The Heparan Sulfate Proteoglycan Syndecan-1 Triggers Breast Cancer Cell-Induced Coagulability by Induced Expression of Tissue Factor
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. siRNA Transfection
2.3. Cytokine, Inhibitor and TFPI Treatment of Breast Cancer Cells
2.4. Quantitative Real-Time PCR
2.5. Protein Extraction and Western Blot
2.6. MTT Metabolic Cell Viability Assay
2.7. Cell Cycle Analysis
2.8. Apoptosis Assay
2.9. Scratch Migration Assay
2.10. Migration Assay
2.11. Light Transmission Aggregometry
2.12. Thrombin Generation Assay
2.13. Flow Cytometric Analysis of TF Expression
2.14. STRING Protein–Protein Interaction Analysis
2.15. Statistical Analysis
3. Results
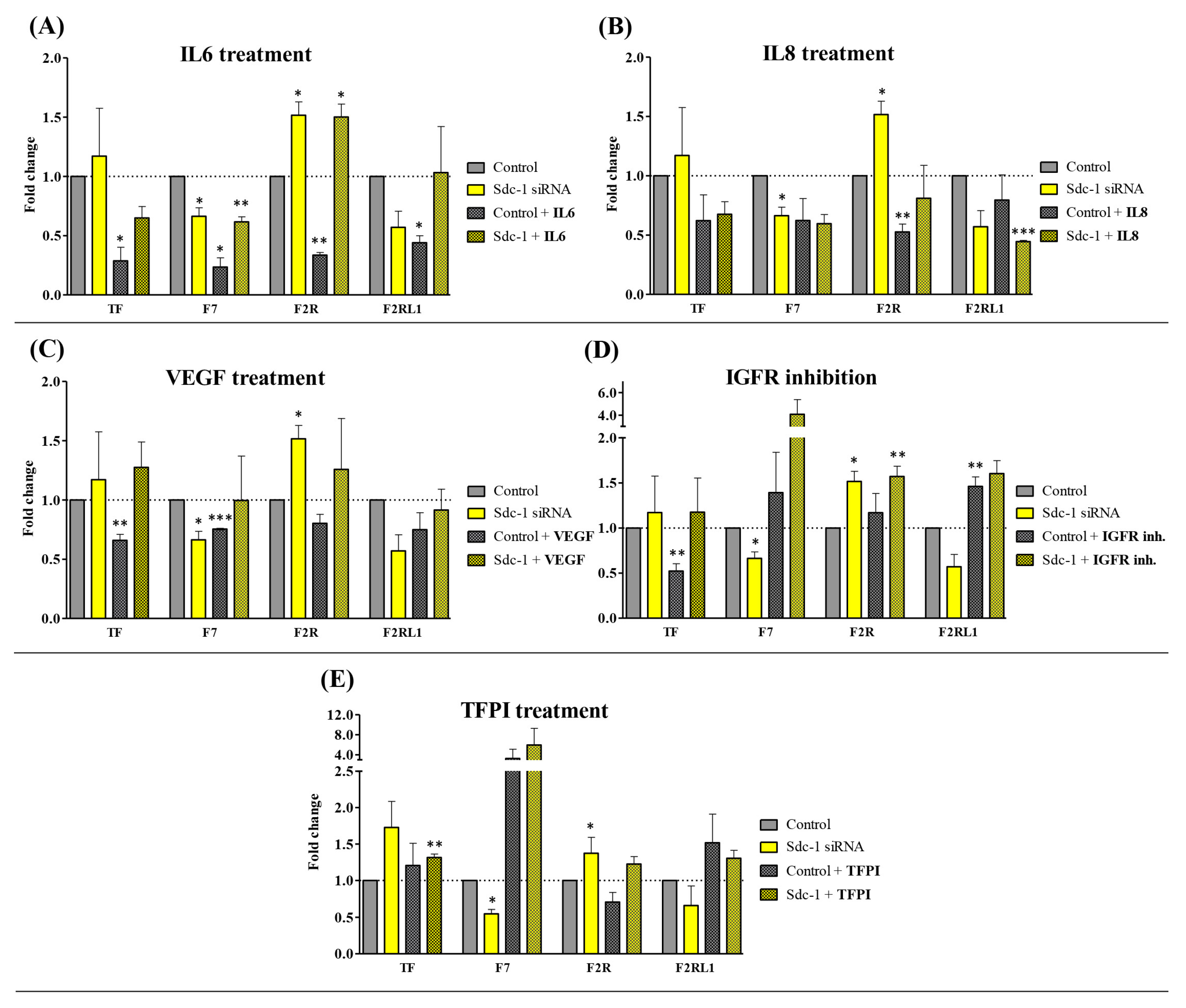
3.1. Sdc-1 Knockdown Modulates Cytokine-Dependent Tissue Factor (TF) Pathway Gene Expression in MDA-MB-231 Breast Cancer Cells
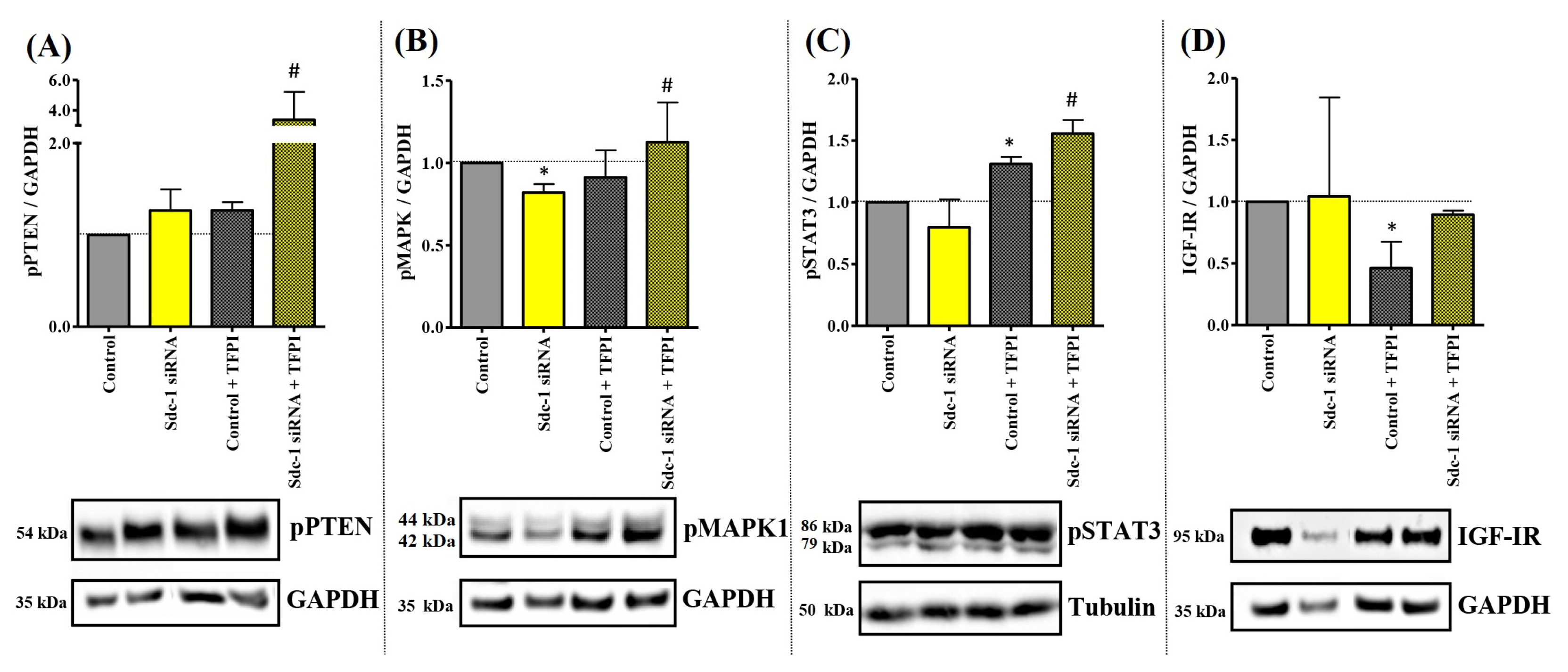
3.2. Sdc-1-KD in MDA-MB-231 Cells Is Associated with a Differential Kinase Activation Profile of Downstream Targets of TF Signaling
3.3. TFPI Treatment Exerts Different Effects on Cell Proliferation, Cell Cycle, and Invasiveness with Respect to Sdc-1-KD in MDA-MB-231 Cells
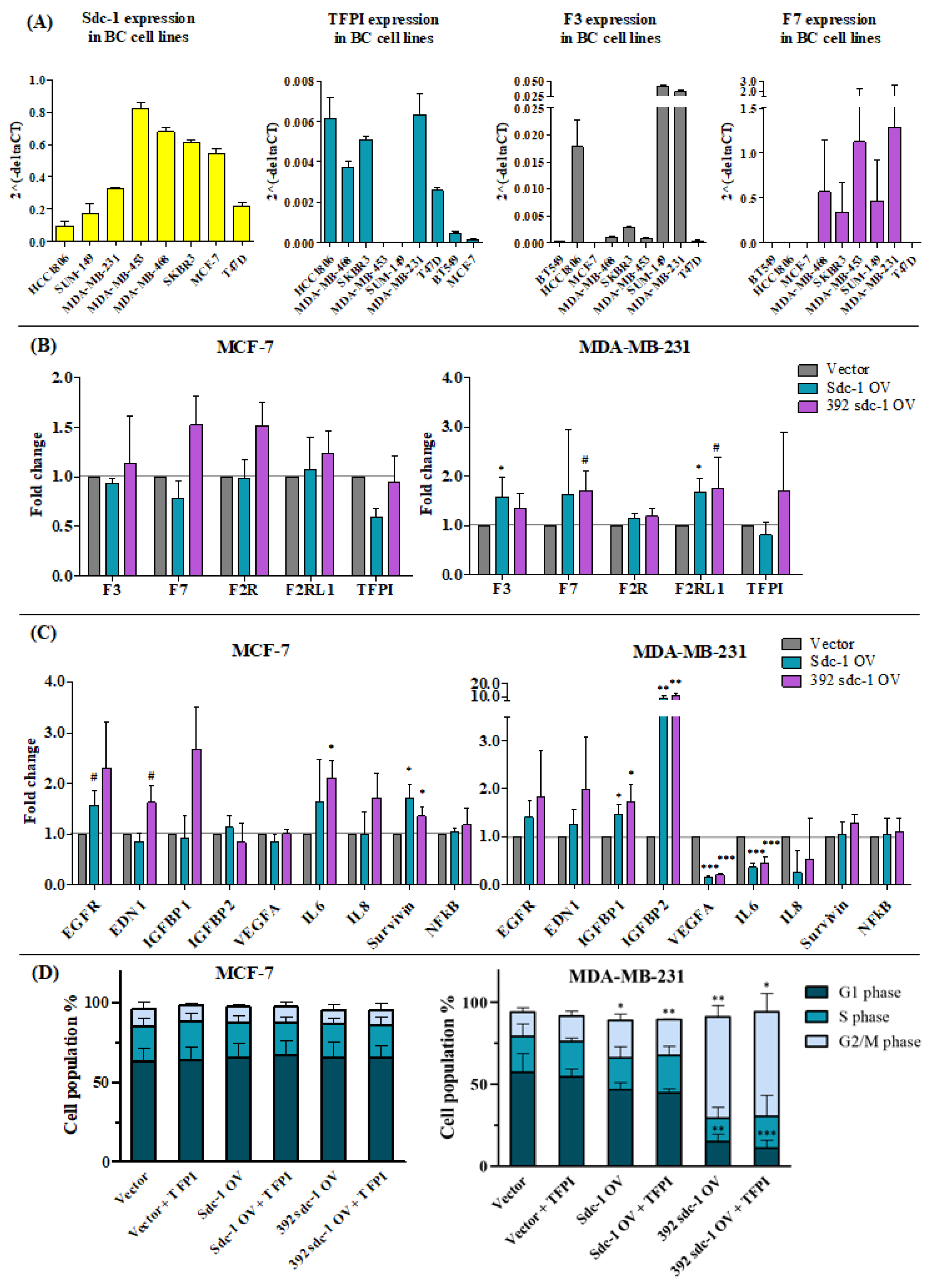
3.4. The Expression of TF and TFPI in Different Breast Cancer Cell Lines Represents Different Molecular Classifications
3.5. Sdc-1 Overexpression Affects Tissue Factor Pathway-Related Genes and Their Downstream Signaling in MCF-7 and MDA-MB-231 Cells
3.6. Sdc-1 Overexpression Influences the Protein Expression of TF, Platelet Aggregation, and Thrombin Generation in MCF-7 and MDA-MB-231 Cells
3.7. Sdc-1 Is Indirectly Linked to the TF Pathway via In Silico Interaction as a Coreceptor with Various Growth Factors, Cytokines, and Chemokines
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Karamanos, N.K.; Piperigkou, Z.; Theocharis, A.D.; Watanabe, H.; Franchi, M.; Baud, S.; Brézillon, S.; Götte, M.; Passi, A.; Vigetti, D.; et al. Proteoglycan Chemical Diversity Drives Multifunctional Cell Regulation and Therapeutics. Chem. Rev. 2018, 118, 9152–9232. [Google Scholar] [CrossRef]
- Hassan, N.; Greve, B.; Espinoza-Sánchez, N.A.; Götte, M. Cell-surface heparan sulfate proteoglycans as multifunctional integrators of signaling in cancer. Cell. Signal. 2021, 77, 109822. [Google Scholar] [CrossRef]
- Ibrahim, S.A.; Gadalla, R.; El-Ghonaimy, E.A.; Samir, O.; Mohamed, H.T.; Hassan, H.; Greve, B.; El-Shinawi, M.; Mohamed, M.M.; Götte, M. Syndecan-1 is a novel molecular marker for triple negative inflammatory breast cancer and modulates the cancer stem cell phenotype via the IL-6/STAT3, Notch and EGFR signaling pathways. Mol. Cancer 2017, 16, 57. [Google Scholar] [CrossRef]
- Ibrahim, S.A.; Yip, G.W.; Stock, C.; Pan, J.-W.; Neubauer, C.; Poeter, M.; Pupjalis, D.; Koo, C.Y.; Kelsch, R.; Schüle, R.; et al. Targeting of syndecan-1 by microRNA miR-10b promotes breast cancer cell motility and invasiveness via a Rho-GTPase- and E-cadherin-dependent mechanism. Int. J. Cancer 2012, 131, E884–E896. [Google Scholar] [CrossRef]
- Nikolova, V.; Koo, C.-Y.; Ibrahim, S.A.; Wang, Z.; Spillmann, D.; Dreier, R.; Kelsch, R.; Fischgräbe, J.; Smollich, M.; Rossi, L.H.; et al. Differential roles for membrane-bound and soluble syndecan-1 (CD138) in breast cancer progression. Carcinogenesis 2009, 30, 397–407. [Google Scholar] [CrossRef]
- Sheta, M.; Götte, M. Syndecan-1 (CD138) as a Pathogenesis Factor and Therapeutic Target in Breast Cancer. Curr. Med. Chem. 2021, 28, 5066–5083. [Google Scholar] [CrossRef]
- Yang, N.; Mosher, R.; Seo, S.; Beebe, D.; Friedl, A. Syndecan-1 in breast cancer stroma fibroblasts regulates extracellular matrix fiber organization and carcinoma cell motility. Am. J. Pathol. 2011, 178, 325–335. [Google Scholar] [CrossRef]
- Chute, C.; Yang, X.; Meyer, K.; Yang, N.; O’Neil, K.; Kasza, I.; Eliceiri, K.; Alexander, C.; Friedl, A. Syndecan-1 induction in lung microenvironment supports the establishment of breast tumor metastases. Breast Cancer Res. 2018, 20, 66. [Google Scholar] [CrossRef]
- Götte, M.; Kersting, C.; Radke, I.; Kiesel, L.; Wülfing, P. An expression signature of syndecan-1 (CD138), E-cadherin and c-met is associated with factors of angiogenesis and lymphangiogenesis in ductal breast carcinoma in situ. Breast Cancer Res. 2007, 9, R8. [Google Scholar] [CrossRef]
- Maeda, T.; Desouky, J.; Friedl, A. Syndecan-1 expression by stromal fibroblasts promotes breast carcinoma growth in vivo and stimulates tumor angiogenesis. Oncogene 2006, 25, 1408–1412. [Google Scholar] [CrossRef]
- Nassar, E.; Hassan, N.; El-Ghonaimy, E.A.; Hassan, H.; Abdullah, M.S.; Rottke, T.V.; Kiesel, L.; Greve, B.; Ibrahim, S.A.; Götte, M. Syndecan-1 Promotes Angiogenesis in Triple-Negative Breast Cancer through the Prognostically Relevant Tissue Factor Pathway and Additional Angiogenic Routes. Cancers 2021, 13, 2318. [Google Scholar] [CrossRef]
- Hassan, N.; Efing, J.; Kiesel, L.; Bendas, G.; Götte, M. The Tissue Factor Pathway in Cancer: Overview and Role of Heparan Sulfate Proteoglycans. Cancers 2023, 15, 1524. [Google Scholar] [CrossRef] [PubMed]
- Gay, L.J.; Felding-Habermann, B. Contribution of platelets to tumour metastasis. Nat. Rev. Cancer 2011, 11, 123–134. [Google Scholar] [CrossRef] [PubMed]
- Kopp, H.-G.; Placke, T.; Salih, H.R. Platelet-derived transforming growth factor-beta down-regulates NKG2D thereby inhibiting natural killer cell antitumor reactivity. Cancer Res. 2009, 69, 7775–7783. [Google Scholar] [CrossRef]
- Gockel, L.M.; Ponert, J.M.; Schwarz, S.; Schlesinger, M.; Bendas, G. The Low Molecular Weight Heparin Tinzaparin Attenuates Platelet Activation in Terms of Metastatic Niche Formation by Coagulation-Dependent and Independent Pathways. Molecules 2018, 23, 2753. [Google Scholar] [CrossRef] [PubMed]
- Hassan, N.; Rutsch, N.; Győrffy, B.; Espinoza-Sánchez, N.A.; Götte, M. SETD3 acts as a prognostic marker in breast cancer patients and modulates the viability and invasion of breast cancer cells. Sci. Rep. 2020, 10, 2262. [Google Scholar] [CrossRef]
- Valla, S.; Hassan, N.; Vitale, D.L.; Madanes, D.; Spinelli, F.M.; Teixeira, F.C.O.B.; Greve, B.; Espinoza-Sánchez, N.A.; Cristina, C.; Alaniz, L.; et al. Syndecan-1 Depletion Has a Differential Impact on Hyaluronic Acid Metabolism and Tumor Cell Behavior in Luminal and Triple-Negative Breast Cancer Cells. Int. J. Mol. Sci. 2021, 22, 5874. [Google Scholar] [CrossRef]
- Schwarz, S.; Schlesinger, M.; Bendas, G. Detection of Tumor Cell-Induced Platelet Aggregation and Granule Secretion. Methods Mol. Biol. 2021, 2294, 181–195. [Google Scholar] [CrossRef]
- Schwarz, S.; Gockel, L.M.; Naggi, A.; Barash, U.; Gobec, M.; Bendas, G.; Schlesinger, M. Glycosaminoglycans as Tools to Decipher the Platelet Tumor Cell Interaction: A Focus on P-Selectin. Molecules 2020, 25, 1039. [Google Scholar] [CrossRef] [PubMed]
- Wan, J.; Konings, J.; de Laat, B.; Hackeng, T.M.; Roest, M. Added Value of Blood Cells in Thrombin Generation Testing. Thromb. Haemost. 2021, 121, 1574–1587. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef]
- Dai, X.; Cheng, H.; Bai, Z.; Li, J. Breast Cancer Cell Line Classification and Its Relevance with Breast Tumor Subtyping. J. Cancer 2017, 8, 3131–3141. [Google Scholar] [CrossRef] [PubMed]
- Hilgers, K.; Ibrahim, S.A.; Kiesel, L.; Greve, B.; Espinoza-Sánchez, N.A.; Götte, M. Differential Impact of Membrane-Bound and Soluble Forms of the Prognostic Marker Syndecan-1 on the Invasiveness, Migration, Apoptosis, and Proliferation of Cervical Cancer Cells. Front. Oncol. 2022, 12, 803899. [Google Scholar] [CrossRef]
- Haschemi, R.; Gockel, L.M.; Bendas, G.; Schlesinger, M. A Combined Activity of Thrombin and P-Selectin Is Essential for Platelet Activation by Pancreatic Cancer Cells. Int. J. Mol. Sci. 2021, 22, 3323. [Google Scholar] [CrossRef]
- Mummery, R.S.; Rider, C.C. Characterization of the heparin-binding properties of IL-6. J. Immunol. 2000, 165, 5671–5679. [Google Scholar] [CrossRef]
- Schneider, C.; Kässens, N.; Greve, B.; Hassan, H.; Schüring, A.N.; Starzinski-Powitz, A.; Kiesel, L.; Seidler, D.G.; Götte, M. Targeting of syndecan-1 by micro-ribonucleic acid miR-10b modulates invasiveness of endometriotic cells via dysregulation of the proteolytic milieu and interleukin-6 secretion. Fertil. Steril. 2013, 99, 871–881.e1. [Google Scholar] [CrossRef] [PubMed]
- Kharabi Masouleh, B.; ten Dam, G.B.; Wild, M.K.; Seelige, R.; van der Vlag, J.; Rops, A.L.; Echtermeyer, F.G.; Vestweber, D.; van Kuppevelt, T.H.; Kiesel, L.; et al. Role of the heparan sulfate proteoglycan syndecan-1 (CD138) in delayed-type hypersensitivity. J. Immunol. 2009, 182, 4985–4993. [Google Scholar] [CrossRef]
- Hassan, H.; Greve, B.; Pavao, M.S.G.; Kiesel, L.; Ibrahim, S.A.; Götte, M. Syndecan-1 modulates β-integrin-dependent and interleukin-6-dependent functions in breast cancer cell adhesion, migration, and resistance to irradiation. FEBS J. 2013, 280, 2216–2227. [Google Scholar] [CrossRef]
- Edgington, T.S.; Mackman, N.; Brand, K.; Ruf, W. The structural biology of expression and function of tissue factor. Thromb. Haemost. 1991, 66, 67–79. [Google Scholar] [CrossRef]
- Siegbahn, A.; Johnell, M.; Nordin, A.; Aberg, M.; Velling, T. TF/FVIIa transactivate PDGFRbeta to regulate PDGF-BB-induced chemotaxis in different cell types: Involvement of Src and PLC. Arterioscler. Thromb. Vasc. Biol. 2008, 28, 135–141. [Google Scholar] [CrossRef]
- Aberg, M.; Johnell, M.; Wickström, M.; Siegbahn, A. Tissue Factor/ FVIIa prevents the extrinsic pathway of apoptosis by regulation of the tumor suppressor Death-Associated Protein Kinase 1 (DAPK1). Thromb. Res. 2011, 127, 141–148. [Google Scholar] [CrossRef] [PubMed]
- Åberg, M.; Eriksson, O.; Mokhtari, D.; Siegbahn, A. Tissue factor/factor VIIa induces cell survival and gene transcription by transactivation of the insulin-like growth factor 1 receptor. Thromb. Haemost. 2014, 111, 748–760. [Google Scholar] [CrossRef]
- Piro, O.; Broze, G.J. Comparison of cell-surface TFPIalpha and beta. J. Thromb. Haemost. 2005, 3, 2677–2683. [Google Scholar] [CrossRef] [PubMed]
- Shimizu, Y.; Suzuki, T.; Yoshikawa, T.; Tsuchiya, N.; Sawada, Y.; Endo, I.; Nakatsura, T. Cancer immunotherapy-targeted glypican-3 or neoantigens. Cancer Sci. 2018, 109, 531–541. [Google Scholar] [CrossRef]
- Kojima, T.; Katsumi, A.; Yamazaki, T.; Muramatsu, T.; Nagasaka, T.; Ohsumi, K.; Saito, H. Human ryudocan from endothelium-like cells binds basic fibroblast growth factor, midkine, and tissue factor pathway inhibitor. J. Biol. Chem. 1996, 271, 5914–5920. [Google Scholar] [CrossRef] [PubMed]
- Hamuro, T.; Kamikubo, Y.; Nakahara, Y.; Miyamoto, S.; Funatsu, A. Human recombinant tissue factor pathway inhibitor induces apoptosis in cultured human endothelial cells. FEBS Lett. 1998, 421, 197–202. [Google Scholar] [CrossRef]
- Feng, C.; Ho, Y.; Sun, C.; Xia, G.; Ding, Q.; Gu, B. TFPI-2 expression is decreased in bladder cancer and is related to apoptosis. J. BUON. 2016, 21, 1518–1523. [Google Scholar] [PubMed]
- Ueno, T.; Toi, M.; Koike, M.; Nakamura, S.; Tominaga, T. Tissue factor expression in breast cancer tissues: Its correlation with prognosis and plasma concentration. Br. J. Cancer 2000, 83, 164–170. [Google Scholar] [CrossRef] [PubMed]
- Hamada, K.; Kuratsu, J.; Saitoh, Y.; Takeshima, H.; Nishi, T.; Ushio, Y. Expression of tissue factor correlates with grade of malignancy in human glioma. Cancer 1996, 77, 1877–1883. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hassan, N.; Bückreiß, N.; Efing, J.; Schulz-Fincke, M.; König, P.; Greve, B.; Bendas, G.; Götte, M. The Heparan Sulfate Proteoglycan Syndecan-1 Triggers Breast Cancer Cell-Induced Coagulability by Induced Expression of Tissue Factor. Cells 2023, 12, 910. https://doi.org/10.3390/cells12060910
Hassan N, Bückreiß N, Efing J, Schulz-Fincke M, König P, Greve B, Bendas G, Götte M. The Heparan Sulfate Proteoglycan Syndecan-1 Triggers Breast Cancer Cell-Induced Coagulability by Induced Expression of Tissue Factor. Cells. 2023; 12(6):910. https://doi.org/10.3390/cells12060910
Chicago/Turabian StyleHassan, Nourhan, Nico Bückreiß, Janes Efing, Marie Schulz-Fincke, Philipp König, Burkhard Greve, Gerd Bendas, and Martin Götte. 2023. "The Heparan Sulfate Proteoglycan Syndecan-1 Triggers Breast Cancer Cell-Induced Coagulability by Induced Expression of Tissue Factor" Cells 12, no. 6: 910. https://doi.org/10.3390/cells12060910
APA StyleHassan, N., Bückreiß, N., Efing, J., Schulz-Fincke, M., König, P., Greve, B., Bendas, G., & Götte, M. (2023). The Heparan Sulfate Proteoglycan Syndecan-1 Triggers Breast Cancer Cell-Induced Coagulability by Induced Expression of Tissue Factor. Cells, 12(6), 910. https://doi.org/10.3390/cells12060910










