The LINC Complex Inhibits Excessive Chromatin Repression
Abstract
:1. Introduction
2. Materials and Methods
2.1. Fly Stocks and Husbandry
2.2. Targeted DamID
2.3. Immunofluorescence and Antibodies
2.4. Live Imaging
2.5. Microscopy and Image Acquisition
2.6. Image Analysis
2.7. Computational Model
2.8. Statistics
3. Results
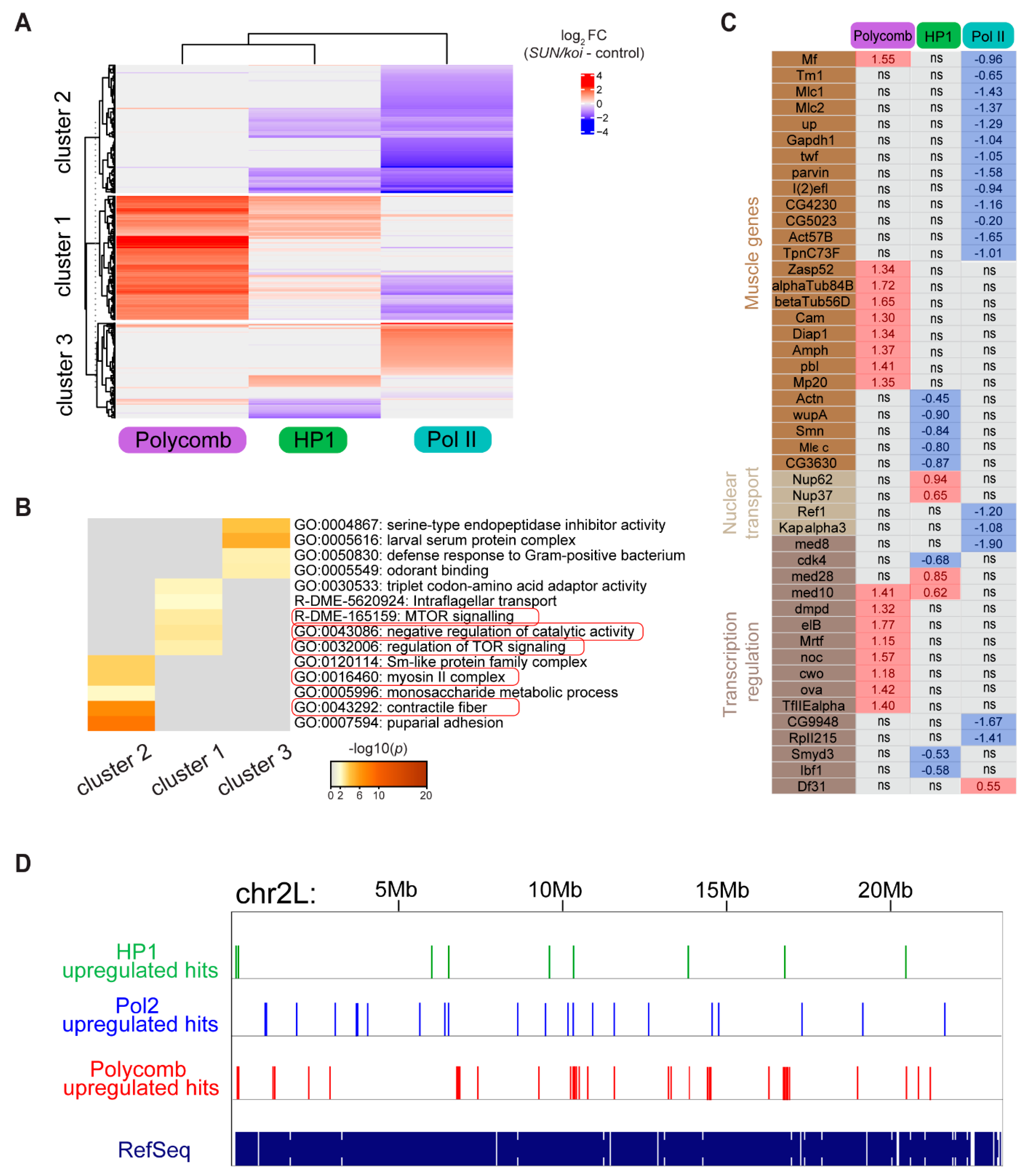
3.1. Genome-Wide Polycomb, HP1a and RNA-Pol II Binding Profiles Reveal Increased Repression and Reduced Activation in SUN/koi Mutant Muscle Fibers
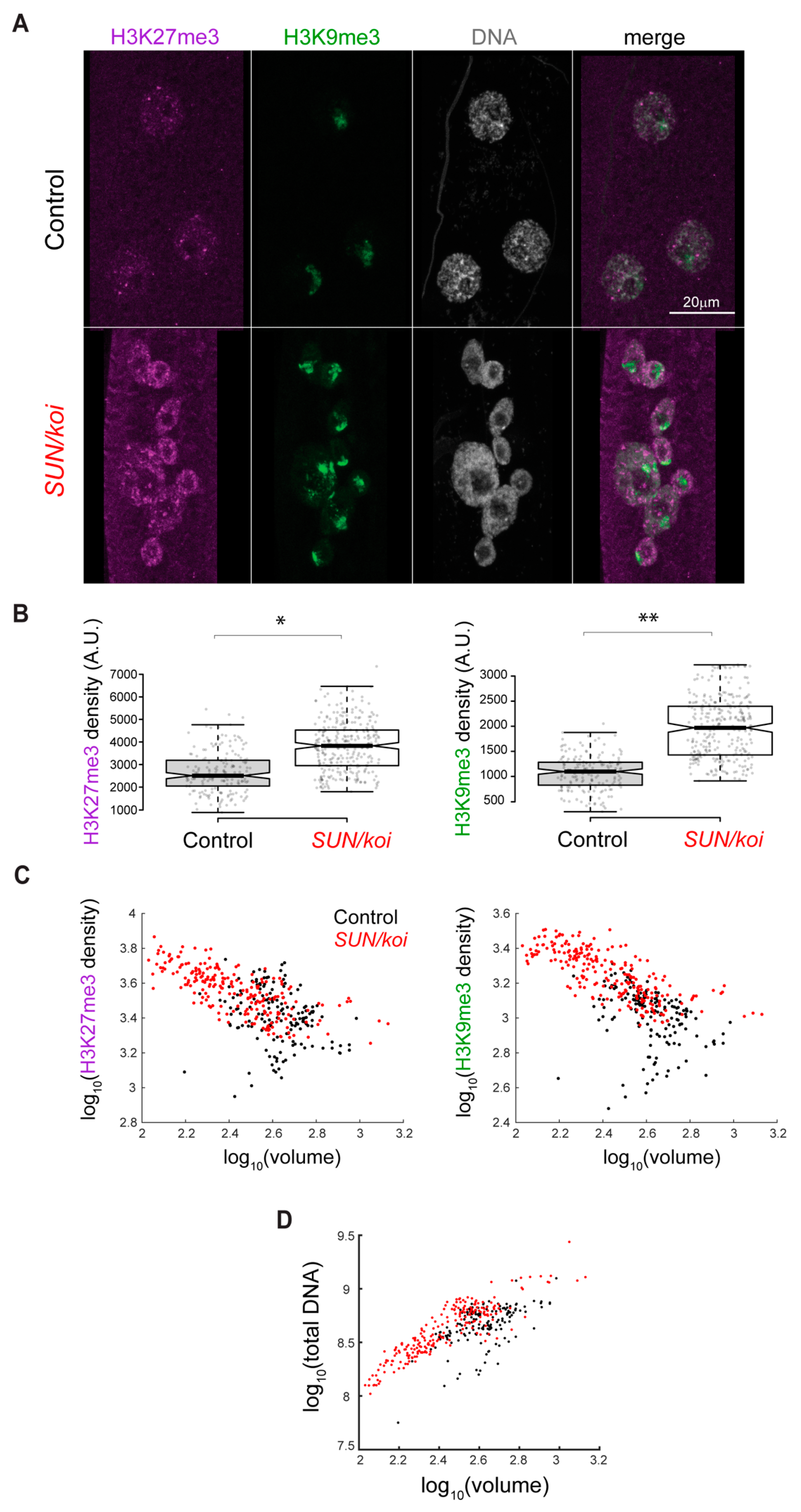
3.2. Perturbed LINC Complex Associates with Increased Epigenetic Chromatin Repression and Reduced Epigenetic Chromatin Activation in Muscle Nuclei
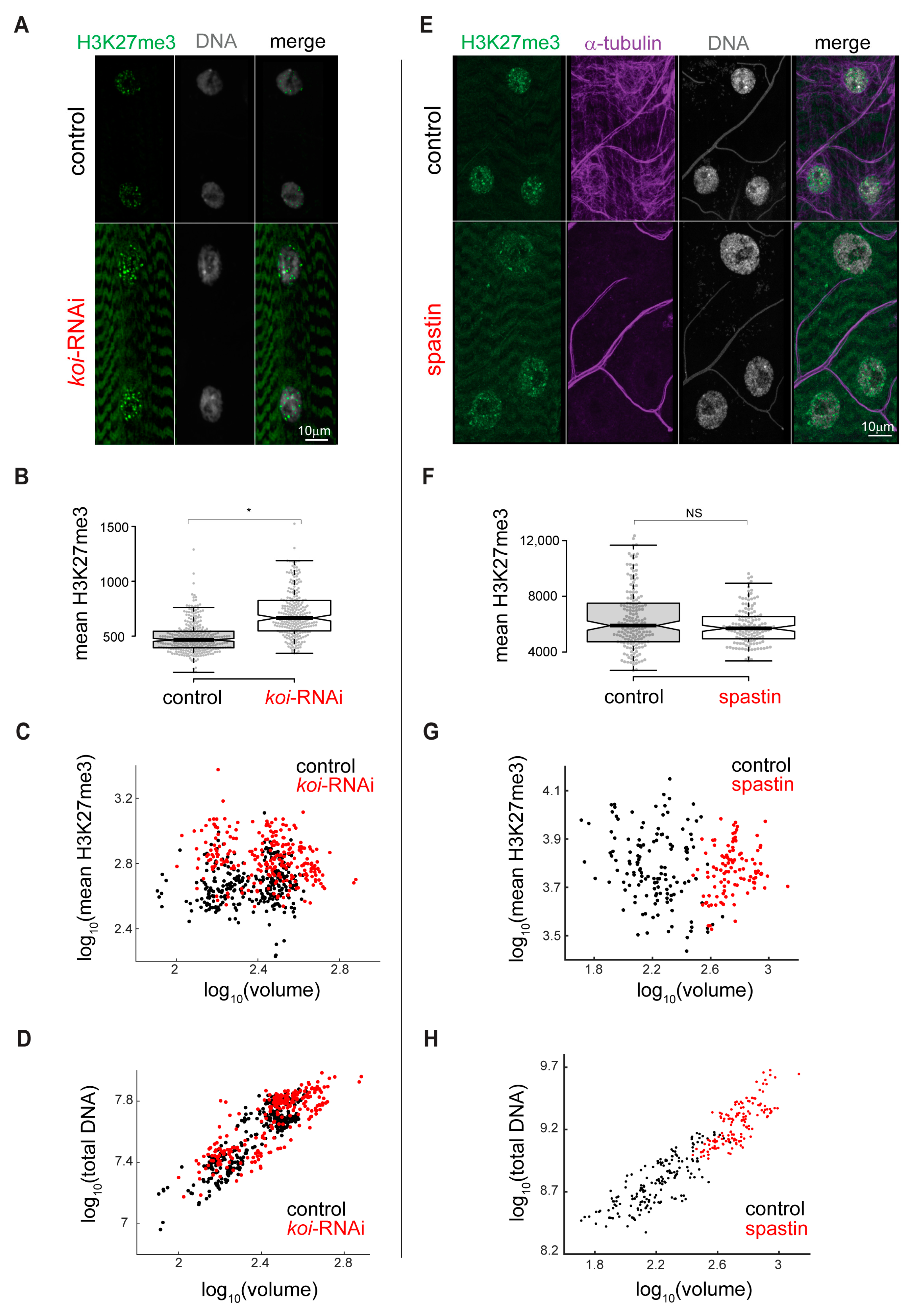
3.3. Increased H3K27me3 Repression Is Specific for Mature LINC Mutant Muscles
3.4. Reduced Epigenetic Gene Acetylation Is Observed in the SUN/koi Mutant Muscles
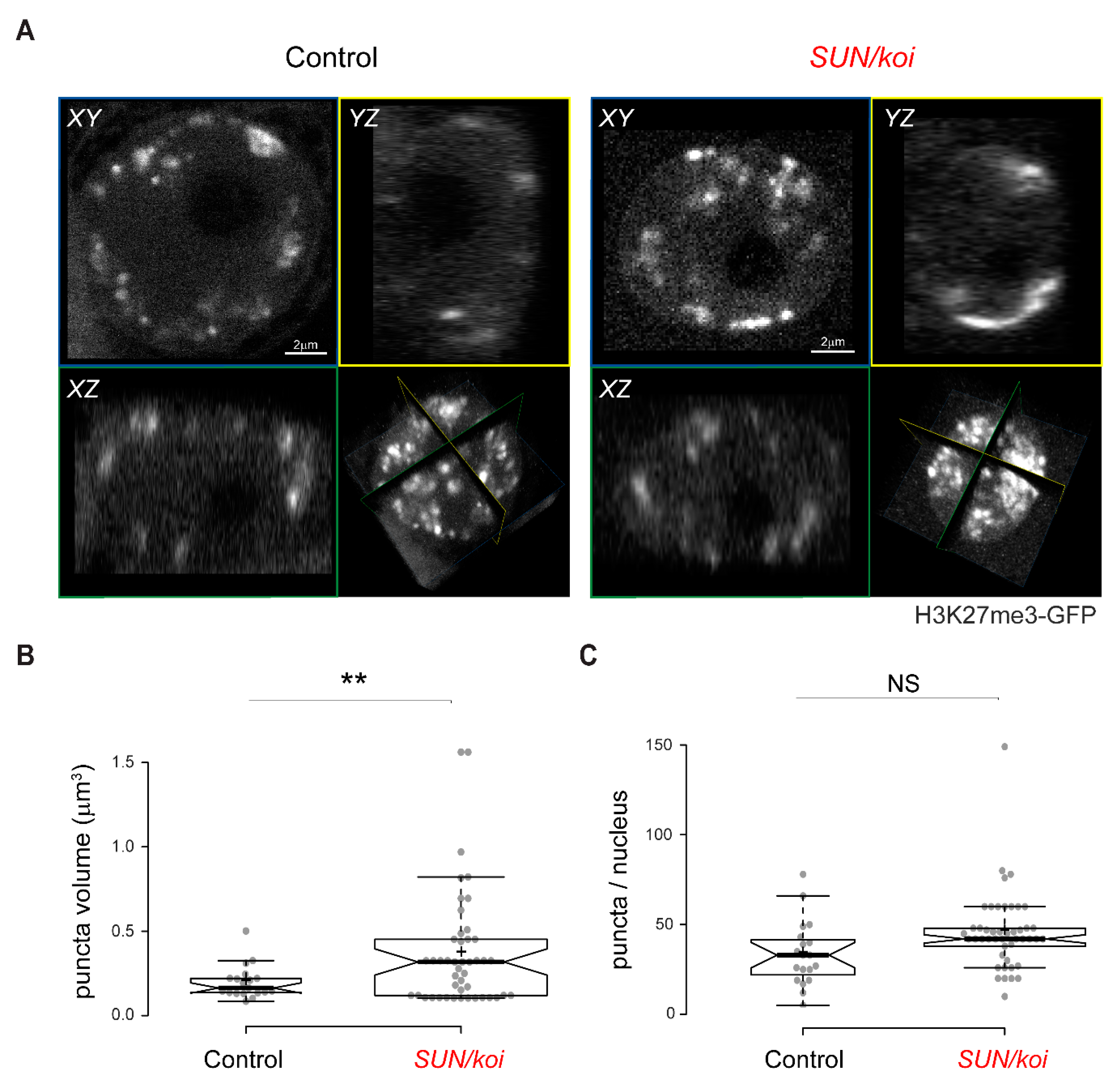
3.5. Live Imaging of the 3D Distribution of H3K27me3 Sites Reveals Increased Cluster Size in SUN/koi Mutated Muscle Fibers
3.6. Dissociation of H3K27me3 Chromatin from the Lamina Is Linked to Increased H3K27me3 Clustering in LINC Mutant Muscle
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kirby, T.J.; Lammerding, J. Emerging Views of the Nucleus as a Cellular Mechanosensor. Nat. Cell Biol. 2018, 20, 373–381. [Google Scholar] [CrossRef]
- Lityagina, O.; Dobreva, G. The LINC Between Mechanical Forces and Chromatin. Front. Physiol. 2021, 12, 1242. [Google Scholar] [CrossRef]
- Poulet, A.; Duc, C.; Voisin, M.; Desset, S.; Tutois, S.; Vanrobays, E.; Benoit, M.; Evans, D.E.; Probst, A.V.; Tatout, C. The LINC Complex Contributes to Heterochromatin Organisation and Transcriptional Gene Silencing in Plants. J. Cell Sci. 2017, 130, 590–601. [Google Scholar] [CrossRef] [Green Version]
- Wagh, K.; Ishikawa, M.; Garcia, D.A.; Stavreva, D.A.; Upadhyaya, A.; Hager, G.L. Mechanical Regulation of Transcription: Recent Advances. Trends Cell Biol. 2021, 31, 457–472. [Google Scholar] [CrossRef]
- Chang, W.; Worman, H.J.; Gundersen, G.G. Accessorizing and Anchoring the LINC Complex for Multifunctionality. J. Cell Biol. 2015, 208, 11–22. [Google Scholar] [CrossRef] [Green Version]
- Razafsky, D.; Hodzic, D. Bringing KASH under the SUN: The Many Faces of Nucleo-Cytoskeletal Connections. J. Cell Biol. 2009, 186, 461–472. [Google Scholar] [CrossRef]
- Sosa, B.A.; Rothballer, A.; Kutay, U.; Schwartz, T.U. LINC Complexes Form by Binding of Three KASH Peptides to Domain Interfaces of Trimeric SUN Proteins. Cell 2012, 149, 1035–1047. [Google Scholar] [CrossRef] [Green Version]
- Tapley, E.C.; Starr, D.A. Connecting the Nucleus to the Cytoskeleton by SUN–KASH Bridges across the Nuclear Envelope. Curr. Opin. Cell Biol. 2013, 25, 57–62. [Google Scholar] [CrossRef] [Green Version]
- Wallrath, L.L.; Bohnekamp, J.; Magin, T.M. Cross Talk between the Cytoplasm and Nucleus during Development and Disease. Curr. Opin. Genet. Dev. 2016, 37, 129–136. [Google Scholar] [CrossRef]
- Cartwright, S.; Karakesisoglou, I. Nesprins in Health and Disease. Semin. Cell Dev. Biol. 2014, 29, 169–179. [Google Scholar] [CrossRef]
- Méjat, A.; Misteli, T. LINC Complexes in Health and Disease. Nucleus 2010, 1, 40–52. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Puckelwartz, M.J.; Kessler, E.; Zhang, Y.; Hodzic, D.; Randles, K.N.; Morris, G.; Earley, J.U.; Hadhazy, M.; Holaska, J.M.; Mewborn, S.K.; et al. Disruption of Nesprin-1 Produces an Emery Dreifuss Muscular Dystrophy-like Phenotype in Mice. Hum. Mol. Genet. 2009, 18, 607–620. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Horn, H.F. LINC Complex Proteins in Development and Disease. Curr. Top. Dev. Biol. 2014, 109, 287–321. [Google Scholar] [CrossRef] [PubMed]
- Heffler, J.; Shah, P.P.; Robison, P.; Phyo, S.; Veliz, K.; Uchida, K.; Bogush, A.; Rhoades, J.; Jain, R.; Prosser, B.L. A Balance Between Intermediate Filaments and Microtubules Maintains Nuclear Architecture in the Cardiomyocyte. Circ. Res. 2019, 126, e10–e26. [Google Scholar] [CrossRef]
- Seelbinder, B.; Ghosh, S.; Schneider, S.E.; Scott, A.K.; Berman, A.G.; Goergen, C.J.; Margulies, K.B.; Bedi, K.C.; Casas, E.; Swearingen, A.R.; et al. Nuclear Deformation Guides Chromatin Reorganization in Cardiac Development and Disease. Nat. Biomed. Eng. 2021, 5, 1500–1516. [Google Scholar] [CrossRef]
- Rashmi, R.N.; Eckes, B.; Glöckner, G.; Groth, M.; Neumann, S.; Gloy, J.; Sellin, L.; Walz, G.; Schneider, M.; Karakesisoglou, I.; et al. The Nuclear Envelope Protein Nesprin-2 Has Roles in Cell Proliferation and Differentiation during Wound Healing. Nucleus 2012, 3, 172–186. [Google Scholar] [CrossRef] [Green Version]
- Carley, E.; Stewart, R.K.; Zieman, A.; Jalilian, I.; King, D.E.; Zubek, A.; Lin, S.; Horsley, V.; King, M.C. The Linc Complex Transmits Integrin-Dependent Tension to the Nuclear Lamina and Represses Epidermal Differentiation. Elife 2021, 10, e58541. [Google Scholar] [CrossRef]
- Hernandez, M.; Patzig, J.; Mayoral, S.R.; Costa, K.D.; Chan, J.R.; Casaccia, P. Mechanostimulation Promotes Nuclear and Epigenetic Changes in Oligodendrocytes. J. Neurosci. 2016, 36, 806–813. [Google Scholar] [CrossRef] [Green Version]
- Ghosh, S.; Gardner, J.M.; Smoyer, C.J.; Friederichs, J.M.; Unruh, J.R.; Slaughter, B.D.; Alexander, R.; Chisholm, R.D.; Lee, K.K.; Workman, J.L.; et al. Acetylation of the SUN Protein Mps3 by Eco1 Regulates Its Function in Nuclear Organization. Mol. Biol. Cell 2012, 23, 2546–2559. [Google Scholar] [CrossRef]
- Elhanany-Tamir, H.; Yu, Y.V.; Shnayder, M.; Jain, A.; Welte, M.; Volk, T. Organelle Positioning in Muscles Requires Cooperation between Two KASH Proteins and Microtubules. J. Cell Biol. 2012, 198, 833–846. [Google Scholar] [CrossRef] [Green Version]
- Wang, S.; Reuveny, A.; Volk, T. Nesprin Provides Elastic Properties to Muscle Nuclei by Cooperating with Spectraplakin and EB1. J. Cell Biol. 2015, 209, 529–538. [Google Scholar] [CrossRef] [Green Version]
- Lorber, D.; Rotkopf, R.; Volk, T. A Minimal Constraint Device for Imaging Nuclei in Live Drosophila Contractile Larval Muscles Reveals Novel Nuclear Mechanical Dynamics. Lab Chip 2020, 20, 2100–2112. [Google Scholar] [CrossRef] [PubMed]
- Kracklauer, M.P.; Banks, S.M.L.; Xie, X.; Wu, Y.; Fischer, J.A. Drosophila Klaroid Encodes a SUN Domain Protein Required for Klarsicht Localization to the Nuclear Envelope and Nuclear Migration in the Eye. Fly 2007, 1, 75–85. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bosveld, F.; Ainslie, A.; Bellaïche, Y. Sequential Activities of Dynein, Mud and Asp in Centrosome-Spindle Coupling Maintain Centrosome Number upon Mitosis. J. Cell Sci. 2017, 130, 3557–3567. [Google Scholar] [CrossRef] [Green Version]
- Marshall, O.J.; Brand, A.H. Chromatin State Changes during Neural Development Revealed by in Vivo Cell-Type Specific Profiling. Nat. Commun. 2017, 8, 2271. [Google Scholar] [CrossRef] [Green Version]
- Marshall, O.J.; Southall, T.D.; Cheetham, S.W.; Brand, A.H. Cell-Type-Specific Profiling of Protein–DNA Interactions without Cell Isolation Using Targeted DamID with next-Generation Sequencing. Nat. Protoc. 2016, 11, 1586–1598. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marshall, O.J.; Brand, A.H. Damidseq_pipeline: An Automated Pipeline for Processing DamID Sequencing Datasets. Bioinformatics 2015, 31, 3371–3373. [Google Scholar] [CrossRef] [Green Version]
- Robinson, J.T.; Thorvaldsdóttir, H.; Winckler, W.; Guttman, M.; Lander, E.S.; Getz, G.; Mesirov, J.P. Integrative Genomics Viewer. Nat. Biotechnol. 2011, 29, 24–26. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Y.; Zhou, B.; Pache, L.; Chang, M.; Khodabakhshi, A.H.; Tanaseichuk, O.; Benner, C.; Chanda, S.K. Metascape Provides a Biologist-Oriented Resource for the Analysis of Systems-Level Datasets. Nat. Commun. 2019, 10, 1523. [Google Scholar] [CrossRef]
- Amiad-Pavlov, D.; Lorber, D.; Bajpai, G.; Reuveny, A.; Roncato, F.; Alon, R.; Safran, S.; Volk, T. Live Imaging of Chromatin Distribution Reveals Novel Principles of Nuclear Architecture and Chromatin Compartmentalization. Sci. Adv. 2021, 7, eabf6251. [Google Scholar] [CrossRef]
- Li, Q.; Tjong, H.; Li, X.; Gong, K.; Zhou, X.J.; Chiolo, I.; Alber, F. The Three-Dimensional Genome Organization of Drosophila Melanogaster through Data Integration. Genome Biol. 2017, 18, 145. [Google Scholar] [CrossRef] [Green Version]
- Lesley Brown, J.; Sun, M.; Kassis, J.A. Global Changes of H3K27me3 Domains and Polycomb Group Protein Distribution in the Absence of Recruiters Spps or Pho. Proc. Natl. Acad. Sci. USA 2018, 115, E1839–E1848. [Google Scholar] [CrossRef]
- Koryakov, D.E.; Walther, M.; Ebert, A.; Lein, S.; Zhimulev, I.F.; Reuter, G. The SUUR Protein Is Involved in Binding of SU(VAR)3-9 and Methylation of H3K9 and H3K27 in Chromosomes of Drosophila Melanogaster. Chromosom. Res. 2011, 19, 235–249. [Google Scholar] [CrossRef]
- Pailles, M.; Hirlemann, M.; Brochard, V.; Chebrout, M.; Oudin, J.F.; Marks, H.; Jouneau, A.; Bonnet-Garnier, A. H3K27me3 at Pericentromeric Heterochromatin Is a Defining Feature of the Early Mouse Blastocyst. Sci. Rep. 2022, 12, 13908. [Google Scholar] [CrossRef]
- Bajpai, G.; Pavlov, D.A.; Lorber, D.; Volk, T.; Safran, S. Mesoscale Phase Separation of Chromatin in the Nucleus. Elife 2021, 10, 549a. [Google Scholar] [CrossRef]
- Dekker, J.; Misteli, T. Long-Range Chromatin Interactions. Cold Spring Harb. Perspect. Biol. 2015, 7, a019356. [Google Scholar] [CrossRef] [Green Version]
- Bajpai, G.; Safran, S. Polymer Dynamics Relates Chromosome Mixing to Temporal Changes in Biological Contact Maps. bioRxiv 2022, 2022.07.20.500905. [Google Scholar] [CrossRef]
- Adame-Arana, O.; Bajpai, G.; Lorber, D.; Volk, T.; Safran, S.A. Regulation of Chromatin Microphase Separation by Adsorbed Protein Complexes. bioRxiv 2022, 2022.09.29.510124. [Google Scholar] [CrossRef]
- Plimpton, S. Fast Parallel Algorithms for Short-Range Molecular Dynamics. J. Comput. Phys. 1995, 117, 1–19. [Google Scholar] [CrossRef] [Green Version]
- Kuznetsova, A.; Brockhoff, P.B.; Christensen, R.H.B. LmerTest Package: Tests in Linear Mixed Effects Models. J. Stat. Softw. 2017, 82, 1–26. [Google Scholar] [CrossRef] [Green Version]
- Spitzer, M.; Wildenhain, J.; Rappsilber, J.; Tyers, M. BoxPlotR: A Web Tool for Generation of Box Plots. Nat. Methods 2014, 11, 121–122. [Google Scholar] [CrossRef] [PubMed]
- Titlow, J.; Robertson, F.; Järvelin, A.; Ish-Horowicz, D.; Smith, C.; Gratton, E.; Davis, I. Syncrip/HnRNP Q Is Required for Activity-Induced Msp300/Nesprin-1 Expression and New Synapse Formation. J. Cell Biol. 2020, 219, e201903135. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Southall, T.D.; Gold, K.S.; Egger, B.; Davidson, C.M.; Caygill, E.E.; Marshall, O.J.; Brand, A.H. Cell-Type-Specific Profiling of Gene Expression and Chromatin Binding without Cell Isolation: Assaying RNA Pol II Occupancy in Neural Stem Cells. Dev. Cell 2013, 26, 101–112. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kwon, S.H.; Workman, J.L. The Changing Faces of HP1: From Heterochromatin Formation and Gene Silencing to Euchromatic Gene Expression. BioEssays 2011, 33, 280–289. [Google Scholar] [CrossRef]
- Yoon, M.S. MTOR as a Key Regulator in Maintaining Skeletal Muscle Mass. Front. Physiol. 2017, 8, 788. [Google Scholar] [CrossRef] [Green Version]
- Wang, S.; Stoops, E.; CP, U.; Markus, B.; Reuveny, A.; Ordan, E.; Volk, T. Mechanotransduction via the LINC Complex Regulates DNA Replication in Myonuclei. J. Cell Biol. 2018, 217, 2005–2018. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xie, X.; Fischer, J.A. On the Roles of the Drosophila KASH Domain Proteins Msp-300 and Klarsicht. Fly 2008, 2, 74–81. [Google Scholar] [CrossRef] [Green Version]
- Schuettengruber, B.; Bourbon, H.M.; Di Croce, L.; Cavalli, G. Genome Regulation by Polycomb and Trithorax: 70 Years and Counting. Cell 2017, 171, 34–57. [Google Scholar] [CrossRef] [Green Version]
- Tjalsma, S.J.D.; Hori, M.; Sato, Y.; Bousard, A.; Ohi, A.; Raposo, A.C.; Roensch, J.; Le Saux, A.; Nogami, J.; Maehara, K.; et al. H4K20me1 and H3K27me3 Are Concurrently Loaded onto the Inactive X Chromosome but Dispensable for Inducing Gene Silencing. EMBO Rep. 2021, 22, e51989. [Google Scholar] [CrossRef]
- Tatavosian, R.; Kent, S.; Brown, K.; Yao, T.; Duc, H.N.; Huynh, T.N.; Zhen, C.Y.; Ma, B.; Wang, H.; Ren, X. Nuclear Condensates of the Polycomb Protein Chromobox 2 (CBX2) Assemble through Phase Separation. J. Biol. Chem. 2019, 294, 1451–1463. [Google Scholar] [CrossRef] [Green Version]
- Matsushita, K.; Nakahara, C.; Kimura, S.; Sakamoto, N.; Ii, S.; Miyoshi, H. Intranuclear Mesoscale Viscoelastic Changes during Osteoblastic Differentiation of Human Mesenchymal Stem Cells. FASEB J. 2021, 35, e22071. [Google Scholar] [CrossRef] [PubMed]
- Heo, S.-J.; Han, W.M.; Szczesny, S.E.; Cosgrove, B.D.; Elliott, D.M.; Lee, D.A.; Duncan, R.L.; Mauck, R.L. Mechanically Induced Chromatin Condensation Requires Cellular Contractility in Mesenchymal Stem Cells. Biophys. J. 2016, 111, 864–874. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Le, H.Q.; Ghatak, S.; Yeung, C.-Y.C.; Tellkamp, F.; Günschmann, C.; Dieterich, C.; Yeroslaviz, A.; Habermann, B.; Pombo, A.; Niessen, C.M.; et al. Mechanical Regulation of Transcription Controls Polycomb-Mediated Gene Silencing during Lineage Commitment. Nat. Cell Biol. 2016, 18, 864–875. [Google Scholar] [CrossRef] [PubMed]
- Fernandez, A.; Bautista, M.; Wu, L.; Pinaud, F. Emerin Self-Assembly and Nucleoskeletal Coupling Regulate Nuclear Envelope Mechanics against Stress. J. Cell Sci. 2022, 135, jcs258969. [Google Scholar] [CrossRef] [PubMed]
- Unnikannan, C.P.; Reuveny, A.; Grunberg, D.; Volk, T. Recruitment of BAF to the Nuclear Envelope Couples the LINC Complex to Endoreplication. Development 2020, 147, dev191304. [Google Scholar] [CrossRef] [PubMed]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Amiad Pavlov, D.; Unnikannan, C.; Lorber, D.; Bajpai, G.; Olender, T.; Stoops, E.; Reuveny, A.; Safran, S.; Volk, T. The LINC Complex Inhibits Excessive Chromatin Repression. Cells 2023, 12, 932. https://doi.org/10.3390/cells12060932
Amiad Pavlov D, Unnikannan C, Lorber D, Bajpai G, Olender T, Stoops E, Reuveny A, Safran S, Volk T. The LINC Complex Inhibits Excessive Chromatin Repression. Cells. 2023; 12(6):932. https://doi.org/10.3390/cells12060932
Chicago/Turabian StyleAmiad Pavlov, Daria, CP Unnikannan, Dana Lorber, Gaurav Bajpai, Tsviya Olender, Elizabeth Stoops, Adriana Reuveny, Samuel Safran, and Talila Volk. 2023. "The LINC Complex Inhibits Excessive Chromatin Repression" Cells 12, no. 6: 932. https://doi.org/10.3390/cells12060932
APA StyleAmiad Pavlov, D., Unnikannan, C., Lorber, D., Bajpai, G., Olender, T., Stoops, E., Reuveny, A., Safran, S., & Volk, T. (2023). The LINC Complex Inhibits Excessive Chromatin Repression. Cells, 12(6), 932. https://doi.org/10.3390/cells12060932







