] [PubMed]
- Zimmermann, K.; Kuehle, J.; Dragon, A.C.; Galla, M.; Kloth, C.; Rudek, L.S.; Sandalcioglu, I.E.; Neyazi, B.; Moritz, T.; Meyer, J.; et al. Design and Characterization of an “All-in-One” Lentiviral Vector System Combining Constitutive Anti-GD2 CAR Expression and Inducible Cytokines. Cancers 2020, 12, 375. [Google Scholar] [CrossRef] [PubMed]
- Castellanos, J.R.; Purvis, I.J.; Labak, C.M.; Guda, M.R.; Tsung, A.J.; Velpula, K.K.; Asuthkar, S. B7-H3 role in the immune landscape of cancer. Am. J. Clin. Exp. Immunol. 2017, 6, 66–75. [Google Scholar] [PubMed]
- Hofmeyer, K.A.; Ray, A.; Zang, X. The contrasting role of B7-H3. Proc. Natl. Acad. Sci. USA 2008, 105, 10277–10278. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Duan, X.; Deng, X.; Huang, Y.; Zhou, X.; Zhang, S.; Zhang, X.; Liu, P.; Yang, C.; Liu, G.; et al. EBV-Upregulated B7-H3 Inhibits NK cell–Mediated Antitumor Function and Contributes to Nasopharyngeal Carcinoma Progression. Cancer Immunol. Res. 2023, 11, 830–846. [Google Scholar] [CrossRef]
- Lee, Y.H.; Martin-Orozco, N.; Zheng, P.; Li, J.; Zhang, P.; Tan, H.; Park, H.J.; Jeong, M.; Chang, S.H.; Kim, B.S.; et al. Inhibition of the B7-H3 immune checkpoint limits tumor growth by enhancing cytotoxic lymphocyte function. Cell Res. 2017, 27, 1034–1045. [Google Scholar] [CrossRef]
- Zhang, Z.; Jiang, C.; Liu, Z.; Yang, M.; Tang, X.; Wang, Y.; Zheng, M.; Huang, J.; Zhong, K.; Zhao, S.; et al. B7-H3-Targeted CAR-T Cells Exhibit Potent Antitumor Effects on Hematologic and Solid Tumors. Mol. Ther. Oncolytics 2020, 17, 180–189. [Google Scholar] [CrossRef]
- Yamato, M.; Hasegawa, J.; Maejima, T.; Hattori, C.; Kumagai, K.; Watanabe, A.; Nishiya, Y.; Shibutani, T.; Aida, T.; Hayakawa, I.; et al. DS-7300a, a DNA Topoisomerase I Inhibitor, DXd-Based Antibody–Drug Conjugate Targeting B7-H3, Exerts Potent Antitumor Activities in Preclinical Models. Mol. Cancer Ther. 2022, 21, 635–646. [Google Scholar] [CrossRef]
- Chaudhry, K.; Geiger, A.; Dowlati, E.; Lang, H.; Sohai, D.K.; Hwang, E.I.; Lazarski, C.A.; Yvon, E.; Holdhoff, M.; Jones, R.; et al. Co-transducing B7H3 CAR-NK cells with the DNR preserves their cytolytic function against GBM in the presence of exogenous TGF-β. Mol. Ther. Methods Clin. Dev. 2022, 27, 415–430. [Google Scholar] [CrossRef] [PubMed]
- Tang, X.; Wang, Y.; Huang, J.; Zhang, Z.; Liu, F.; Xu, J.; Guo, G.; Wang, W.; Tong, A.; Zhou, L. Administration of B7-H3 targeted chimeric antigen receptor-T cells induce regression of glioblastoma. Signal Transduct. Target. Ther. 2021, 6, 125. [Google Scholar] [CrossRef] [PubMed]
- Majzner, R.G.; Theruvath, J.L.; Nellan, A.; Heitzeneder, S.; Cui, Y.; Mount, C.W.; Rietberg, S.P.; Linde, M.H.; Xu, P.; Rota, C.; et al. CAR T Cells Targeting B7-H3, a Pan-Cancer Antigen, Demonstrate Potent Preclinical Activity Against Pediatric Solid Tumors and Brain Tumors. Clin. Cancer Res. 2019, 25, 2560–2574. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Zhang, M.; Wang, M.; Wang, H.; Wu, H.; Mao, L.; Zhang, M.; Li, H.; Zheng, J.; Ma, P.; et al. B7-H3 specific CAR-T cells exhibit potent activity against prostate cancer. Cell Death Discov. 2023, 9, 147. [Google Scholar] [CrossRef] [PubMed]
- Molhoek, K.R.; Erdag, G.; Rasamny, J.; Murphy, C.; Deacon, D.; Patterson, J.W.; Slingluff, C.L.; Brautigan, D.L. VEGFR-2 expression in human melanoma: Revised assessment. Int. J. Cancer 2011, 129, 2807–2815. [Google Scholar] [CrossRef] [PubMed]
- Kufe, D.W. MUC1-C oncoprotein as a target in breast cancer: Activation of signaling pathways and therapeutic approaches. Oncogene 2013, 32, 1073–1081. [Google Scholar] [CrossRef] [PubMed]
- Wei, J.; Sun, H.; Zhang, A.; Wu, X.; Li, Y.; Liu, J.; Duan, Y.; Xiao, F.; Wang, H.; Lv, M.; et al. A novel AXL chimeric antigen receptor endows T cells with anti-tumor effects against triple negative breast cancers. Cell Immunol. 2018, 331, 49–58. [Google Scholar] [CrossRef]
- DeRenzo, C. B7-H3-Specific Chimeric Antigen Receptor Autologous T-Cell Therapy for Pediatric Patients with Solid Tumors (3CAR). ClinicalTrials.gov identifier: NCT04897321. Updated 8 August 2023. Available online: https://clinicaltrials.gov/study/NCT04897321 (accessed on 10 October 2023).
- Yang, M.; Tang, X.; Zhang, Z.; Gu, L.; Wei, H.; Zhao, S.; Zhong, K.; Mu, M.; Huang, C.; Jiang, C.; et al. Tandem CAR-T cells targeting CD70 and B7-H3 exhibit potent preclinical activity against multiple solid tumors. Theranostics 2020, 10, 7622–7634. [Google Scholar] [CrossRef]
- Mishra, A.K.; Kemler, I.; Dingli, D. Preclinical development of CD126 CAR-T cells with broad antitumor activity. Blood Cancer J. 2021, 11, 3. [Google Scholar] [CrossRef]
- Rataj, F.; Jacobi, S.J.; Stoiber, S.; Asang, F.; Ogonek, J.; Tokarew, N.; Cadilha, B.L.; van Puijenbroek, E.; Heise, C.; Duewell, P.; et al. High-affinity CD16-polymorphism and Fc-engineered antibodies enable activity of CD16-chimeric antigen receptor-modified T cells for cancer therapy. Br. J. Cancer 2019, 120, 79–87. [Google Scholar] [CrossRef]
- Lu, Y.; Huang, H.; Chen, C.; Wang, H.; Wu, H.; Liu, X.; Zhao, W.; Gao, C.; Tan, S. Developing CD70-Targeted CAR-T Cells for the Treatment of Hematological and Solid Malignancies. Blood 2022, 140 (Suppl. S1), 12695. [Google Scholar] [CrossRef]
- Yang, J. Administering Peripheral Blood Lymphocytes Transduced with a CD70-Binding Chimeric Antigen Receptor to People with CD70 Expressing Cancers. ClinicalTrials.gov identifier: NCT02830724. Updated 1 September 2023. Available online: https://clinicaltrials.gov/study/NCT02830724 (accessed on 10 October 2023).
- Shah, P.D.; Huang, A.C.; Xu, X.; Orlowski, R.; Amaravadi, R.K.; Schuchter, L.M.; Zhang, P.; Tchou, J.; Matlawski, T.; Cervini, A.; et al. Phase I Trial of Autologous RNA-electroporated cMET-directed CAR T Cells Administered Intravenously in Patients with Melanoma and Breast Carcinoma. Cancer Res. Commun. 2023, 3, 821–829. [Google Scholar] [CrossRef] [PubMed]
- Kang, C.H.; Kim, Y.; Lee, D.Y.; Choi, S.U.; Lee, H.K.; Park, C.H. c-Met-Specific Chimeric Antigen Receptor T Cells Demonstrate Anti-Tumor Effect in c-Met Positive Gastric Cancer. Cancers 2021, 13, 5738. [Google Scholar] [CrossRef] [PubMed]
- Simon, B.; Harrer, D.C.; Schuler-Thurner, B.; Schuler, G.; Uslu, U. Arming T Cells with a gp100-Specific TCR and a CSPG4-Specific CAR Using Combined DNA- and RNA-Based Receptor Transfer. Cancers 2019, 11, 696. [Google Scholar] [CrossRef] [PubMed]
- Krug, C.; Birkholz, K.; Paulus, A.; Schwenkert, M.; Schmidt, P.; Hoffmann, N.; Hombach, A.; Fey, G.; Abken, H.; Schuler, G.; et al. Stability and activity of MCSP-specific chimeric antigen receptors (CARs) depend on the scFv antigen-binding domain and the protein backbone. Cancer Immunol. Immunother. 2015, 64, 1623–1635. [Google Scholar] [CrossRef]
- Wiesinger, M.; März, J.; Kummer, M.; Schuler, G.; Dörrie, J.; Schuler-Thurner, B.; Schaft, N. Clinical-Scale Production of CAR-T Cells for the Treatment of Melanoma Patients by mRNA Transfection of a CSPG4-Specific CAR under Full GMP Compliance. Cancers 2019, 11, 1198. [Google Scholar] [CrossRef] [PubMed]
- Fan, J.; Yu, Y.; Yan, L.; Yuan, Y.; Sun, B.; Yang, D.; Liu, N.; Guo, J.; Zhang, J.; Zhao, X. GAS6-based CAR-T cells exhibit potent antitumor activity against pancreatic cancer. J. Hematol. Oncol. 2023, 16, 77. [Google Scholar] [CrossRef]
- Gargett, T.; Yu, W.; Dotti, G.; Yvon, E.S.; Christo, S.N.; Hayball, J.D.; Lewis, I.D.; Brenner, M.K.; Brown, M.P. GD2-specific CAR T Cells Undergo Potent Activation and Deletion Following Antigen Encounter but can be Protected from Activation-induced Cell Death by PD-1 Blockade. Mol. Ther. 2016, 24, 1135–1149. [Google Scholar] [CrossRef]
- Kaplan, R. A Phase I Trial of T Cells Expressing an Anti-GD2 Chimeric Antigen Receptor in Children and Young Adults with GD2+ Solid Tumors. ClinicalTrials.gov identifier: NCT02107963. Updated 24 August 2023. Available online: https://clinicaltrials.gov/study/NCT02107963 (accessed on 10 October 2023).
- Omer, B. C7R-GD2.CART Cells for Patients With Relapsed or Refractory Neuroblastoma and Other GD2 Positive Cancers (GAIL-N). ClinicalTrial.gov identifier: NCT03635632. Updated 2 August 2023. Available online: https://clinicaltrials.gov/study/NCT03635632 (accessed on 10 October 2023).
- Lo, A.S.Y.; Ma, Q.; Liu, D.L.; Junghans, R.P. Anti-GD3 Chimeric sFv-CD28/T-Cell Receptor ζ Designer T Cells for Treatment of Metastatic Melanoma and Other Neuroectodermal Tumors. Clin. Cancer Res. 2010, 16, 2769–2780. [Google Scholar] [CrossRef]
- Yun, C.O.; Nolan, K.F.; Beecham, E.J.; Reisfeld, R.A.; Junghans, P. Targeting of T Lymphocytes to Melanoma Cells through Chimeric Anti-GD3 Immunoglobulin T-Cell Receptors. Neoplasia 2000, 2, 449–459. [Google Scholar] [CrossRef]
- Forsberg, E.M.; Lindberg, M.F.; Jespersen, H.; Alsén, S.; Bagge, R.O.; Donia, M.; Svane, I.M.; Nilsson, O.; Ny, L.; Nilsson, L.M.; et al. HER2 CAR-T Cells Eradicate Uveal Melanoma and T-cell Therapy–Resistant Human Melanoma in IL2 Transgenic NOD/SCID IL2 Receptor Knockout Mice. Cancer Res. 2019, 79, 899–904. [Google Scholar] [CrossRef] [PubMed]
- Park, J.; Talukder, A.H.; Lim, S.A.; Kim, K.; Pan, K.; Melendez, B.; Bradley, S.D.; Jackson, K.R.; Khalili, J.S.; Wang, J.; et al. SLC45A2: A Melanoma Antigen with High Tumor Selectivity and Reduced Potential for Autoimmune Toxicity. Cancer Immunol. Res. 2017, 5, 618–629. [Google Scholar] [CrossRef] [PubMed]
- Shivde, R.; Jaishankar, D.; Thomas, A.; Le Poole, I. 549 Design and in vitro efficacy of TRP-1 CAR T cells to target melanoma. J. Investig. Dermatol. 2021, 141, S95. [Google Scholar] [CrossRef]
- Lanitis, E.; Kosti, P.; Ronet, C.; Cribioli, E.; Rota, G.; Spill, A.; Reichenbach, P.; Zoete, V.; Laniti, D.D.; Coukos, G.; et al. VEGFR-2 redirected CAR-T cells are functionally impaired by soluble VEGF-A competition for receptor binding. J. Immunother. Cancer 2021, 9, e002151. [Google Scholar] [CrossRef]
- Cobb, D.A.; de Rossi, J.; Liu, L.; An, E.; Lee, D.W. Targeting of the alphavbeta3 integrin complex by CAR-T cells leads to rapid regression of diffuse intrinsic pontine glioma and glioblastoma. J. Immunother. Cancer 2022, 10, e003816. [Google Scholar] [CrossRef] [PubMed]
- Wallstabe, L.; Mades, A.; Frenz, S.; Einsele, H.; Rader, C.; Hudecek, M. CAR T cells targeting αvβ3 integrin are effective against advanced cancer in preclinical models. Adv. Cell Gene Ther. 2018, 1, e11. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Lan, H.; Li, J.; Su, Y.; Xu, L. Muc1 promotes migration and lung metastasis of melanoma cells. Am. J. Cancer Res. 2015, 5, 2590–2604. [Google Scholar] [PubMed]
- Zhang, C.; Guo, C.; Li, Y.; Liu, K.; Zhao, Q.; Ouyang, L. Identification of Claudin-6 as a Molecular Biomarker in Pan-Cancer Through Multiple Omics Integrative Analysis. Front. Cell Dev. Biol. 2021, 9, 726656. [Google Scholar] [CrossRef]
- Carubia, J.M.; Yu, R.K.; Macala, L.J.; Kirkwood, J.M.; Varga, J.M. Gangliosides of normal and neoplastic human melanocytes. Biochem. Biophys. Res. Commun. 1984, 120, 500–504. [Google Scholar] [CrossRef]
- Hamamura, K.; Furukawa, K.; Hayashi, T.; Hattori, T.; Nakano, J.; Nakashima, H.; Okuda, T.; Mizutani, H.; Hattori, H.; Ueda, M.; et al. Ganglioside GD3 promotes cell growth and invasion through p130Cas and paxillin in malignant melanoma cells. Proc. Natl. Acad. Sci. USA 2005, 102, 11041–11046. [Google Scholar] [CrossRef]
- Ohkawa, Y.; Miyazaki, S.; Hamamura, K.; Kambe, M.; Miyata, M.; Tajima, O.; Ohmi, Y.; Yamauchi, Y.; Furukawa, K.; Furukawa, K. Ganglioside GD3 Enhances Adhesion Signals and Augments Malignant Properties of Melanoma Cells by Recruiting Integrins to Glycolipid-enriched Microdomains. J. Biol. Chem. 2010, 285, 27213–27223. [Google Scholar] [CrossRef] [PubMed]
- Ramos, R.I.; Bustos, M.A.; Wu, J.; Jones, P.; Chang, S.C.; Kiyohara, E.; Tran, K.; Zhang, X.; Stern, S.L.; Izraely, S.; et al. Upregulation of cell surface GD3 ganglioside phenotype is associated with human melanoma brain metastasis. Mol. Oncol. 2020, 14, 1760–1778. [Google Scholar] [CrossRef] [PubMed]
- Huang, R.; Rofstad, E.K. Integrins as therapeutic targets in the organ-specific metastasis of human malignant melanoma. J. Exp. Clin. Cancer Res. 2018, 37, 92. [Google Scholar] [CrossRef] [PubMed]
- Seftor, R.E.; Seftor, E.A.; Gehlsen, K.R.; Stetler-Stevenson, W.G.; Brown, P.D.; Ruoslahti, E.; Hendrix, M.J. Role of the alpha v beta 3 integrin in human melanoma cell invasion. Proc. Natl. Acad. Sci. USA 1992, 89, 1557–1561. [Google Scholar] [CrossRef] [PubMed]
- Pocheć, E.; Bubka, M.; Rydlewska, M.; Janik, M.; Pokrywka, M.; Lityńska, A. Aberrant glycosylation of αvβ3 integrin is associated with melanoma progression. Anticancer Res. 2015, 35, 2093–2103. [Google Scholar] [PubMed]
- Scheller, J.; Chalaris, A.; Schmidt-Arras, D.; Rose-John, S. The pro- and anti-inflammatory properties of the cytokine interleukin-6. Biochim. Biophys. Acta Mol. Cell Res. 2011, 1813, 878–888. [Google Scholar] [CrossRef] [PubMed]
- Zuo, Q.; Liu, J.; Huang, L.; Qin, Y.; Hawley, T.; Seo, C.; Merlino, G.; Yu, Y. AXL/AKT axis mediated-resistance to BRAF inhibitor depends on PTEN status in melanoma. Oncogene 2018, 37, 3275–3289. [Google Scholar] [CrossRef]
- Tworkoski, K.A.; Platt, J.T.; Bacchiocchi, A.; Bosenberg, M.; Boggon, T.J.; Stern, D.F. MERTK controls melanoma cell migration and survival and differentially regulates cell behavior relative to AXL. Pigment. Cell Melanoma Res. 2013, 26, 527–541. [Google Scholar] [CrossRef]
- Tizpa, E.; Young, H.J.; Bonjoc, K.-J.C.; Chang, C.-W.; Liu, Y.; Foulks, J.M.; Chaudhry, A. Role of AXL in metastatic melanoma and impact of TP-0903 as a novel therapeutic option for melanoma brain metastasis. J. Clin. Oncol. 2020, 38 (Suppl. S15), e22021. [Google Scholar] [CrossRef]
- Boshuizen, J.; Koopman, L.A.; Krijgsman, O.; Shahrabi, A.; van den Heuvel, E.G.; A Ligtenberg, M.A.; Vredevoogd, D.W.; Kemper, K.; Kuilman, T.; Song, J.-Y.; et al. Cooperative targeting of melanoma heterogeneity with an AXL antibody-drug conjugate and BRAF/MEK inhibitors. Nat. Med. 2018, 24, 203–212. [Google Scholar] [CrossRef]
- Nyakas, M.; Fleten, K.G.; Haugen, M.H.; Engedal, N.; Sveen, C.; Farstad, I.N.; Flørenes, V.A.; Prasmickaite, L.; Mælandsmo, G.M.; Seip, K. AXL inhibition improves BRAF-targeted treatment in melanoma. Sci. Rep. 2022, 12, 5076. [Google Scholar] [CrossRef]
- Nicolè, L.; Cappello, F.; Cappellesso, R.; Piccin, L.; Ventura, L.; Guzzardo, V.; Del Fiore, P.; Chiarion-Sileni, V.; Tos, A.P.D.; Mocellin, S.; et al. RIPK3 and AXL Expression Study in Primary Cutaneous Melanoma Unmasks AXL as Predictor of Sentinel Node Metastasis: A Pilot Study. Front. Oncol. 2021, 11, 728319. [Google Scholar] [CrossRef]
- Shao, H.; Teramae, D.; Wells, A. Axl contributes to efficient migration and invasion of melanoma cells. PLoS ONE 2023, 18, e0283749. [Google Scholar] [CrossRef] [PubMed]
- Xue, G.; Kohler, R.; Tang, F.; Hynx, D.; Wang, Y.; Orso, F.; Prêtre, V.; Ritschard, R.; Hirschmann, P.; Cron, P.; et al. mTORC1/autophagy-regulated MerTK in mutant BRAFV600 melanoma with acquired resistance to BRAF inhibition. Oncotarget 2017, 8, 69204–69218. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Garcia, A.; Lynn, R.C.; Poussin, M.; Eiva, M.A.; Shaw, L.C.; O’connor, R.S.; Minutolo, N.G.; Casado-Medrano, V.; Lopez, G.; Matsuyama, T.; et al. CAR-T cell-mediated depletion of immunosuppressive tumor-associated macrophages promotes endogenous antitumor immunity and augments adoptive immunotherapy. Nat. Commun. 2021, 12, 877. [Google Scholar] [CrossRef]
- Schlegel, J.; Sambade, M.J.; Sather, S.; Moschos, S.J.; Tan, A.-C.; Winges, A.; DeRyckere, D.; Carson, C.C.; Trembath, D.G.; Tentler, J.J.; et al. MERTK receptor tyrosine kinase is a therapeutic target in melanoma. J. Clin. Investig. 2013, 123, 2257–2267. [Google Scholar] [CrossRef] [PubMed]
- Sinik, L.; Minson, K.A.; Tentler, J.J.; Carrico, J.; Bagby, S.M.; Robinson, W.A.; Kami, R.; Burstyn-Cohen, T.; Eckhardt, S.G.; Wang, X.; et al. Inhibition of MERTK Promotes Suppression of Tumor Growth in BRAF Mutant and BRAF Wild-Type Melanoma. Mol. Cancer Ther. 2019, 18, 278–288. [Google Scholar] [CrossRef] [PubMed]
- Boshuizen, J.; Vredevoogd, D.W.; Krijgsman, O.; Ligtenberg, M.A.; Blankenstein, S.; de Bruijn, B.; Frederick, D.T.; Kenski, J.C.N.; Parren, M.; Brüggemann, M.; et al. Reversal of pre-existing NGFR-driven tumor and immune therapy resistance. Nat. Commun. 2020, 11, 3946. [Google Scholar] [CrossRef]
- Benboubker, V.; Boivin, F.; Dalle, S.; Caramel, J. Cancer Cell Phenotype Plasticity as a Driver of Immune Escape in Melanoma. Front. Immunol. 2022, 13, 873116. [Google Scholar] [CrossRef]
- Radke, J.; Roßner, F.; Redmer, T. CD271 determines migratory properties of melanoma cells. Sci. Rep. 2017, 7, 9834. [Google Scholar] [CrossRef]
- Furuta, J.; Inozume, T.; Harada, K.; Shimada, S. CD271 on Melanoma Cell Is an IFN-γ-Inducible Immunosuppressive Factor that Mediates Downregulation of Melanoma Antigens. J. Investig. Dermatol. 2014, 134, 1369–1377. [Google Scholar] [CrossRef] [PubMed]
- Lehmann, J.; Caduff, N.; Krzywińska, E.; Stierli, S.; Salas-Bastos, A.; Loos, B.; Levesque, M.P.; Dummer, R.; Stockmann, C.; Münz, C.; et al. Escape from NK cell tumor surveillance by NGFR-induced lipid remodeling in melanoma. Sci. Adv. 2023, 9, eadc8825. [Google Scholar] [CrossRef] [PubMed]
- Teppert, K.; Wang, X.; Anders, K.; Evaristo, C.; Lock, D.; Künkele, A. Joining Forces for Cancer Treatment: From “TCR versus CAR” to “TCR and CAR. ” Int. J. Mol. Sci. 2022, 23, 14563. [Google Scholar] [CrossRef] [PubMed]
- Khawar, M.B.; Sun, H. CAR-NK Cells: From Natural Basis to Design for Kill. Front. Immunol. 2021, 12, 707542. [Google Scholar] [CrossRef] [PubMed]
- Oberschmidt, O.; Kloess, S.; Koehl, U. Redirected Primary Human Chimeric Antigen Receptor Natural Killer Cells as an “Off-the-Shelf Immunotherapy” for Improvement in Cancer Treatment. Front. Immunol. 2017, 8, 654. [Google Scholar] [CrossRef] [PubMed]
- Ruppel, K.E.; Fricke, S.; Köhl, U.; Schmiedel, D. Taking Lessons from CAR-T Cells and Going Beyond: Tailoring Design and Signaling for CAR-NK Cells in Cancer Therapy. Front. Immunol. 2022, 13, 822298. [Google Scholar] [CrossRef] [PubMed]
- Zenere, G.; Olwenyi, O.A.; Byrareddy, S.N.; Braun, S.E. Optimizing intracellular signaling domains for CAR NK cells in HIV immunotherapy: A comprehensive review. Drug Discov. Today 2019, 24, 983–991. [Google Scholar] [CrossRef]
- Voynova, E.; Hawk, N.; Flomerfelt, F.A.; Telford, W.G.; Gress, R.E.; Kanakry, J.A.; Kovalovsky, D. Increased Activity of a NK-Specific CAR-NK Framework Targeting CD3 and CD5 for T-Cell Leukemias. Cancers 2022, 14, 524. [Google Scholar] [CrossRef]
- Lee, Y.E.; Ju, A.; Choi, H.W.; Kim, J.-C.; Kim, E.E.; Kim, T.S.; Kang, H.J.; Kim, S.-Y.; Jang, J.-Y.; Ku, J.-L.; et al. Rationally designed redirection of natural killer cells anchoring a cytotoxic ligand for pancreatic cancer treatment. J. Control. Release 2020, 326, 310–323. [Google Scholar] [CrossRef]
- Li, Y.; Hermanson, D.L.; Moriarity, B.S.; Kaufman, D.S. Human iPSC-Derived Natural Killer Cells Engineered with Chimeric Antigen Receptors Enhance Anti-Tumor Activity. Cell Stem Cell 2018, 23, 181–192.e5. [Google Scholar] [CrossRef]
- Blake, S.J.; Dougall, W.C.; Miles, J.J.; Teng, M.W.; Smyth, M.J. Molecular Pathways: Targeting CD96 and TIGIT for Cancer Immunotherapy. Clin. Cancer Res. 2016, 22, 5183–5188. [Google Scholar] [CrossRef] [PubMed]
- Jan, C.I.; Huang, S.W.; Canoll, P.; Bruce, J.N.; Lin, Y.C.; Pan, C.M.; Lu, H.M.; Chiu, S.C.; Cho, D.Y. Targeting human leukocyte antigen G with chimeric antigen receptors of natural killer cells convert immunosuppression to ablate solid tumors. J. Immunother. Cancer 2021, 9, e003050. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Li, H.; Xu, C. Structural understanding of T cell receptor triggering. Cell Mol. Immunol. 2020, 17, 193–202. [Google Scholar] [CrossRef]
- Ferrara, J.; Reddy, P.; Paczesny, S.; Reimann, C.; Cortivo, L.D.; Hacein-Bey-Abina, S.; Fischer, A.; André-Schmutz, I.; Cavazzana-Calvo, M.; Ribas, A.; et al. Immunotherapy through T-cell receptor gene transfer induces severe graft-versus-host disease. Immunotherapy 2010, 2, 791–794. [Google Scholar] [CrossRef] [PubMed]
- Bijen, H.M.; van der Steen, D.M.; Hagedoorn, R.S.; Wouters, A.K.; Wooldridge, L.; Falkenburg, J.F.; Heemskerk, M.H. Preclinical Strategies to Identify Off-Target Toxicity of High-Affinity TCRs. Mol. Ther. 2018, 26, 1206–1214. [Google Scholar] [CrossRef] [PubMed]
- Parlar, A.; Sayitoglu, E.C.; Ozkazanc, D.; Georgoudaki, A.M.; Pamukcu, C.; Aras, M.; Josey, B.J.; Chrobok, M.; Branecki, S.; Zahedimaram, P.; et al. Engineering antigen-specific NK cell lines against the melanoma-associated antigen tyrosinase via TCR gene transfer. Eur. J. Immunol. 2019, 49, 1278–1290. [Google Scholar] [CrossRef] [PubMed]
- Walseng, E.; Köksal, H.; Sektioglu, I.M.; Fåne, A.; Skorstad, G.; Kvalheim, G.; Gaudernack, G.; Inderberg, E.M.; Wälchli, S. A TCR-based Chimeric Antigen Receptor. Sci. Rep. 2017, 7, 10713. [Google Scholar] [CrossRef] [PubMed]
- Wellbrock, C.; Arozarena, I. The Complexity of the ERK/MAP-Kinase Pathway and the Treatment of Melanoma Skin Cancer. Front. Cell Dev. Biol. 2016, 4, 33. [Google Scholar] [CrossRef]
- Rauch, N.; Rukhlenko, O.S.; Kolch, W.; Kholodenko, B.N. MAPK kinase signalling dynamics regulate cell fate decisions and drug resistance. Curr. Opin. Struct. Biol. 2016, 41, 151–158. [Google Scholar] [CrossRef]
- Kunz, M.; Vera, J. Modelling of Protein Kinase Signaling Pathways in Melanoma and Other Cancers. Cancers 2019, 11, 465. [Google Scholar] [CrossRef]
- Guo, W.; Wang, H.; Li, C. Signal pathways of melanoma and targeted therapy. Signal Transduct. Target. Ther. 2021, 6, 424. [Google Scholar] [CrossRef]
- Wang, Y.; Poulin, E.J.; Coffey, R.J. LRIG1 is a triple threat: ERBB negative regulator, intestinal stem cell marker and tumour suppressor. Br. J. Cancer 2013, 108, 1765–1770. [Google Scholar] [CrossRef]
- Johansson, M.; Oudin, A.; Tiemann, K.; Bernard, A.; Golebiewska, A.; Keunen, O.; Fack, F.; Stieber, D.; Wang, B.; Hedman, H.; et al. The soluble form of the tumor suppressor Lrig1 potently inhibits in vivo glioma growth irrespective of EGF receptor status. Neuro Oncol. 2013, 15, 1200–1211. [Google Scholar] [CrossRef]
- Billing, O.; Holmgren, Y.; Nosek, D.; Hedman, H.; Hemmingsson, O. LRIG1 is a conserved EGFR regulator involved in melanoma development, survival and treatment resistance. Oncogene 2021, 40, 3707–3718. [Google Scholar] [CrossRef]
- Wu, C.H.; Liu, I.J.; Lu, R.M.; Wu, H.C. Advancement and applications of peptide phage display technology in biomedical science. J. Biomed. Sci. 2016, 23, 8. [Google Scholar] [CrossRef]
- Garner, H.; de Visser, K.E. Immune crosstalk in cancer progression and metastatic spread: A complex conversation. Nat. Rev. Immunol. 2020, 20, 483–497. [Google Scholar] [CrossRef]
- Kim, J.; Kim, J.S.; Lee, H.K.; Kim, H.S.; Park, E.J.; Choi, J.E.; Choi, Y.J.; Shin, B.R.; Kim, E.Y.; Hong, J.T.; et al. CXCR3-deficient natural killer cells fail to migrate to B16F10 melanoma cells. Int. Immunopharmacol. 2018, 63, 66–73. [Google Scholar] [CrossRef] [PubMed]
- Mgrditchian, T.; Arakelian, T.; Paggetti, J.; Noman, M.Z.; Viry, E.; Moussay, E.; Van Moer, K.; Kreis, S.; Guerin, C.; Buart, S.; et al. Targeting autophagy inhibits melanoma growth by enhancing NK cells infiltration in a CCL5-dependent manner. Proc. Natl. Acad. Sci. USA 2017, 114, E9271–E9279. [Google Scholar] [CrossRef]
- Jobe, N.P.; Rösel, D.; Dvořánková, B.; Kodet, O.; Lacina, L.; Mateu, R.; Smetana, K.; Brábek, J. Simultaneous blocking of IL-6 and IL-8 is sufficient to fully inhibit CAF-induced human melanoma cell invasiveness. Histochem. Cell Biol. 2016, 146, 205–217. [Google Scholar] [CrossRef] [PubMed]
- Straussman, R.; Morikawa, T.; Shee, K.; Barzily-Rokni, M.; Qian, Z.R.; Du, J.; Davis, A.; Mongare, M.M.; Gould, J.; Frederick, D.T.; et al. Tumour micro-environment elicits innate resistance to RAF inhibitors through HGF secretion. Nature 2012, 487, 500–504. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Zhou, L.; Xiao, Y.; Andl, T.; Zhang, Y. BRAF Inhibitors Reprogram Cancer-Associated Fibroblasts to Drive Matrix Remodeling and Therapeutic Escape in Melanoma. Cancer Res. 2022, 82, 419–432. [Google Scholar] [CrossRef] [PubMed]
- Morales, D.; Vigneron, P.; Ferreira, I.; Hamitou, W.; Magnano, M.; Mahenthiran, L.; Lok, C.; Vayssade, M. Fibroblasts Influence Metastatic Melanoma Cell Sensitivity to Combined BRAF and MEK Inhibition. Cancers 2021, 13, 4761. [Google Scholar] [CrossRef]
- Zhou, L.; Yang, K.; Wickett, R.R.; Kadekaro, A.L.; Zhang, Y. Targeted deactivation of cancer-associated fibroblasts by β-catenin ablation suppresses melanoma growth. Tumor Biol. 2016, 37, 14235–14248. [Google Scholar] [CrossRef]
- Balsamo, M.; Scordamaglia, F.; Pietra, G.; Manzini, C.; Cantoni, C.; Boitano, M.; Queirolo, P.; Vermi, W.; Facchetti, F.; Moretta, A.; et al. Melanoma-associated fibroblasts modulate NK cell phenotype and antitumor cytotoxicity. Proc. Natl. Acad. Sci. USA 2009, 106, 20847–20852. [Google Scholar] [CrossRef] [PubMed]
- Kahounová, Z.; Kurfürstová, D.; Bouchal, J.; Kharaishvili, G.; Navrátil, J.; Remšík, J.; Šimečková, K.; Študent, V.; Kozubík, A.; Souček, K. The fibroblast surface markers FAP, anti-fibroblast, and FSP are expressed by cells of epithelial origin and may be altered during epithelial-to-mesenchymal transition. Cytom. Part A 2018, 93, 941–951. [Google Scholar] [CrossRef] [PubMed]
- Vázquez-Villa, F.; García-Ocaña, M.; Galván, J.A.; García-Martínez, J.; García-Pravia, C.; Menéndez-Rodríguez, P.; González-del Rey, C.; Barneo-Serra, L.; de los Toyos, J.R. COL11A1/(pro)collagen 11A1 expression is a remarkable biomarker of human invasive carcinoma-associated stromal cells and carcinoma progression. Tumor Biol. 2015, 36, 2213–2222. [Google Scholar] [CrossRef] [PubMed]
- Mezawa, Y.; Orimo, A. The roles of tumor- and metastasis-promoting carcinoma-associated fibroblasts in human carcinomas. Cell Tissue Res. 2016, 365, 675–689. [Google Scholar] [CrossRef]
- Gunaydin, G. CAFs Interacting With TAMs in Tumor Microenvironment to Enhance Tumorigenesis and Immune Evasion. Front. Oncol. 2021, 11, 668349. [Google Scholar] [CrossRef]
- Chen, P.; Huang, Y.; Bong, R.; Ding, Y.; Song, N.; Wang, X.; Song, X.; Luo, Y. Tumor-Associated Macrophages Promote Angiogenesis and Melanoma Growth via Adrenomedullin in a Paracrine and Autocrine Manner. Clin. Cancer Res. 2011, 17, 7230–7239. [Google Scholar] [CrossRef]
- Filippi, L.; Bruno, G.; Domazetovic, V.; Favre, C.; Calvani, M. Current Therapies and New Targets to Fight Melanoma: A Promising Role for the β3-Adrenoreceptor. Cancers 2020, 12, 1415. [Google Scholar] [CrossRef]
- Zhang, P.; Zhao, S.; Wu, C.; Li, J.; Li, Z.; Wen, C.; Hu, S.; An, G.; Meng, H.; Zhang, X.; et al. Effects of CSF1R-targeted chimeric antigen receptor-modified NK92MI & T cells on tumor-associated macrophages. Immunotherapy 2018, 10, 935–949. [Google Scholar] [CrossRef]
- Wang, L.; He, T.; Liu, J.; Tai, J.; Wang, B.; Chen, Z.; Quan, Z. Pan-cancer analysis reveals tumor-associated macrophage communication in the tumor microenvironment. Exp. Hematol. Oncol. 2021, 10, 31. [Google Scholar] [CrossRef] [PubMed]
- Dixon, M.L.; Luo, L.; Ghosh, S.; Grimes, J.M.; Leavenworth, J.D.; Leavenworth, J.W. Remodeling of the tumor microenvironment via disrupting Blimp1+ effector Treg activity augments response to anti-PD-1 blockade. Mol. Cancer 2021, 20, 150. [Google Scholar] [CrossRef] [PubMed]
- Salmi, S.; Lin, A.; Hirschovits-Gerz, B.; Valkonen, M.; Aaltonen, N.; Sironen, R.; Siiskonen, H.; Pasonen-Seppänen, S. The role of FoxP3+ regulatory T cells and IDO+ immune and tumor cells in malignant melanoma—An immunohistochemical study. BMC Cancer 2021, 21, 641. [Google Scholar] [CrossRef]
- Watson, M.J.; Vignali, P.D.A.; Mullett, S.J.; Overacre-Delgoffe, A.E.; Peralta, R.M.; Grebinoski, S.; Menk, A.V.; Rittenhouse, N.L.; DePeaux, K.; Whetstone, R.D.; et al. Metabolic support of tumour-infiltrating regulatory T cells by lactic acid. Nature 2021, 591, 645–651. [Google Scholar] [CrossRef] [PubMed]
- Lainé, A.; Labiad, O.; Hernandez-Vargas, H.; This, S.; Sanlaville, A.; Léon, S.; Dalle, S.; Sheppard, D.; Travis, M.A.; Paidassi, H.; et al. Regulatory T cells promote cancer immune-escape through integrin αvβ8-mediated TGF-β activation. Nat. Commun. 2021, 12, 6228. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Jiang, P.; Wei, S.; Xu, X.; Wang, J. Regulatory T cells in tumor microenvironment: New mechanisms, potential therapeutic strategies and future prospects. Mol. Cancer 2020, 19, 116. [Google Scholar] [CrossRef] [PubMed]
- Dehbashi, M.; Hojati, Z.; Motovali-Bashi, M.; Ganjalikhany, M.R.; Cho, W.C.; Shimosaka, A.; Navabi, P.; Ganjalikhani-Hakemi, M. A Novel CAR Expressing NK Cell Targeting CD25 with the Prospect of Overcoming Immune Escape Mechanism in Cancers. Front. Oncol. 2021, 11, 649710. [Google Scholar] [CrossRef]
- Kageshita, T.; Kashio, Y.; Yamauchi, A.; Seki, M.; Abedin, M.J.; Nishi, N.; Shoji, H.; Nakamura, T.; Ono, T.; Hirashima, M. Possible role of galectin-9 in cell aggregation and apoptosis of human melanoma cell lines and its clinical significance. Int. J. Cancer 2002, 99, 809–816. [Google Scholar] [CrossRef]
- Francisco, L.M.; Salinas, V.H.; Brown, K.E.; Vanguri, V.K.; Freeman, G.J.; Kuchroo, V.K.; Sharpe, A.H. PD-L1 regulates the development, maintenance, and function of induced regulatory T cells. J. Exp. Med. 2009, 206, 3015–3029. [Google Scholar] [CrossRef]
- Unger, W.W.J.; Laban, S.; Kleijwegt, F.S.; van der Slik, A.R.; Roep, B.O. Induction of Treg by monocyte-derived DC modulated by vitamin D3 or dexamethasone: Differential role for PD-L1. Eur. J. Immunol. 2009, 39, 3147–3159. [Google Scholar] [CrossRef] [PubMed]
- Kararoudi, M.N.; Likhite, S.; Elmas, E.; Yamamoto, K.; Schwartz, M.; Sorathia, K.; Pereira, M.d.S.F.; Sezgin, Y.; Devine, R.D.; Lyberger, J.M.; et al. Optimization and validation of CAR transduction into human primary NK cells using CRISPR and AAV. Cell Rep. Methods 2022, 2, 100236. [Google Scholar] [CrossRef] [PubMed]
- de Andrade, L.F.; Ngiow, S.F.; Stannard, K.; Rusakiewicz, S.; Kalimutho, M.; Khanna, K.K.; Tey, S.-K.; Takeda, K.; Zitvogel, L.; Martinet, L.; et al. Natural Killer Cells Are Essential for the Ability of BRAF Inhibitors to Control BRAFV600E-Mutant Metastatic Melanoma. Cancer Res. 2014, 74, 7298–7308. [Google Scholar] [CrossRef] [PubMed]
- Frazao, A.; Rethacker, L.; Jeudy, G.; Colombo, M.; Pasmant, E.; Avril, M.-F.; Toubert, A.; Moins-Teisserenc, H.; Roelens, M.; Dalac, S.; et al. BRAF inhibitor resistance of melanoma cells triggers increased susceptibility to natural killer cell-mediated lysis. J. Immunother. Cancer 2020, 8, e000275. [Google Scholar] [CrossRef]
- Sottile, R.; Pangigadde, P.N.; Tan, T.; Anichini, A.; Sabbatino, F.; Trecroci, F.; Favoino, E.; Orgiano, L.; Roberts, J.; Ferrone, S.; et al. HLA class I downregulation is associated with enhanced NK-cell killing of melanoma cells with acquired drug resistance to BRAF inhibitors. Eur. J. Immunol. 2016, 46, 409–419. [Google Scholar] [CrossRef]


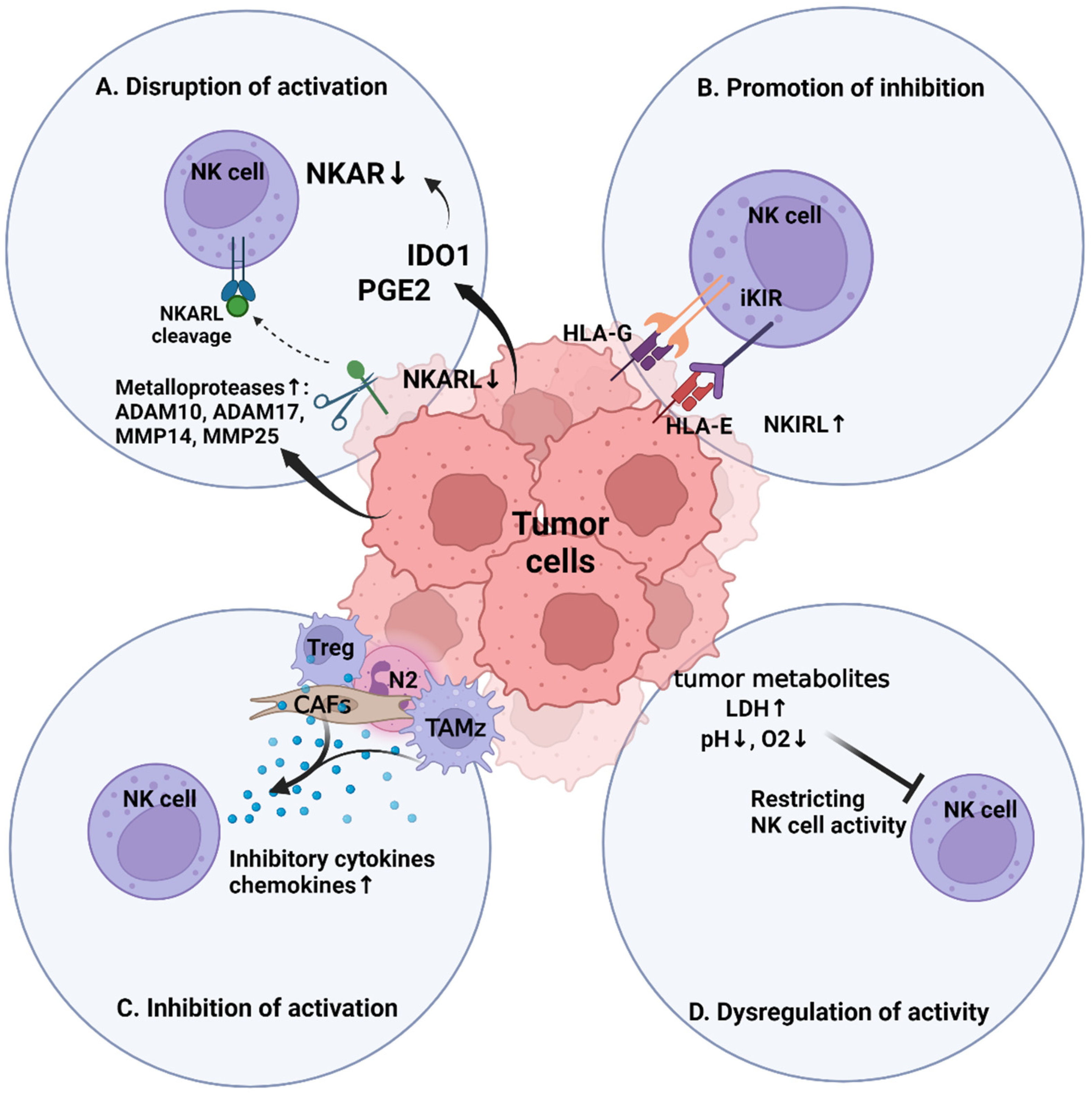
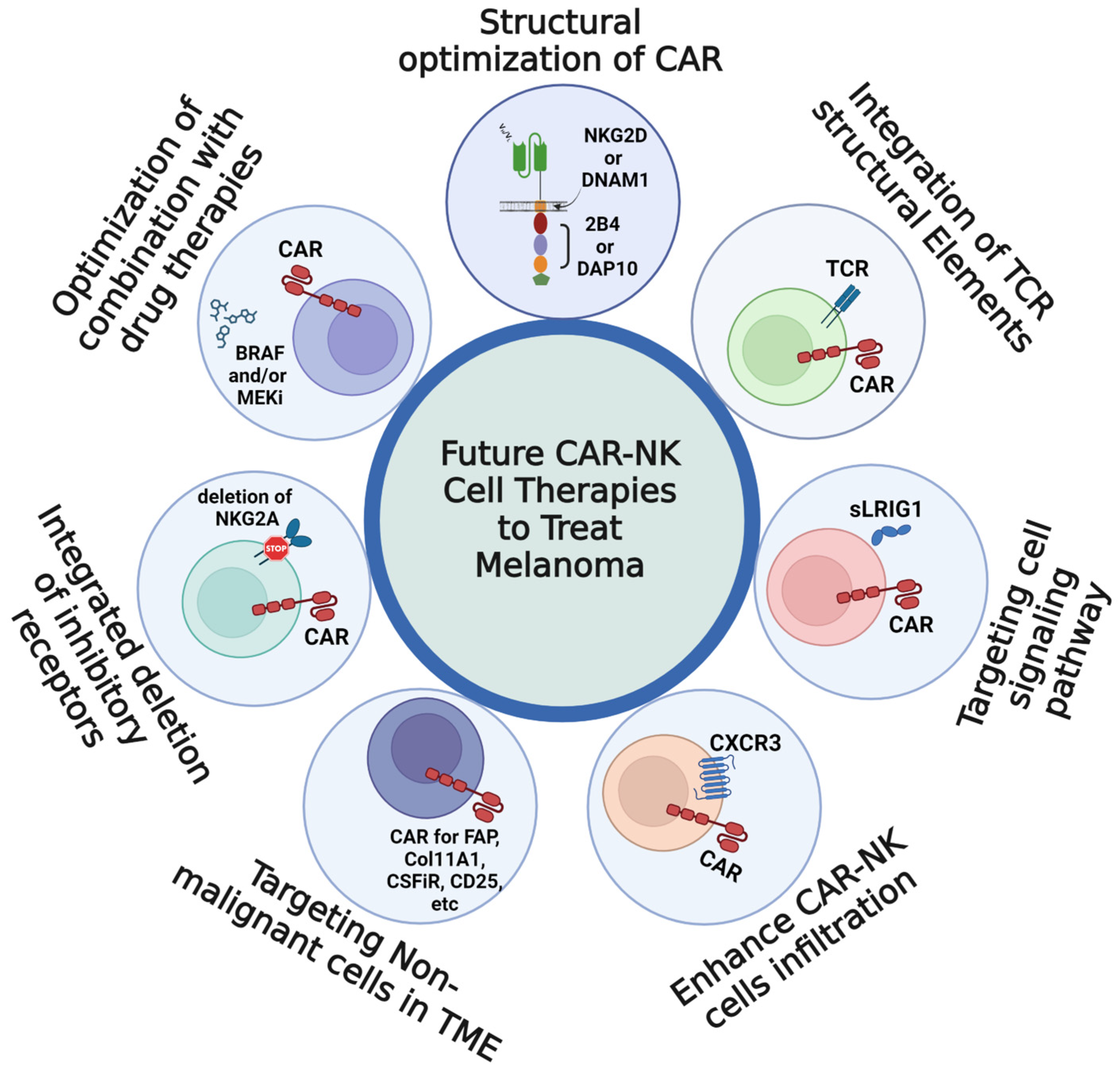
| Target | Study Type | Clinical Trial Status | Cancer Type | Reference |
|---|---|---|---|---|
| 5T4 | Clinical | Active/Recruiting | Solid tumors | NCT05194709 |
| B7-H3 | Preclinical | - | Non-small cell lung cancer | [161] |
| B7-H6 | Preclinical | - | Solid tumors | [162] |
| CD24 | Preclinical | - | Ovarian cancer | [163] |
| CD276 | Preclinical | - | Solid tumors | [164] |
| CD44 | Preclinical | - | Ovarian cancer | [165] |
| CLDN6 | Clinical | Active/Recruiting | Solid tumors | [166] |
| c-Met | Preclinical | - | Lung adenocarcinoma | [160] |
| EGFR/EGFRvIII | Preclinical | - | Glioblastoma | [167] |
| - | Glioblastoma | [168] | ||
| - | Glioblastoma | [169] | ||
| - | Glioblastoma | [170] | ||
| EpCAM | Preclinical | - | Solid tumors | [171] |
| GD2 | Preclinical | - | GD2+ solid tumors | [172] |
| - | GD2+ solid tumors | [173] | ||
| - | Ewing sarcoma | [174] | ||
| gp100 | Preclinical | - | melanoma | [175] |
| HER2 | Preclinical | - | Rhabdomyosarcoma | [146] |
| - | Glioblastoma | [176] | ||
| - | Solid tumors | [177] | ||
| - | Rhabdomyosarcoma | [178] | ||
| Mesothelin | Preclinical | - | Gastric cancer | [158] |
| MUC1 | Clinical | Completed | Solid tumors | [179] |
| NKG2DL | Clinical | Active/Recruiting | Solid tumors | NCT05528341 |
| Recruiting | Solid tumors | NCT03415100 | ||
| ROBO1 | Clinical | Unknown | Solid tumors | NCT03940820 |
| VEGFR2 | Preclinical | - | Solid tumors | [180] |
| Target | Study Type | Clinical Trial Status | Cancer Type | Reference |
|---|---|---|---|---|
| AXL | Preclinical | - | Triple-negative breast cancers | [202] |
| B7-H3 | Clinical | Active, Not Recruiting | Solid tumors | NCT04483778 |
| Recruiting | Solid tumors | NCT04897321 | ||
| Recruiting | Solid tumors | NCT05190185 | ||
| Preclinical | - | Solid tumors | [194] | |
| - | Glioblastoma | [197] | ||
| - | Solid tumors | [198] | ||
| - | Prostate cancer | [199] | ||
| - | Solid tumors | [203] | ||
| B7-H3/CD70 | Preclinical | - | Solid tumors | [204] |
| CD126 | Preclinical | - | Solid tumors | [205] |
| CD16 | Preclinical | - | Solid tumors | [206] |
| CD70 | Clinical | Recruiting | Solid tumors | NCT02830724 |
| Preclinical | - | CD70+ cancers | [207] | |
| - | CD70+ cancers | [208] | ||
| c-Met | Clinical | Completed | Melanoma, breast carcinoma | [209] |
| Preclinical | - | Gastric cancer | [210] | |
| CSPG4 | Preclinical | - | Solid tumors | [211] |
| - | Solid tumors | [212] | ||
| - | Melanoma | [213] | ||
| GAS6 | Preclinical | - | Pancreatic cancer | [214] |
| GD2 | Clinical | Completed | Melanoma | [186,215] |
| Completed | Solid tumors | [216] | ||
| Completed | Neuroblastoma | [217] | ||
| Recruiting | Solid tumors | NCT03635632 | ||
| Completed | Solid tumors | NCT02107963 | ||
| Preclinical | - | Lung cancer | [185] | |
| - | Solid tumors | [189] | ||
| - | Solid tumors | [215] | ||
| GD3 | Preclinical | - | Melanoma | [218] |
| - | Melanoma | [219] | ||
| HER2 | Preclinical | - | HER2+ sarcomas | [29] |
| - | HER2+ melanoma subtypes | [220] | ||
| IL13Ra2 | Clinical | Recruiting | Solid tumors | NCT04119024 |
| Multiple targets (NY-ESO-1, DR5, EGFRvIII, c-Met) | Clinical | Unknown | Solid tumors | NCT03638206 |
| SLC45A2 | Preclinical | - | Melanoma | [221] |
| TRP-1 | Preclinical | - | Melanoma | [222] |
| VEGFR2 | Clinical | Terminated | Solid tumors | NCT01218867 |
| Preclinical | - | Solid tumors | [180] | |
| - | Solid tumors | [223] | ||
| αvβ3 | Preclinical | - | Glioma and glioblastoma | [224] |
| - | Advanced cancers | [225] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hibler, W.; Merlino, G.; Yu, Y. CAR NK Cell Therapy for the Treatment of Metastatic Melanoma: Potential & Prospects. Cells 2023, 12, 2750. https://doi.org/10.3390/cells12232750
Hibler W, Merlino G, Yu Y. CAR NK Cell Therapy for the Treatment of Metastatic Melanoma: Potential & Prospects. Cells. 2023; 12(23):2750. https://doi.org/10.3390/cells12232750
Chicago/Turabian StyleHibler, Winston, Glenn Merlino, and Yanlin Yu. 2023. "CAR NK Cell Therapy for the Treatment of Metastatic Melanoma: Potential & Prospects" Cells 12, no. 23: 2750. https://doi.org/10.3390/cells12232750
APA StyleHibler, W., Merlino, G., & Yu, Y. (2023). CAR NK Cell Therapy for the Treatment of Metastatic Melanoma: Potential & Prospects. Cells, 12(23), 2750. https://doi.org/10.3390/cells12232750







