Oxalic Acid Inhibits Feeding Behavior of the Brown Planthopper via Binding to Gustatory Receptor Gr23a
Abstract
:1. Introduction
2. Material and Methods
2.1. Insect and Rice Culture
2.2. Chemical Sources
2.3. Ligand Determination for NlGr23a
2.3.1. NlGr23a cDNA Cloning and Cell Line Construction
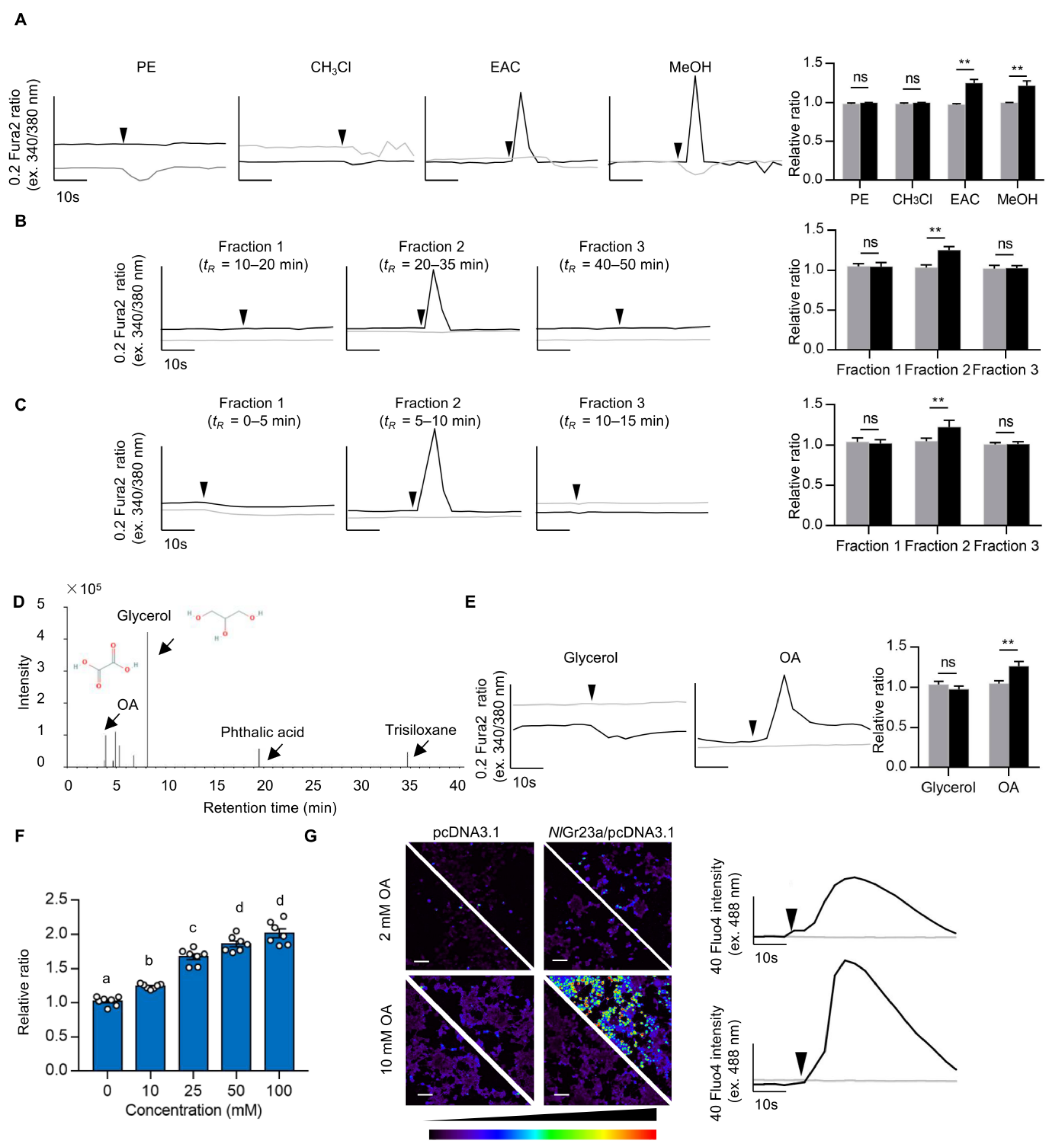
2.3.2. Isolation and Identification of Active Compounds in Rice
2.4. Calcium Imaging Assay
2.5. RNAi Treatments
2.5.1. dsRNA Preparation and Injection into the BPH
2.5.2. qRT-PCR Analysis
2.6. Behavioral Experiments
2.6.1. Feeding Bioassays of BPHs on Rice Plants
2.6.2. Feeding Bioassays of BPHs on Artificial Diets
2.7. Surgeries
2.8. Immunohistochemistry and Microscopy
2.9. Statistics
3. Results
3.1. Oxalic Acid Is a Ligand of NlGr23a
3.2. NlGr23a Is Required for Perception of Oxalic Acid in Rice Plants
3.3. NlGr23a Mediated the Antifeedant Activity of Oxalic Acid in Artificial Diets
3.4. Localization of NlGr23a in the Labium
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Prokopy, R.J.; Collier, R.H.; Finch, S. Leaf color used by cabbage root flies to distinguish among host plants. Science 1983, 221, 190–192. [Google Scholar] [CrossRef] [PubMed]
- Chapman, R.F. Contact chemoreception in feeding by phytophagous insects. Annu. Rev. Entomol. 2003, 48, 455–484. [Google Scholar] [CrossRef] [PubMed]
- Khan, Z.R.; Midega, C.A.O.; Pittchar, J.O.; Murage, A.W.; Birkett, M.A.; Bruce, T.J.A.; Pickett, J.A. Achieving food security for one million sub-Saharan African poor through push-pull innovation by 2020. Phil. Trans. R. Soc. B 2014, 369, 20120284. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cook, S.M.; Khan, Z.R.; Pickett, J.A. The use of push-pull strategies in integrated pest management. Annu. Rev. Entomol. 2007, 52, 375–400. [Google Scholar] [CrossRef] [Green Version]
- Douglas, A.E. Strategies for enhanced crop resistance to insect pests. Annu. Rev. Plant Biol. 2018, 69, 637–660. [Google Scholar] [CrossRef] [Green Version]
- Yoshihara, T.; Sogawa, K.; Pathak, M.D.; Juliano, B.O.; Sakamura, S. Oxalic acid as a sucking inhibitor of the brown planthopper in rice (Delphacidae, Homoptera). Entomol. Exp. Appl. 1980, 27, 149–155. [Google Scholar] [CrossRef]
- Massonie, G. Breeding of a biotype of Myzus persicae Sulzer on a synthetic medium. V. Influence of oxalic and gentisic acids on the nutritive value of a synthetic medium. Ann. Nutr. Aliment. 1980, 34, 139–146. [Google Scholar]
- Karatolos, N.; Hatcher, P.E. The effect of acetylsalicylic acid and oxalic acid on Myzus persicae and Aphidius colemani. Entomol. Exp. Appl. 2009, 130, 98–105. [Google Scholar] [CrossRef]
- Zhang, Z.F.; Cui, B.Y.; Zhang, Y. Electrical penetration graphs indicate that tricin is a key secondary metabolite of rice, inhibiting phloem feeding of brown planthopper, Nilaparvata lugens. Entomol. Exp. Appl. 2015, 156, 14–27. [Google Scholar] [CrossRef]
- Scott, K. Gustatory processing in Drosophila melanogaster. Annu. Rev. Entomol. 2018, 63, 15–30. [Google Scholar] [CrossRef]
- Liman, E.R.; Zhang, Y.V.; Montell, C. Peripheral coding of taste. Neuron 2014, 81, 984–1000. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.C.D.; Dahanukar, A. Recent advances in the genetic basis of taste detection in Drosophila. Cell. Mol. Life Sci. 2020, 77, 1087–1101. [Google Scholar] [CrossRef]
- Kim, H.; Kim, H.; Kwon, J.Y.; Seo, J.T.; Shin, D.M.; Moon, S.J. Drosophila Gr64e mediates fatty acid sensing via the phospholipase C pathway. PLoS Genet. 2018, 14, e1007229. [Google Scholar] [CrossRef] [Green Version]
- Stanley, M.; Ghosh, B.; Weiss, Z.F.; Christiaanse, J.; Gordon, M.D. Mechanisms of lactic acid gustatory attraction in Drosophila. Curr. Biol. 2021, 31, 3525–3537.e6. [Google Scholar] [CrossRef]
- Mi, T.W.; Mack, J.O.; Lee, C.M.; Zhang, Y.V. Molecular and cellular basis of acid taste sensation in Drosophila. Nat. Commun. 2021, 12, 3730. [Google Scholar] [CrossRef]
- Ganguly, A.; Chandel, A.; Turner, H.; Wang, S.; Liman, E.R.; Montell, C. Requirement for an Otopetrin-like protein for acid taste in Drosophila. Proc. Natl. Acad. Sci. USA 2021, 118, e2110641118. [Google Scholar] [CrossRef]
- Sogawa, K. The rice brown planthopper: Feeding physiology and host plant interactions. Annu. Rev. Entomol. 1982, 27, 49–73. [Google Scholar] [CrossRef]
- Foster, S.; Goodman, L.J.; Duckett, J.G. Ultrastructure of sensory receptors on the labium of the rice brown planthopper. Cell Tissue Res. 1983, 230, 353–366. [Google Scholar] [CrossRef]
- Foster, S.; Goodman, L.J.; Duckett, J.G. Sensory receptors associated with the stylets and cibarium of the rice brown planthopper, Nilapavarta lugens. Cell Tissue Res. 1983, 232, 111–119. [Google Scholar] [CrossRef]
- Kang, K.; Yang, P.; Chen, L.E.; Pang, R.; Yu, L.J.; Zhou, W.W.; Zhu, Z.R.; Zhang, W.Q. Identification of putative fecundity-related gustatory receptor genes in the brown planthopper Nilaparvata lugens. BMC Genom. 2018, 19, 970. [Google Scholar] [CrossRef]
- Chen, W.W.; Kang, K.; Lv, J.; Yue, L.; Zhang, W.Q. Galactose-NlGr11 inhibits AMPK phosphorylation by activating the PI3K-AKT-PKF-ATP signaling cascade via insulin receptor and Gβγ. Insect Sci. 2021, 28, 735–745. [Google Scholar] [CrossRef] [PubMed]
- Ojha, A.; Zhang, W. Characterization of gustatory receptor 7 in the brown planthopper reveals functional versatility. Insect Biochem. Mol. Biol. 2021, 132, 103567. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Chen, L.; Li, D.; Kang, K.; Liu, K.; Yue, L.; Zhang, W. Two alternative splicing variants of a sugar gustatory receptor modulate fecundity through different signalling pathways in the brown planthopper, Nilaparvata lugens. J. Insect Physiol. 2019, 119, 103966. [Google Scholar] [CrossRef]
- Kang, K.; Yue, L.; Xia, X.; Liu, K.; Zhang, W.Q. Comparative metabolomics analysis of different resistant rice varieties in response to the brown planthopper Nilaparvata lugens Hemiptera: Delphacidae. Metabolomics 2019, 15, 62. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nelson, G.; Hoon, M.A.; Chandrashekar, J.; Zhang, Y.F.; Ryba, N.J.P.; Zuker, C.S. Mammalian sweet taste receptors. Cell 2001, 106, 381–390. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sato, K.; Tanaka, K.; Touhara, K. Sugar-regulated cation channel formed by an insect gustatory receptor. Proc. Natl. Acad. Sci. USA 2011, 108, 11680–11685. [Google Scholar] [CrossRef] [Green Version]
- Chen, J.; Liang, Z.K.; Liang, Y.K.; Pang, R.; Zhang, W.Q. Conserved microRNAs miR-8-5p and miR-2a-3p modulate chitin biosynthesis in response to 20-hydroxyecdysone signaling in the brown planthopper, Nilaparvata lugens. Insect Biochem. Mol. Biol. 2013, 43, 839–848. [Google Scholar] [CrossRef]
- Sogawa, K.; Pathak, M.D. Mechanisms of brown planthopper resistance in Mudgo variety of rice (Hemiptera: Delphacidae). Appl. Entomol. Zool. 1970, 5, 145–158. [Google Scholar] [CrossRef] [Green Version]
- Yu, L.; Jiang, J.Z.; Zhang, C.; Jiang, L.R.; Ye, N.H.; Lu, Y.S.; Yang, G.Z.; Liu, E.; Peng, C.L.; He, Z.H.; et al. Glyoxylate rather than ascorbate is an efficient precursor for oxalate biosynthesis in rice. J. Exp. Bot. 2010, 61, 1625–1634. [Google Scholar] [CrossRef] [Green Version]
- Fu, Q.; Zhang, Z.T.; Hu, C.; Lai, F.X.; Sun, Z.X. A chemically defined diet enables continuous rearing of the brown planthopper, Nilaparvata lugens (Stal) (Homoptera: Delphacidae). Appl. Entomol. Zool. 2001, 36, 111–116. [Google Scholar] [CrossRef]
- Sang, J.; Rimal, S.; Lee, Y. Gustatory receptor 28b is necessary for avoiding saponin in Drosophila melanogaster. EMBO Rep. 2019, 20, e47328. [Google Scholar] [CrossRef]
- Lee, Y.; Kim, S.H.; Montell, C. Avoiding DEET through insect gustatory receptors. Neuron 2010, 67, 555–561. [Google Scholar] [CrossRef] [Green Version]
- Aljunid, S.F.; Anderson, M. Ultrastructure of sensilla on the antennal pedicel of the brown planthopper Nilaparvata lugens Stal (Insecta: Homoptera). Cell Tissue Res. 1983, 228, 313–322. [Google Scholar] [CrossRef]
- Mang, D.Z.; Shu, M.; Endo, H.; Yoshizawa, Y.; Nagata, S.; Kikuta, S.; Sato, R. Expression of a sugar clade gustatory receptor, BmGr6, in the oral sensory organs, midgut, and central nervous system of larvae of the silkworm Bombyx mori. Insect Biochem. Mol. Biol. 2016, 70, 85–98. [Google Scholar] [CrossRef]
- Zindler-Frank, E. Oxalate biosynthesis in relation to photosynthetic pathway and plant productivity: A survey. Z. Pflanzenphysiol. 1976, 80, 1–13. [Google Scholar] [CrossRef]
- Zhang, G.R.; Zhang, W.Q.; Lian, B.; Gu, L.Q.; Zhou, Q.; Liu, T.X. Insecticidal effects of extracts from two rice varieties to brown planthopper, Nilaparvata lugens. J. Chem. Ecol. 1999, 25, 1843–1853. [Google Scholar] [CrossRef]
- Wu, B.Q.; Huang, S.S.; Li, C.; Sun, Z.X.; Zhou, J.L.; Ling, Y.; Jiang, X.B.; Huang, Q.; Long, L.P.; Huang, F.K. Effect of environmental factor on secondary substances of rice planting and relationship with resistance to brown planthopper (Homoptera: Delphacidae). Southwest China J. Agric. Sci. 2016, 29, 2371–2378. [Google Scholar]
- Moon, S.J.; Kottgen, M.; Jiao, Y.C.; Xu, H.; Montell, C. A taste receptor required for the caffeine response in vivo. Curr. Biol. 2006, 16, 1812–1817. [Google Scholar] [CrossRef] [Green Version]
- Poudel, S.; Kim, Y.; Kim, Y.T.; Lee, Y. Gustatory receptors required for sensing umbelliferone in Drosophila melanogaster. Insect Biochem. Mol. Biol. 2015, 66, 110–118. [Google Scholar] [CrossRef]
- Poudel, S.; Kim, Y.; Kwak, J.S.; Jeong, S.; Lee, Y. Gustatory receptor 22e is essential for sensing chloroquine and strychnine in Drosophila melanogaster. Insect Biochem. Mol. Biol. 2017, 88, 30–36. [Google Scholar] [CrossRef]
- Shim, J.; Lee, Y.; Jeong, Y.T.; Kim, Y.; Lee, M.G.; Montell, C.; Moon, S.J. The full repertoire of Drosophila gustatory receptors for detecting an aversive compound. Nat. Commun. 2015, 6, 8867. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ozaki, K.; Ryuda, M.; Yamada, A.; Utoguchi, A.; Ishimoto, H.; Calas, D.; Marion-Poll, F.; Tanimura, T.; Yoshikawa, H. A gustatory receptor involved in host plant recognition for oviposition of a swallowtail butterfly. Nat. Commun. 2011, 2, 542. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, W.; Papanicolaou, A.; Zhang, H.J.; Anderson, A. Expansion of a bitter taste receptor family in a polyphagous insect herbivore. Sci. Rep. 2016, 6, 23666. [Google Scholar] [CrossRef] [Green Version]
- Rimal, S.; Sang, J.; Poudel, S.; Thakur, D.; Montell, C.; Lee, Y. Mechanism of acetic acid gustatory repulsion in Drosophila. Cell Rep. 2019, 26, 1432–1442.e4. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.; Amrein, H. Ionotropic receptors mediate Drosophila oviposition preference through sour gustatory receptor neurons. Curr. Biol. 2017, 27, 2741–2750.e4. [Google Scholar] [CrossRef] [Green Version]
- Ahn, J.E.; Chen, Y.; Amrein, H. Molecular basis of fatty acid taste in Drosophila. Elife 2017, 6, e30115. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kang, K.; Zhang, M.; Yue, L.; Chen, W.; Dai, Y.; Lin, K.; Liu, K.; Lv, J.; Guan, Z.; Xiao, S.; et al. Oxalic Acid Inhibits Feeding Behavior of the Brown Planthopper via Binding to Gustatory Receptor Gr23a. Cells 2023, 12, 771. https://doi.org/10.3390/cells12050771
Kang K, Zhang M, Yue L, Chen W, Dai Y, Lin K, Liu K, Lv J, Guan Z, Xiao S, et al. Oxalic Acid Inhibits Feeding Behavior of the Brown Planthopper via Binding to Gustatory Receptor Gr23a. Cells. 2023; 12(5):771. https://doi.org/10.3390/cells12050771
Chicago/Turabian StyleKang, Kui, Mengyi Zhang, Lei Yue, Weiwen Chen, Yangshuo Dai, Kai Lin, Kai Liu, Jun Lv, Zhanwen Guan, Shi Xiao, and et al. 2023. "Oxalic Acid Inhibits Feeding Behavior of the Brown Planthopper via Binding to Gustatory Receptor Gr23a" Cells 12, no. 5: 771. https://doi.org/10.3390/cells12050771
APA StyleKang, K., Zhang, M., Yue, L., Chen, W., Dai, Y., Lin, K., Liu, K., Lv, J., Guan, Z., Xiao, S., & Zhang, W. (2023). Oxalic Acid Inhibits Feeding Behavior of the Brown Planthopper via Binding to Gustatory Receptor Gr23a. Cells, 12(5), 771. https://doi.org/10.3390/cells12050771





