Functional Analysis of Pheromone Biosynthesis Activating Neuropeptide Receptor Isoforms in Maruca vitrata
Abstract
1. Introduction
2. Materials and Methods
2.1. Insects
2.2. Cloning of PBANR Isoforms
2.3. Phylogenetic Analysis of Lepidopteran PBANR Isoforms
2.4. Developmental Stage and Tissue-Specific Expression of PBANR Isoforms
2.5. Transformation for In Vitro Heterologous Expression of MviPBANR Isoforms
2.6. Confocal Microscopy for Calcium Assay
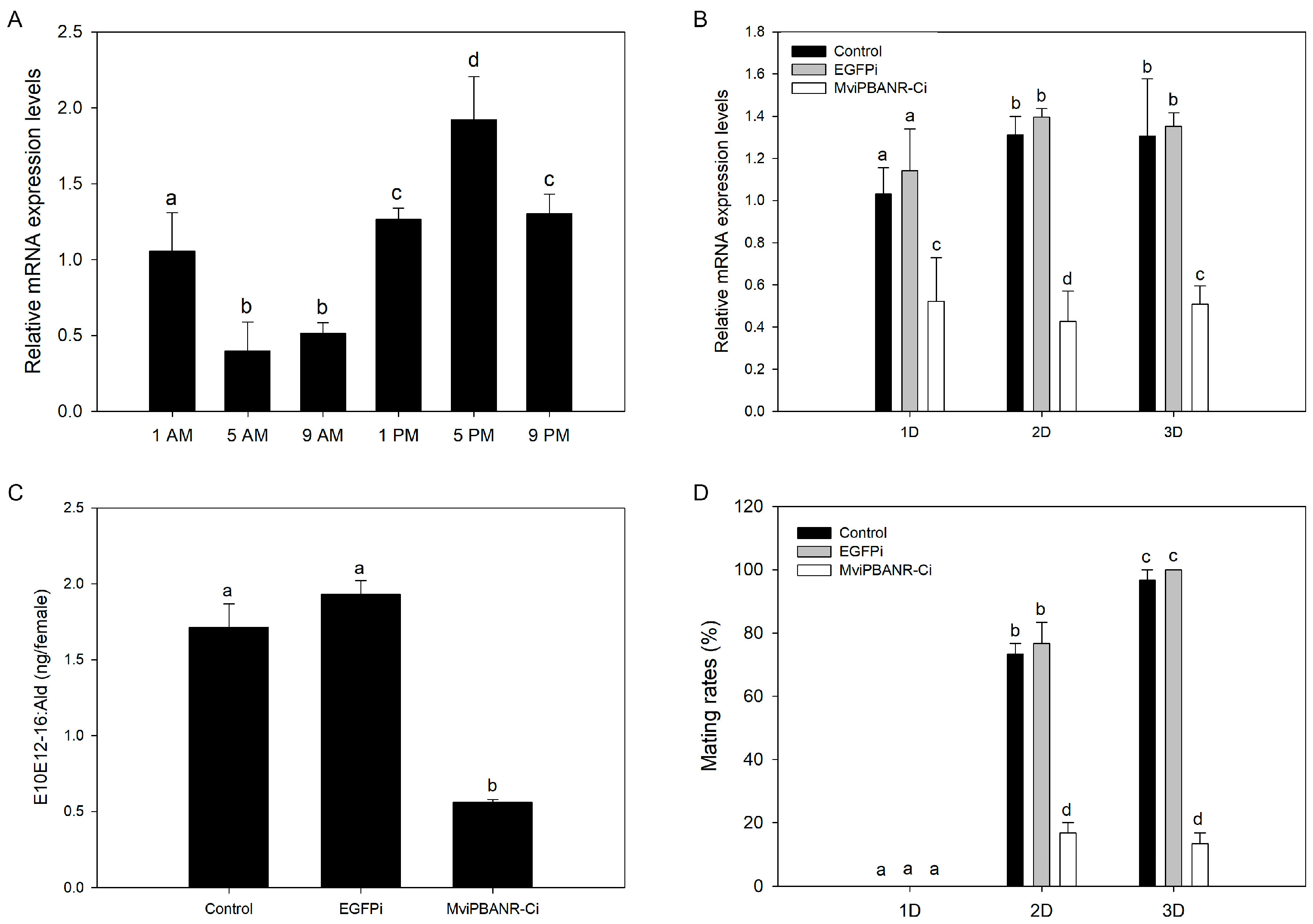
2.7. Measurement of the Diurnal Expression Level of MviPBANR-C
2.8. Suppression of MviPBANR-C by RNAi
2.9. Gas Chromatography (GC) Analysis
2.10. Mating Rates after Suppression of MviPBANR-C by RNAi
2.11. Statistical Analysis
3. Results
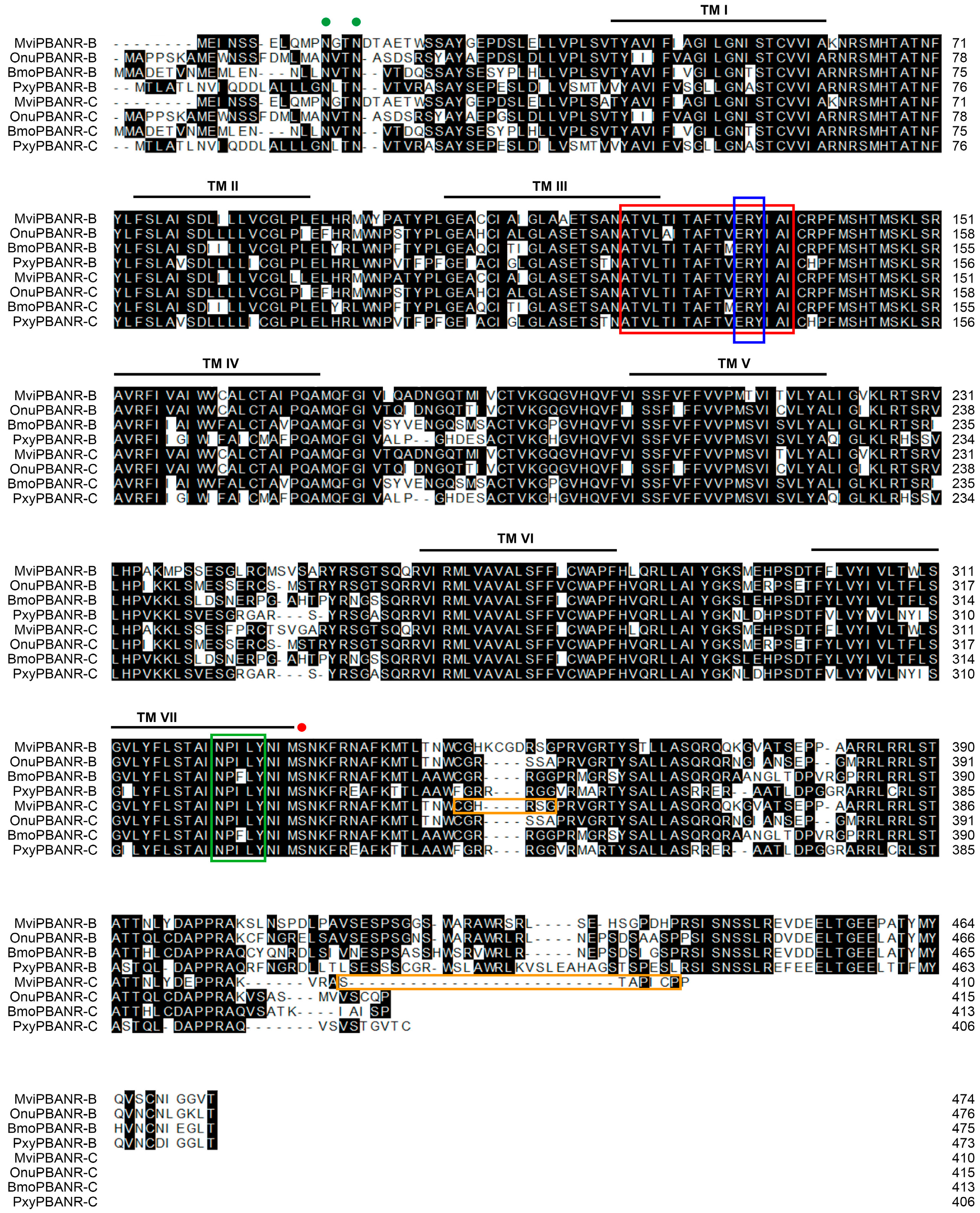
3.1. Cloning of PBANR Isoforms from Pheromone Gland of M. vitrata
3.2. Developmental Stage and Tissue-Specific Expression of PBANR Isoforms in M. vitrata
3.3. In Vitro Expression of PBANR Isoforms
3.4. Change of Pheromone Production and Mating Behavior by Suppression of MviPBANR-C in M. vitrata
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- De Pasqual, C.; Groot, A.T.; Mappes, J.; Burdfield-Steel, E. Evolutionary importance of intraspecific variation in sex pheromones. Trends Ecol. Evol. 2021, 36, 848–859. [Google Scholar] [CrossRef] [PubMed]
- Stökl, J.; Steiger, S. Evolutionary origin of insect pheromones. Curr. Opin. Insect Sci. 2017, 24, 36–42. [Google Scholar] [CrossRef] [PubMed]
- Jurenka, R. Regulation of pheromone biosynthesis in moths. Curr. Opin. Insect Sci. 2017, 24, 29–35. [Google Scholar] [CrossRef]
- Fujii, T.; Yasukochi, Y.; Rong, Y.; Matsuo, T.; Ishikawa, Y. Multiple Δ11-desaturase genes selectively used for sex pheromone biosynthesis are conserved in Ostrinia moth genomes. Insect Biochem. Mol. Biol. 2015, 61, 62–68. [Google Scholar] [CrossRef]
- He, P.; Zhang, Y.F.; Hong, D.Y.; Wang, J.; Wang, X.L.; Zuo, L.H.; Tang, X.F.; Xu, W.M.; He, M. A reference gene set for sex pheromone biosynthesis and degradation genes from the diamondback moth, Plutella xylostella, based on genome and transcriptome digital gene expression analyses. BMC Genom. 2017, 18, 219. [Google Scholar] [CrossRef][Green Version]
- Závodská, R.; Fexová, S.; Wowern, G.; Han, G.B.; Dolezel, D.; Sauman, I. Is the sex communication of two pyralid moths, Plodia interpunctella and Ephestia kuehniella, under circadian clock regulation? J. Biol. Rhythm. 2012, 27, 206–216. [Google Scholar] [CrossRef]
- Foster, S.P. Reinvestigation of sex pheromone biosynthesis in the moth Trichoplusia ni reveals novel quantitative control mechanisms. Insect Biochem. Mol. Biol. 2022, 140, 103700. [Google Scholar] [CrossRef] [PubMed]
- Rafaeli, A. Pheromone biosynthesis activating neuropeptide (PBAN): Regulatory role and mode of action. Gen. Comp. Endocrinol. 2009, 162, 69–78. [Google Scholar] [CrossRef]
- Jurenka, R.; Rafaeli, A. Regulatory role of PBAN in sex pheromone biosynthesis of Heliothine moths. Front. Endocrinol. 2011, 2, 46. [Google Scholar] [CrossRef][Green Version]
- Tsfadia, O.; Azrielli, A.; Falach, L.; Zada, A.; Roelofs, W.; Rafaeli, A. Pheromone biosynthetic pathways: PBAN-regulated rate-limiting steps and differential expression of desaturase genes in moth species. Insect Biochem. Mol. Biol. 2008, 38, 552–567. [Google Scholar] [CrossRef]
- Choi, M.Y.; Jurenka, R.A. Role of extracellular Ca2+ and calcium channel activated by a G protein-coupled receptor regulating pheromone production in Helicoverpa zea (Lepidoptera: Noctuidae). Ann. Entomol. Soc. Am. 2006, 99, 905–909. [Google Scholar] [CrossRef][Green Version]
- Choi, M.Y.; Fuerst, E.J.; Rafaeli, A.; Jurenka, R. Role of extracellular domains in PBAN/pyrokinin GPCRs from insects using chimera receptors. Insect Biochem. Mol. Biol. 2007, 37, 296–306. [Google Scholar] [CrossRef] [PubMed]
- Choi, M.Y.; Fuerst, E.J.; Rafaeli, A.; Jurenka, R. Identification of a G protein coupled receptor for pheromone biosynthesis activating neuropeptide from pheromone glands of the moth Helicoverpa zea. Proc. Natl. Acad. Sci. USA 2003, 100, 9721–9726. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Hull, J.J.; Ohnishi, A.; Moto, K.; Kawasaki, Y.; Kurata, R.; Suzuki, M.G.; Matsumoto, S. Cloning and characterization of the pheromone biosynthesis activating neuropeptide receptor from the silkmoth, Bombyx mori. Significance of the carboxyl terminus in receptor internalization. J. Biol. Chem. 2004, 279, 51500–51507. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Cheng, Y.; Luo, L.; Jiang, X.; Zhang, L.; Niu, C. Expression of pheromone biosynthesis activating neuropeptide and its receptor (PBANR) mRNA in adult female Spodoptera exigua (Lepidoptera: Noctuidae). Arch. Insect Biochem. Physiol. 2010, 75, 13–27. [Google Scholar] [CrossRef]
- Lee, D.W.; Shrestha, S.; Kim, A.Y.; Park, S.J.; Yang, C.Y.; Kim, Y.; Koh, Y.H. RNA interference of pheromone biosynthesis-activating neuropeptide receptor suppresses mating behavior by inhibiting sex pheromone production in Plutella xylostella (L.). Insect Biochem. Mol. Biol. 2011, 41, 236–243. [Google Scholar] [CrossRef]
- Jiang, L.; Zhang, F.; Hou, Y.; Thakur, K.; Hu, F.; Zhang, J.G.; Jiang, X.F.; Liu, Y.Q.; Wei, Z.J. Isolation and functional characterization of the pheromone biosynthesis activating neuropeptide receptor of Chinese oak silkworm, Antheraea pernyi. Int. J. Biol. Macromol. 2018, 117, 42–50. [Google Scholar] [CrossRef]
- Kim, Y.-J.; Nachman, R.J.; Aimanova, K.; Gill, S.; Adams, M.E. The pheromone biosynthesis activating neuropeptide (PBAN) receptor of Heliothis virescens: Identification, functional expression, and structure-activity relationships of ligand analogs. Peptides 2008, 29, 268–275. [Google Scholar] [CrossRef][Green Version]
- Lee, J.M.; Hull, J.J.; Kawai, T.; Goto, C.; Kurihara, M.; Tanokura, M.; Nagata, K.; Nagasawa, H.; Matsumoto, S. Re-evaluation of the PBAN receptor molecule: Characterization of PBANR variants expressed in the pheromone glands of moths. Front. Endocrinol. 2012, 3, 6. [Google Scholar] [CrossRef][Green Version]
- Fodor, J.; Hull, J.J.; Köblös, G.; Jacquin-Joly, E.; Szlanka, T.; Fónagy, A. Identification and functional characterization of the pheromone biosynthesis activating neuropeptide receptor isoforms from Mamestra brassicae. Gen. Comp. Endocrinol. 2018, 258, 60–693. [Google Scholar] [CrossRef][Green Version]
- Zhou, J.; Zhang, N.; Wang, P.; Zhang, S.; Li, D.; Liu, K.; Wang, G.; Wang, X.; Ai, H. Identification of host-plant volatiles and characterization of two novel general odorant-binding proteins from the legume pod borer, Maruca vitrata Fabricius (Lepidoptera: Crambidae). PLoS ONE 2015, 10, e0141208. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Srinivasan, R.; Tamò, M.; Malini, P. Emergence of Maruca vitrata as a major pest of food legumes and evolution of management practices in Asia and Africa. Annu. Rev. Entomol. 2021, 66, 141–161. [Google Scholar] [CrossRef]
- Schläger, S.; Beran, F.; Groot, A.T.; Ulrichs, C.; Veit, D.; Paetz, C.; Karumuru, B.R.; Srinivasan, R.; Schreiner, M.; Mewis, I. Pheromone blend analysis and cross-attraction among populations of Maruca vitrata from Asia and West Africa. J. Chem. Ecol. 2015, 41, 1155–1162. [Google Scholar] [CrossRef]
- Cha, W.H.; Kim, W.J.; Jung, J.K.; Lee, D.-W. Putative pheromone biosynthesis pathway in Maruca vitrata by transcriptomic analysis. J. Asia-Pac. Entomol. 2017, 20, 165–173. [Google Scholar] [CrossRef]
- Cha, W.H.; Jung, J.K.; Kim, Y.; Lee, D.-W. Identification and pheromonotropic activity of pheromone biosynthesis activating neuropeptide in Maruca vitrata. J. Asia-Pac. Entomol. 2018, 21, 156–160. [Google Scholar] [CrossRef]
- Cha, W.H.; Lee, D.-W. Suppression of pheromone biosynthesis and mating behavior by RNA interference of pheromone gland-specific fatty acyl reductase in Maruca vitrata. Insect Sci. 2022, 29, 1135–1144. [Google Scholar] [CrossRef] [PubMed]
- Jung, J.K.; Seo, B.Y.; Park, J.H.; Moon, J.K.; Choi, B.S.; Lee, Y.H. Developmental characteristics of soybean podworm, Matsumuraeses phaseoli (Lepidoptera: Tortricidae) and legume pod borer, Maruca vitrata (Lepidoptera: Pyraidae) on semi-synthetic artificial diets. Korean J. Appl. Entomol. 2007, 46, 393–399. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knvaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across computing platform. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2002, 25, 402–408. [Google Scholar] [CrossRef]
- Yang, S.; Hu, Y.; Cheng, Z.; Rice, J.H.; Miao, L.; Ma, J.; Hewezi, T.; Li, Y.; Gai, J. An efficient Agrobacterium-mediated soybean transformation method using green fluorescent protein as a selectable marker. Plant Signal. Behav. 2019, 14, 1612682. [Google Scholar] [CrossRef]
- Závodská, R.; von Wowern, G.; Löfstedt, C.; Rosén, W.; Sauman, I. The release of a pheromonotropic neuropeptide, PBAN, in the turnip moth Agrotis segetum, exhibits a circadian rhythm. J. Insect Physiol. 2009, 55, 435–440. [Google Scholar] [CrossRef]
- Wheatley, M.; Hawtin, S.R.; Wesley, V.J.; Howard, H.C.; Simms, J.; Miles, A.; McEwan, K.; Parslow, R.A. Agonist binding to peptide hormone receptors. Biochem. Soc. Trans. 2003, 31, 35–39. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Hull, J.J.; Lee, J.M.; Matsumoto, S. Identification of specific sites in the thirdintracellular loop and carboxyl terminus of the Bombyx mori pheromone biosynthesis activating neuropeptide receptor crucial for ligand-inducedinternalization. Insect Mol. Biol. 2011, 20, 801–811. [Google Scholar] [CrossRef] [PubMed]
- Matsumoto, S.; Ozawa, R.; Nagamine, T.; Kim, G.H.; Uchiumi, K.; Shono, T.; Mitsui, T. Intracellular transduction in the regulation of pheromone biosynthesis of the silkworm, Bombyx mori: Suggested involvement of calmodulin and phosphoprotein phosphatase. Biosci. Biotechnol. Biochem. 1995, 59, 560–562. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Sakurai, N.; Moriya, K.; Suzuki, T.; Sofuku, K.; Mochiki, H.; Nishimura, O.; Utsumi, T. Detection of co- and posttranslational protein N-myristoylation by metabolic labeling in an insect cell-free protein synthesis system. Anal. Biochem. 2007, 362, 236–244. [Google Scholar] [CrossRef]
- Ramasamy, M.; Damaj, M.B.; Vargas-Bautista, C.; Mora, V.; Liu, J.; Padilla, C.S.; Irigoyen, S.; Saini, T.; Sahoo, N.; DaSilva, J.A.; et al. A Sugarcane G-protein-coupled receptor, ShGPCR1, confers tolerance to multiple abiotic stresses. Front. Plant. Sci. 2021, 12, 745891. [Google Scholar] [CrossRef]
- Luo, M.; Zhou, X.C.; Wang, Z.; Chen, J.X.; Chung, H.; Wei, H.Y. Identification and gene expression analysis of the pheromone biosynthesis activating neuropeptide receptor (PBANR) from the Ostrinia furnacalis (Lepidoptera: Pyralidae). J. Insect Sci. 2019, 19, 25. [Google Scholar] [CrossRef]
- Nusawardani, T.; Kroemer, J.A.; Choi, M.Y.; Jurenka, R.A. Identification and characterization of the pyrokinin/pheromone biosynthesis activating neuropeptide family of G protein-coupled receptors from Ostrinia nubilalis. Insect Mol. Biol. 2013, 22, 331–340. [Google Scholar] [CrossRef]
- Bober, R.; Rafaeli, A. Gene-silencing reveals the functional significance of pheromone biosynthesis activating neuropeptide receptor (PBAN-R) in a male moth. Proc. Natl. Acad. Sci. USA 2010, 107, 16858–16862. [Google Scholar] [CrossRef][Green Version]
- Zheng, L.; Lytle, C.; Njauw, C.N.; Altstein, M.; Martins-Green, M. Cloning and characterization of the pheromone biosynthesis activating neuropeptide receptor gene in Spodoptera littoralis larvae. Gene 2007, 393, 20–30. [Google Scholar] [CrossRef]
- Gu, S.H.; Chow, Y.S.; O’Reilly, D.R. Role of calcium in the stimulation of ecdysteroidogenesis by recombinant prothoracicotropic hormone in the prothoracic glands of the silkworm, Bombyx mori. Insect Biochem. Mol. Biol. 1998, 28, 861–867. [Google Scholar] [CrossRef]
- Lin, J.L.; Gu, S.H. In vitro and in vivo stimulation of extracellular signalregulated kinase (ERK) by the prothoracicotropic hormone in prothoracic gland cells and its developmental regulation in the silkworm, Bombyx mori. J. Insect Physiol. 2007, 53, 622–631. [Google Scholar] [CrossRef]
- Iga, M.; Nakaoka, T.; Suzuki, Y.; Kataoka, H. Pigment dispersing factor regulates ecdysone biosynthesis via bombyx neuropeptide G protein coupled receptor-B2 in the prothoracic glands of Bombyx mori. PLoS ONE 2014, 9, e103239. [Google Scholar] [CrossRef] [PubMed]
- Dhyani, V.; Gare, S.; Gupta, R.K.; Swain, S.; Venkatesh, K.V.; Giri, L. GPCR mediated control of calcium dynamics: A systems perspective. Cell Signal. 2020, 74, 109717. [Google Scholar] [CrossRef]
- Matsumoto, S.; Hull, J.J.; Ohnishi, A. Molecular mechanisms underlying PBAN signaling in the silkmoth Bombyx mori. Ann. N. Y. Acad. Sci. 2009, 1163, 464–468. [Google Scholar] [CrossRef] [PubMed]
- Zhu, K.Y.; Palli, S.R. Mechanisms, Applications, and Challenges of Insect RNA Interference. Annu. Rev. Entomol. 2020, 65, 293–311. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Park, Y.; Vatanparast, M. Suppression of PBAN receptor expression reduces fecundity in the fall armyworm, Spodoptera frugiperda. Arch. Insect Biochem. Physiol. 2022, 110, e21897. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cha, W.H.; Kim, B.; Lee, D.-W. Functional Analysis of Pheromone Biosynthesis Activating Neuropeptide Receptor Isoforms in Maruca vitrata. Cells 2023, 12, 1410. https://doi.org/10.3390/cells12101410
Cha WH, Kim B, Lee D-W. Functional Analysis of Pheromone Biosynthesis Activating Neuropeptide Receptor Isoforms in Maruca vitrata. Cells. 2023; 12(10):1410. https://doi.org/10.3390/cells12101410
Chicago/Turabian StyleCha, Wook Hyun, Boyun Kim, and Dae-Weon Lee. 2023. "Functional Analysis of Pheromone Biosynthesis Activating Neuropeptide Receptor Isoforms in Maruca vitrata" Cells 12, no. 10: 1410. https://doi.org/10.3390/cells12101410
APA StyleCha, W. H., Kim, B., & Lee, D.-W. (2023). Functional Analysis of Pheromone Biosynthesis Activating Neuropeptide Receptor Isoforms in Maruca vitrata. Cells, 12(10), 1410. https://doi.org/10.3390/cells12101410







