Effect of Thyroxine on the Structural and Dynamic Features of Cardiac Mitochondria and Mitophagy in Rats
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals
2.2. Electron Microscopy
2.3. Quantification of the mRNA Expression of Mitochondrial Dynamics, Mitochondrial Biogenesis, and Mitophagy Genes Using Quantitative Real-Time PCR
2.4. Immunoblotting Analysis
2.5. Statistical Analysis
3. Results
3.1. Characterization of the Animals with Experimentally Induced Hyperthyroidism
3.2. Ultrastructural Features of Heart Tissue in the Control and Hyperthyroid Rats
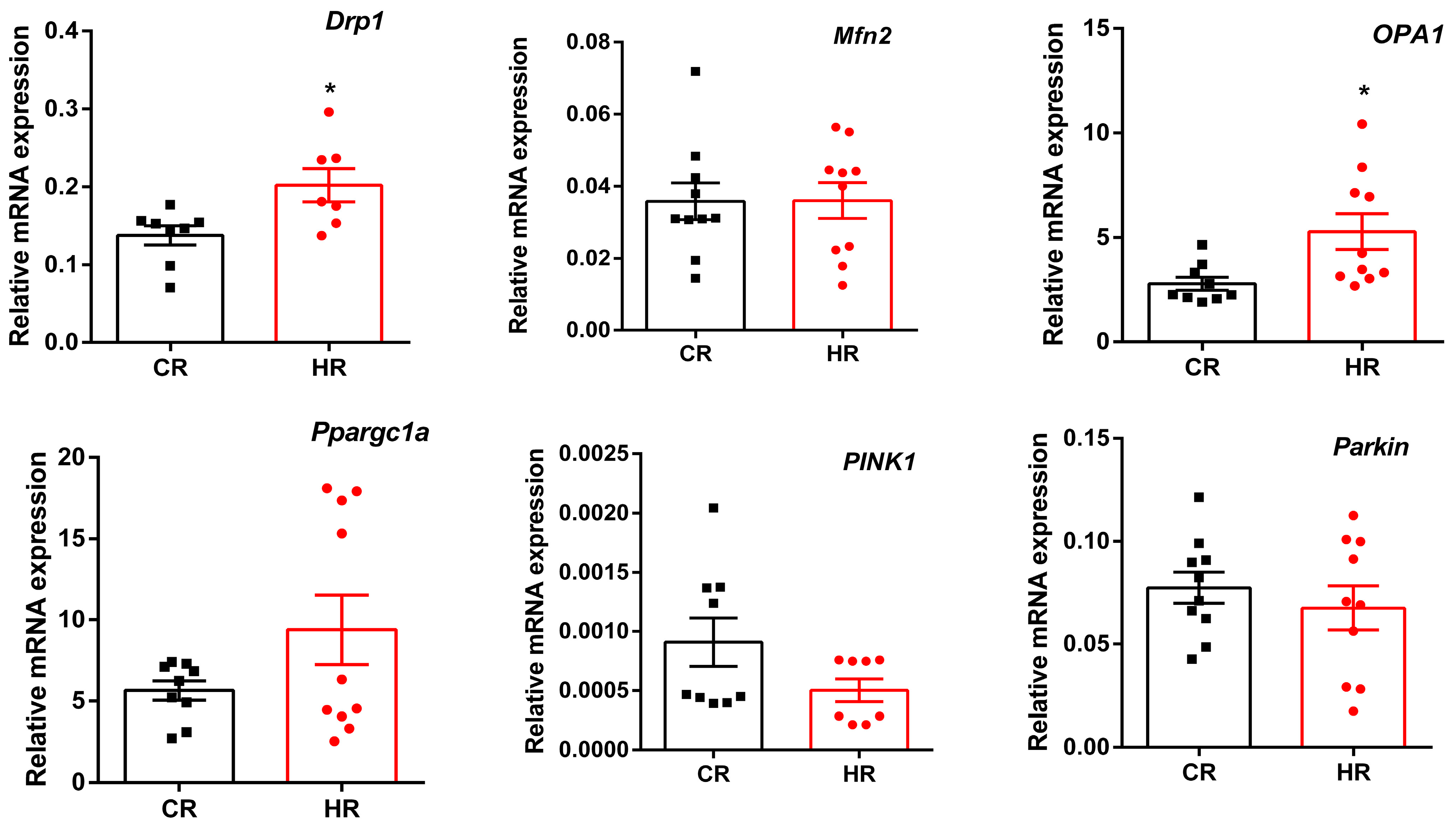
3.3. Gene Expression Differences in Heart Tissue of the Control and Hyperthyroid Rats
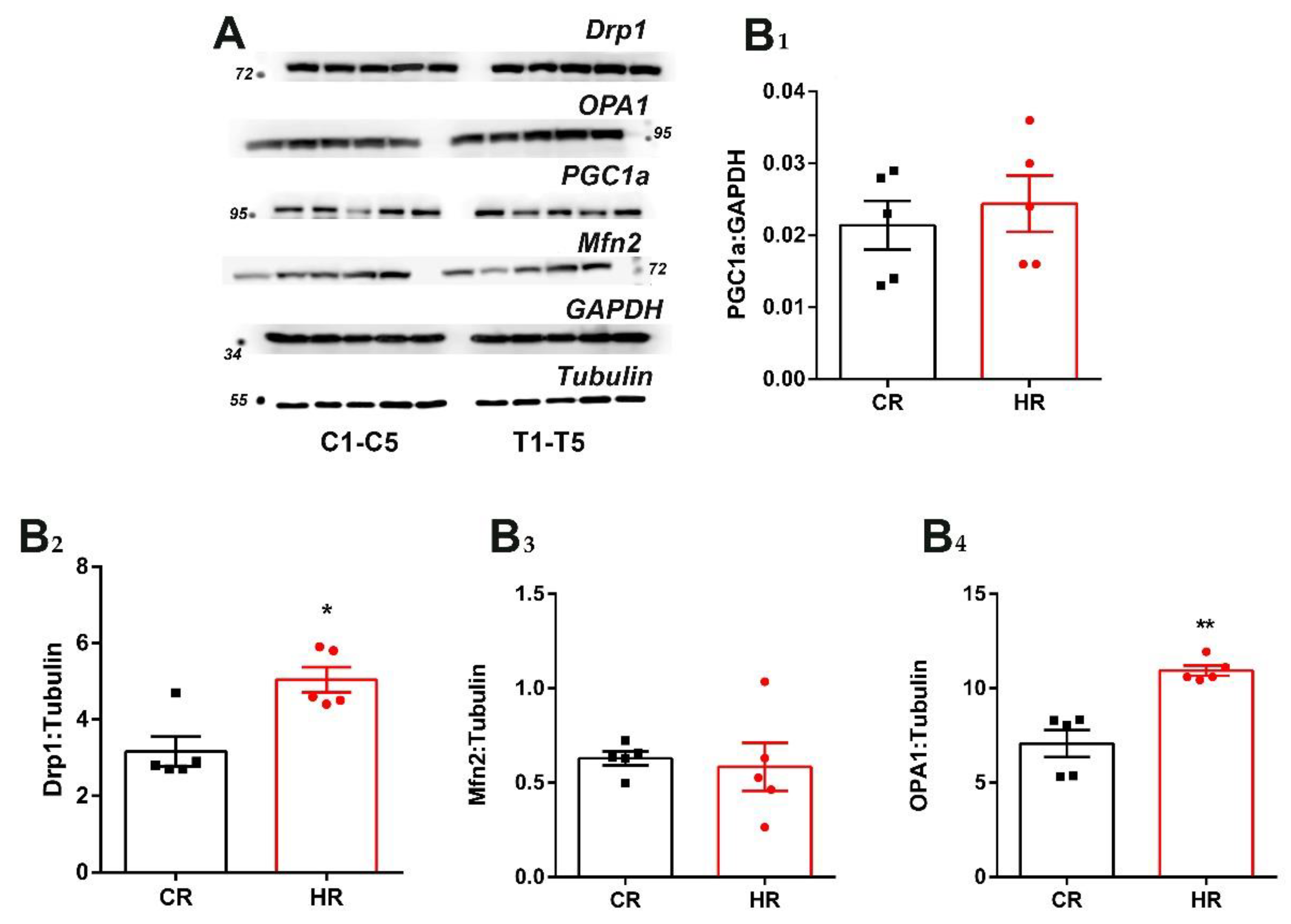
3.4. Immunoblotting Analysis of Heart Proteins of the Control and Hyperthyroid Rats
4. Discussion
- The effect of TH on the uncoupling of mitochondrial respiration.
- The effect of TH on the activity of mitochondrial respiratory chain complexes.
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cioffi, F.; Giacco, A.; Goglia, F.; Silvestri, E. Bioenergetic Aspects of Mitochondrial Actions of Thyroid Hormones. Cells 2022, 11, 997. [Google Scholar] [CrossRef] [PubMed]
- Twig, G.; Shirihai, O.S. The Interplay Between Mitochondrial Dynamics and Mitophagy. Antioxid. Redox Signal. 2011, 14, 1939–1951. [Google Scholar] [CrossRef] [PubMed]
- Aon, M.A.; Cortassa, S.; O’Rourke, B. Percolation and criticality in a mitochondrial network. Proc. Natl. Acad. Sci. USA 2004, 101, 4447–4452. [Google Scholar] [CrossRef]
- Frieden, M.; James, D.; Castelbou, C.; Danckaert, A.; Martinou, J.-C.; Demaurex, N. Ca2+ Homeostasis during Mitochondrial Fragmentation and Perinuclear Clustering Induced by hFis1. J. Biol. Chem. 2004, 279, 22704–22714. [Google Scholar] [CrossRef]
- Lee, Y.-J.; Jeong, S.-Y.; Karbowski, M.; Smith, C.L.; Youle, R.J. Roles of the Mammalian Mitochondrial Fission and Fusion Mediators Fis1, Drp1, and Opa1 in Apoptosis. Mol. Biol. Cell 2004, 15, 5001–5011. [Google Scholar] [CrossRef]
- Westermann, B. Bioenergetic role of mitochondrial fusion and fission. Biochim. Biophys. Acta (BBA) Bioenerg. 2012, 1817, 1833–1838. [Google Scholar] [CrossRef]
- Gao, F.; Zhang, J. Mitochondrial quality control and neurodegenerative diseases. Neuronal Signal. 2018, 2, NS20180062. [Google Scholar] [CrossRef]
- Silvestri, E.; Cioffi, F.; De Matteis, R.; Senese, R.; De Lange, P.; Coppola, M.; Salzano, A.M.; Scaloni, A.; Ceccarelli, M.; Goglia, F.; et al. 3,5-Diiodo-L-Thyronine Affects Structural and Metabolic Features of Skeletal Muscle Mitochondria in High-Fat-Diet Fed Rats Producing a Co-adaptation to the Glycolytic Fiber Phenotype. Front. Physiol. 2018, 9, 194. [Google Scholar] [CrossRef]
- Truban, D.; Hou, X.; Caulfield, T.R.; Fiesel, F.C.; Springer, W. PINK1, Parkin, and Mitochondrial Quality Control: What can we Learn about Parkinson’s Disease Pathobiology? J. Park. Dis. 2017, 7, 13–29. [Google Scholar] [CrossRef]
- Wang, Z.; Figueiredo-Pereira, C.; Oudot, C.; Vieira, H.; Brenner, C. Mitochondrion: A Common Organelle for Distinct Cell Deaths? Int. Rev. Cell Mol. Biol. 2017, 331, 245–287. [Google Scholar] [CrossRef]
- Ni, H.-M.; Williams, J.A.; Ding, W.-X. Mitochondrial dynamics and mitochondrial quality control. Redox Biol. 2015, 4, 6–13. [Google Scholar] [CrossRef] [PubMed]
- Venediktova, N.; Solomadin, I.; Starinets, V.; Mironova, G. Structural and Dynamic Features of Liver Mitochondria and Mitophagy in Rats with Hyperthyroidism. Int. J. Mol. Sci. 2022, 23, 14327. [Google Scholar] [CrossRef] [PubMed]
- Ye, J.; Coulouris, G.; Zaretskaya, I.; Cutcutache, I.; Rozen, S.; Madden, T.L. Primer-BLAST: A tool to design target-specific primers for polymerase chain reaction. BMC Bioinform. 2012, 13, 134. [Google Scholar] [CrossRef] [PubMed]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef] [PubMed]
- Lowry, O.H.; Rosebrough, N.J.; Farr, A.L.; Randall, R.J. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 1951, 193, 265–275. [Google Scholar] [CrossRef]
- Venediktova, N.; Solomadin, I.; Nikiforova, A.; Starinets, V.; Mironova, G. Functional State of Rat Heart Mitochondria in Experimental Hyperthyroidism. Int. J. Mol. Sci. 2021, 22, 11744. [Google Scholar] [CrossRef]
- Ferreira, P.J.; L’Abbate, C.; Abrahamsohn, P.A.; Gouveia, C.A.; Moriscot, A.S. Temporal and topographic ultrastructural alterations of rat heart myofibrils caused by thyroid hormone. Microsc. Res. Tech. 2003, 62, 451–459. [Google Scholar] [CrossRef]
- Dupont, N.; Orhon, I.; Bauvy, C.; Codogno, P. Autophagy and Autophagic Flux in Tumor Cells. Methods Enzymol. 2014, 543, 73–88. [Google Scholar] [CrossRef]
- Venditti, P.; Bari, A.; Di Stefano, L.; Di Meo, S. Role of mitochondria in exercise-induced oxidative stress in skeletal muscle from hyperthyroid rats. Arch. Biochem. Biophys. 2007, 463, 12–18. [Google Scholar] [CrossRef]
- Paradies, G.; Ruggiero, F.; Petrosillo, G.; Quagliariello, E. Enhanced cytochrome oxidase activity and modification of lipids in heart mitochondria from hyperthyroid rats. Biochim. Biophys. Acta 1994, 1225, 165–170. [Google Scholar] [CrossRef]
- Nesci, S.; Trombetti, F.; Pagliarani, A.; Ventrella, V.; Algieri, C.; Tioli, G.; Lenaz, G. Molecular and Supramolecular Structure of the Mitochondrial Oxidative Phosphorylation System: Implications for Pathology. Life 2021, 11, 242. [Google Scholar] [CrossRef] [PubMed]
- Lapuente-Brun, E.; Moreno-Loshuertos, R.; Acín-Pérezet, R.; Latorre-Pellicer, A.; Colás, C.; Balsa, E.; Perales-Clemente, E.; Quirós, P.M.; Calvo, E.; Rodríguez-Hernández, M.A.; et al. Supercomplex assembly determines electron flux in the mitochondrial electron transport chain. Science 2013, 340, 1567–1570. [Google Scholar] [CrossRef] [PubMed]
- Maranzana, E.; Barbero, G.; Falasca, A.I.; Lenaz, G.; Genova, M.L. Mitochondrial Respiratory Supercomplex Association Limits Production of Reactive Oxygen Species from Complex I. Antioxid. Redox Signal. 2013, 19, 1469–1480. [Google Scholar] [CrossRef]
- Schäfer, E.; Seelert, H.; Reifschneider, N.H.; Krause, F.; Dencher, N.A.; Vonck, J. Architecture of Active Mammalian Respiratory Chain Supercomplexes. J. Biol. Chem. 2006, 281, 15370–15375. [Google Scholar] [CrossRef] [PubMed]
- Pfeiffer, K.; Gohil, V.; Stuart, R.A.; Hunte, C.; Brandt, U.; Greenberg, M.L.; Schägger, H. Cardiolipin Stabilizes Respiratory Chain Supercomplexes. J. Biol. Chem. 2003, 278, 52873–52880. [Google Scholar] [CrossRef]
- Mileykovskaya, E.; Dowhan, W. Cardiolipin-dependent formation of mitochondrial respiratory supercomplexes. Chem. Phys. Lipids 2014, 179, 42–48. [Google Scholar] [CrossRef]
- Araujo, A.S.R.; Schenkel, P.; Enzveiler, A.T.; Fernandes, T.R.G.; A Partata, W.; Llesuy, S.; Ribeiro, M.F.M.; Khaper, N.; Singal, P.K.; Belló-Klein, A. The role of redox signaling in cardiac hypertrophy induced by experimental hyperthyroidism. J. Mol. Endocrinol. 2008, 41, 423–430. [Google Scholar] [CrossRef]
- Venditti, P.; Bari, A.; Di Stefano, L.; Di Meo, S. Tri-iodothyronine treatment differently affects liver metabolic response and oxidative stress in sedentary and trained rats. J. Endocrinol. 2008, 197, 65–74. [Google Scholar] [CrossRef]
- Venediktova, N.I.; Mashchenko, O.V.; Talanov, E.Y.; Belosludtseva, N.V.; Mironova, G.D. Energy metabolism and oxidative status of rat liver mitochondria in conditions of experimentally induced hyperthyroidism. Mitochondrion 2020, 52, 190–196. [Google Scholar] [CrossRef]
- Trofimova, A.V.; Chizh, N.A.; Belochkina, I.; Marchenko, L.; Govorukha, T.; Repin, N.; Sandomirsky, B.P. Cardiomyocyte Ultrastructure of Rats with Experimental Myocardial Infarction After Therapeutic Hypothermia and Mesenchymal Stromal Cell Administration. Probl. Cryobiol. Cryomed. 2017, 27, 334–347. [Google Scholar] [CrossRef]
- Oztay, F.; Ergin, B.; Ustunova, S.; Balci, H.; Kapucu, A.; Caner, M.; Demirci, C. Effects of coenzyme Q10 on the heart ultrastructure and nitric oxide synthase during hyperthyroidism. Chin. J. Physiol. 2007, 50, 217–224. [Google Scholar] [PubMed]
- Hsieh, C.; Li, C.; Hsu, C.; Chen, H.; Chen, Y.; Liu, Y.; Kuo, H.; Liu, P. Mitochondrial protection by simvastatin against angiotensin II-mediated heart failure. Br. J. Pharmacol. 2019, 176, 3791–3804. [Google Scholar] [CrossRef] [PubMed]
- Cook, K.L.; Soto-Pantoja, D.R.; Abu-Asab, M.; Clarke, P.A.; Roberts, D.D.; Clarke, R. Mitochondria directly donate their membrane to form autophagosomes during a novel mechanism of parkin-associated mitophagy. Cell Biosci. 2014, 4, 16. [Google Scholar] [CrossRef]
- Eldarov, C.M.; Vangely, I.M.; Vays, V.B.; Sheval, E.V.; Holtze, S.; Hildebrandt, T.B.; Kolosova, N.G.; Popkov, V.A.; Plotnikov, E.Y.; Zorov, D.B.; et al. Mitochondria in the Nuclei of Rat Myocardial Cells. Cells 2020, 9, 712. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Hu, W.; Song, X.; Zhao, Y. Generation of Multipotent Stem Cells from Adult Human Peripheral Blood Following the Treatment with Platelet-Derived Mitochondria. Cells 2020, 9, 1350. [Google Scholar] [CrossRef] [PubMed]
- Santillo, A.; Burrone, L.; Falvo, S.; Senese, R.; Lanni, A.; Baccari, G.C. Triiodothyronine induces lipid oxidation and mitochondrial biogenesis in rat Harderian gland. J. Endocrinol. 2013, 219, 69–78. [Google Scholar] [CrossRef]
- Bahi, L.; Garnier, A.; Fortin, D.; Serrurier, B.; Veksler, V.; Bigard, A.; Ventura-Clapier, R. Differential effects of thyroid hormones on energy metabolism of rat slow- and fast-twitch muscles. J. Cell. Physiol. 2005, 203, 589–598. [Google Scholar] [CrossRef]
- Tondera, D.; Grandemange, S.; Jourdain, A.; Karbowski, M.; Mattenberger, Y.; Herzig, S.; Da Cruz, S.; Clerc, P.; Raschke, I.; Merkwirth, C.; et al. SLP-2 is required for stress-induced mitochondrial hyperfusion. EMBO J. 2009, 28, 1589–1600. [Google Scholar] [CrossRef]
- Rambold, A.S.; Kostelecky, B.; Elia, N.; Lippincott-Schwartz, J. Tubular network formation protects mitochondria from autophagosomal degradation during nutrient starvation. Proc. Natl. Acad. Sci. USA 2011, 108, 10190–10195. [Google Scholar] [CrossRef]
- Gomes, L.C.; Di Benedetto, G.; Scorrano, L. During autophagy mitochondria elongate, are spared from degradation and sustain cell viability. Nat. Cell Biol. 2011, 13, 589–598. [Google Scholar] [CrossRef]
- Cadete, V.J.J.; Deschênes, S.; Cuillerier, A.; Brisebois, F.; Sugiura, A.; Vincent, A.; Turnbull, D.; Picard, M.; McBride, H.M.; Burelle, Y. Formation of mitochondrial-derived vesicles is an active and physiologically relevant mitochondrial quality control process in the cardiac system. J. Physiol. 2016, 594, 5343–5362. [Google Scholar] [CrossRef] [PubMed]
- Peng, T.; Xie, Y.; Sheng, H.; Wang, C.; Lian, Y.; Xie, N. Mitochondrial-derived vesicles: Gatekeepers of mitochondrial response to oxidative stress. Free Radic. Biol. Med. 2022, 188, 185–193. [Google Scholar] [CrossRef] [PubMed]
- Lewis, P.; Pérez-Carrión, M.; Piccoli, G. Chapter 6: Autophagy. In Parkinson’s Disease: Molecular Mechanisms Underlying Pathology; Academic Press: Cambridge, MA, USA, 2017; pp. 179–206. [Google Scholar] [CrossRef]




| Gene | Forward (5′→3′) | Reverse (5′→3′) |
|---|---|---|
| Drp1 | GATCCAGATGGGCGCAGAAC | ATGTCCAGTTGGCTCCTGTT |
| Mfn2 | AGCGTCCTCTCCCTCTGACA | TTCCACACCACTCCTCCGAC |
| OPA1 | GCAGAAGACAGCTTGAGGGT | TGCGTCCCACTGTTGCTTAT |
| PINK1 | GATGTGGAATATCTCGGCAGGA | TGTTTGCTGAACCCAAGGCT |
| Parkin | GGCCAGAGGAAAGTCACCTG | CACCCGGTATGCCTGAGAAG |
| Ppargc1α | TGACATAGAGTGTGCTGCCC | GCTGTCTGTGTCCAGGTCAT |
| Actb | GACCCAGATCATGTTTGAGACCT | CCAGAGGCATACAGGGACAAC |
| CR | HR | |
|---|---|---|
| T3 free, pmol/L | 5.2 ± 0.1 | 9.3 ± 1.2 *** |
| T4 free, pmol/L | 19.2 ± 1.0 | 66.2 ± 4.4 *** |
| Body weight, g | 259 ± 2.3 | 235 ± 3.1 *** |
| Body weight gain, g | 37 ± 2.0 | 15 ± 2 *** |
| Heart weight, g | 0.9 ± 0.02 | 1.25 ± 0.03 *** |
| Heart/body weight (×103) | 3.5 ± 0.06 | 5 ± 0.13 *** |
| CR | HR | |
|---|---|---|
| Number of mitochondria per image (48 µm2) | 38.3 ± 3.5 | 30.3 ± 2.2 |
| Number of swollen mitochondria relative to the total number of mitochondria, % | 8 | 55 |
| Number of swollen mitochondria with MLBs relative to the total number of mitochondria, % | - | 3 |
| Number of damaged mitochondria relative to the total number of mitochondria, % | 2.5 | 13 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Venediktova, N.; Solomadin, I.; Starinets, V. Effect of Thyroxine on the Structural and Dynamic Features of Cardiac Mitochondria and Mitophagy in Rats. Cells 2023, 12, 396. https://doi.org/10.3390/cells12030396
Venediktova N, Solomadin I, Starinets V. Effect of Thyroxine on the Structural and Dynamic Features of Cardiac Mitochondria and Mitophagy in Rats. Cells. 2023; 12(3):396. https://doi.org/10.3390/cells12030396
Chicago/Turabian StyleVenediktova, Natalya, Ilya Solomadin, and Vlada Starinets. 2023. "Effect of Thyroxine on the Structural and Dynamic Features of Cardiac Mitochondria and Mitophagy in Rats" Cells 12, no. 3: 396. https://doi.org/10.3390/cells12030396
APA StyleVenediktova, N., Solomadin, I., & Starinets, V. (2023). Effect of Thyroxine on the Structural and Dynamic Features of Cardiac Mitochondria and Mitophagy in Rats. Cells, 12(3), 396. https://doi.org/10.3390/cells12030396






