Bromodomain Protein Inhibitors Reorganize the Chromatin of Synovial Fibroblasts
Abstract
1. Introduction
2. Materials and Methods
2.1. Patient Samples and Cell Preparation
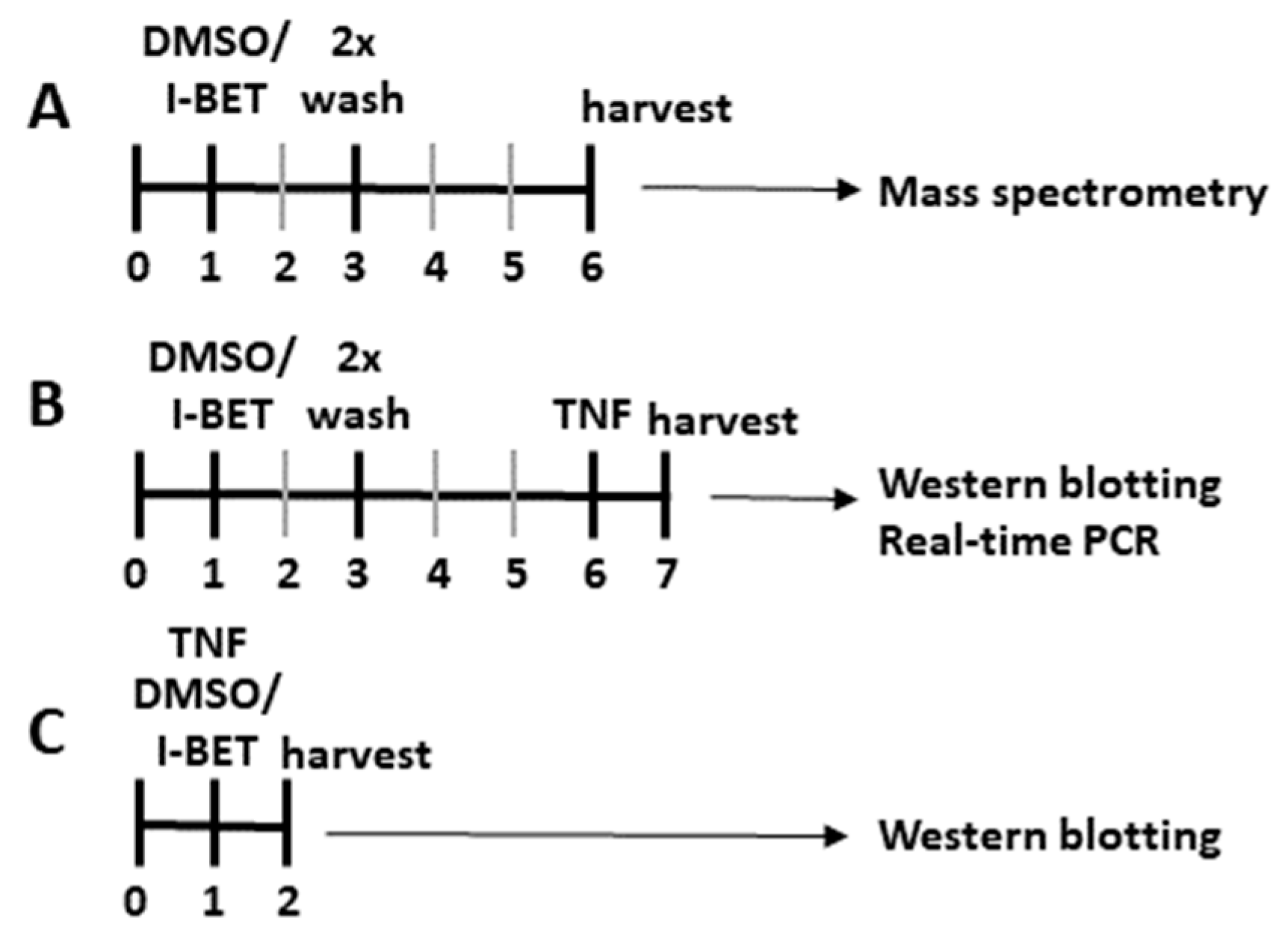
2.2. Stimulation Experiments
2.3. Mass Spectrometry
2.4. Western Blotting
2.5. Real-Time PCR
2.6. Assay for Transposase-Accessible Chromatin Using Sequencing (ATAC-Seq)
2.7. Statistical Analysis
3. Results
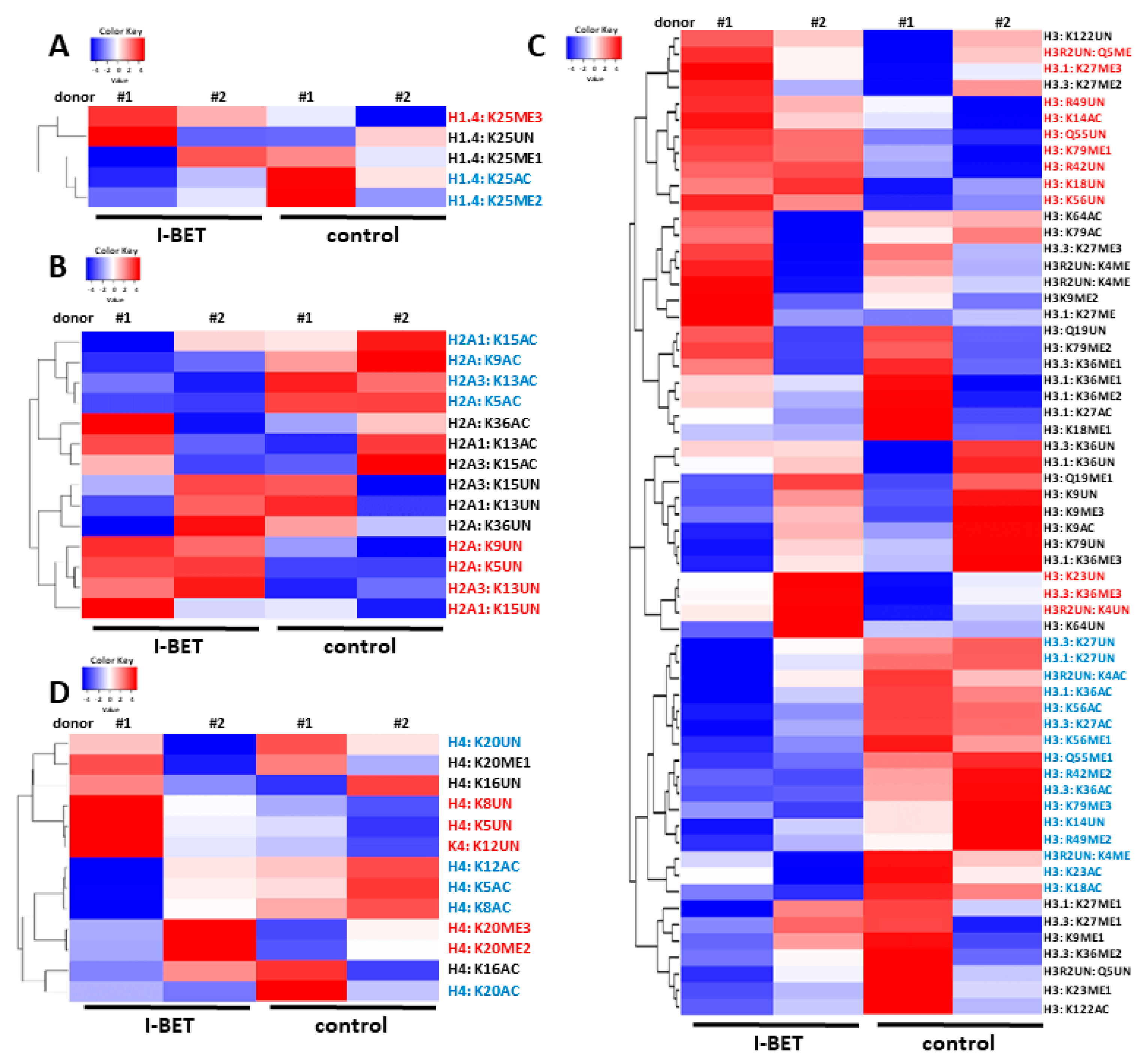
3.1. I-BET151 Induces Global Changes in Post-Translational Histone Modifications
3.2. I-BET151 Suppresses TNF-Induced Changes in Histone Acetylation
3.3. Long-Lasting Effects of I-BET151 on TNF-Induced Target Genes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Choudhary, C.; Weinert, B.T.; Nishida, Y.; Verdin, E.; Mann, M. The growing landscape of lysine acetylation links metabolism and cell signalling. Nat. Rev. Mol. Cell Biol. 2014, 15, 536–550. [Google Scholar] [CrossRef] [PubMed]
- Filippakopoulos, P.; Picaud, S.; Mangos, M.; Keates, T.; Lambert, J.-P.; Barsyte-Lovejoy, D.; Felletar, I.; Volkmer, R.; Müller, S.; Pawson, T.; et al. Histone recognition and large-scale structural analysis of the human bromodomain family. Cell 2012, 149, 214–231. [Google Scholar] [CrossRef] [PubMed]
- Frank-Bertoncelj, M.; Trenkmann, M.; Klein, K.; Karouzakis, E.; Rehrauer, H.; Bratus, A.; Kolling, C.; Armaka, M.; Filer, A.; Michel, B.A.; et al. Epigenetically-driven anatomical diversity of synovial fibroblasts guides joint-specific fibroblast functions. Nat. Commun. 2017, 8, 14852. [Google Scholar] [CrossRef] [PubMed]
- Ge, X.; Frank-Bertoncelj, M.; Klein, K.; McGovern, A.; Kuret, T.; Houtman, M.; Burja, B.; Micheroli, R.; Shi, C.; Marks, M.; et al. Functional genomics atlas of synovial fibroblasts defining rheumatoid arthritis heritability. Genome Biol. 2021, 22, 247. [Google Scholar] [PubMed]
- Ospelt, C. Synovial fibroblasts in 2017. RMD Open 2017, 3, e000471. [Google Scholar] [CrossRef]
- Friscic, J.; Bottcher, M.; Reinwald, C.; Bruns, H.; Wirth, B.; Popp, S.J.; Walker, K.I.; Ackermann, J.A.; Chen, X.; Turner, J.; et al. The complement system drives local inflammatory tissue priming by metabolic reprogramming of synovial fibroblasts. Immunity 2021, 54, 1002–1021.e1010. [Google Scholar] [CrossRef]
- Klein, K.; Gay, S. Epigenetics in rheumatoid arthritis. Curr. Opin. Rheumatol. 2015, 27, 76–82. [Google Scholar] [CrossRef]
- Creyghton, M.P.; Cheng, A.W.; Welstead, G.G.; Kooistra, T.; Carey, B.W.; Steine, E.J.; Hanna, J.; Lodato, M.A.; Frampton, G.M.; Sharp, P.A.; et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc. Natl. Acad. Sci. USA 2010, 107, 21931–21936. [Google Scholar] [CrossRef]
- Ai, R.; Laragione, T.; Hammaker, D.; Boyle, D.L.; Wildberg, A.; Maeshima, K.; Palescandolo, E.; Krishna, V.; Pocalyko, D.; Whitaker, J.W.; et al. Comprehensive epigenetic landscape of rheumatoid arthritis fibroblast-like synoviocytes. Nat. Commun. 2018, 9, 1921. [Google Scholar] [CrossRef]
- Klein, K.; Frank-Bertoncelj, M.; Karouzakis, E.; Gay, R.E.; Kolling, C.; Ciurea, A.; Bostanci, N.; Belibasakis, G.N.; Lin, L.-L.; Distler, O.; et al. The epigenetic architecture at gene promoters determines cell type-specific LPS tolerance. J. Autoimmun. 2017, 83, 122–133. [Google Scholar] [CrossRef]
- Loh, C.; Park, S.H.; Lee, A.; Yuan, R.; Ivashkiv, L.B.; Kalliolias, G.D. TNF-induced inflammatory genes escape repression in fibroblast-like synoviocytes: Transcriptomic and epigenomic analysis. Ann. Rheum. Dis. 2019, 78, 1205–1214. [Google Scholar] [CrossRef] [PubMed]
- Klein, K. Bromodomain protein inhibition: A novel therapeutic strategy in rheumatic diseases. RMD Open 2018, 4, e000744. [Google Scholar] [CrossRef] [PubMed]
- Fujisawa, T.; Filippakopoulos, P. Functions of bromodomain-containing proteins and their roles in homeostasis and cancer. Nat. Rev. Mol. Cell Biol. 2017, 18, 246–262. [Google Scholar] [CrossRef] [PubMed]
- Nicodeme, E.; Jeffrey, K.L.; Schaefer, U.; Beinke, S.; Dewell, S.; Chung, C.-W.; Chandwani, R.; Marazzi, I.; Wilson, P.; Coste, H.; et al. Suppression of inflammation by a synthetic histone mimic. Nature 2010, 468, 1119–1123. [Google Scholar] [CrossRef]
- Kanno, T.; Kanno, Y.; LeRoy, G.; Campos, E.I.; Sun, H.-W.; Brooks, S.R.; Vahedi, G.; Heightman, T.D.; A Garcia, B.; Reinberg, D.; et al. BRD4 assists elongation of both coding and enhancer RNAs by interacting with acetylated histones. Nat. Struct. Mol. Biol. 2014, 21, 1047–1057. [Google Scholar] [CrossRef]
- Krishna, V.; Yin, X.; Song, Q.; Walsh, A.; Pocalyko, D.; Bachman, K.; Anderson, I.; Madakamutil, L.; Nagpal, S. Integration of the Transcriptome and Genome-Wide Landscape of BRD2 and BRD4 Binding Motifs Identifies Key Superenhancer Genes and Reveals the Mechanism of Bet Inhibitor Action in Rheumatoid Arthritis Synovial Fibroblasts. J. Immunol. 2021, 206, 422–431. [Google Scholar] [CrossRef]
- Garcia, B.A.; Mollah, S.; Ueberheide, B.M.; Busby, S.A.; Muratore, T.L.; Shabanowitz, J.; Hunt, D.F. Chemical derivatization of histones for facilitated analysis by mass spectrometry. Nat. Protoc. 2007, 2, 933–938. [Google Scholar] [CrossRef]
- MacLean, B.; Tomazela, D.M.; Shulman, N.; Chambers, M.; Finney, G.L.; Frewen, B.; Kern, R.; Tabb, D.L.; Liebler, D.C.; MacCoss, M.J. Skyline: An open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics 2010, 26, 966–968. [Google Scholar] [CrossRef]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef]
- Friscic, J.; Reinwald, C.; Bottcher, M.; Houtman, M.; Euler, M.; Chen, X.; Walker, K.I.; Kirchner, P.; Zhu, H.; Wirth, B.; et al. Reset of inflammatory priming of joint tissue and reduction of the severity of arthritis flares by bromodomain inhibition. Arthritis Rheumatol. 2022, 75, 517–532. [Google Scholar] [CrossRef]
- Krosel, M.; Gabathuler, M.; Maciukiewicz, M.; Moser, L.; Lee, G.I.; Marks, M.; Tomsic, M.; Distler, O.; Ospelt, C.; Klein, K. Individual functions of the histone acetyl transferases CBP and p300 in regulating the inflammatory response of synovial fibroblasts. J. Autoimmun. 2021, 123, 102709. [Google Scholar] [CrossRef] [PubMed]
- Klein, K.; Kabala, P.A.; Grabiec, A.M.; Gay, R.E.; Kolling, C.; Lin, L.L.; Gay, S.; Tak, P.P.; Prinjha, R.K.; Ospelt, C.; et al. The bromodomain protein inhibitor I-BET151 suppresses expression of inflammatory genes and matrix degrading enzymes in rheumatoid arthritis synovial fibroblasts. Ann. Rheum. Dis. 2016, 75, 422–429. [Google Scholar] [CrossRef] [PubMed]
- Huang, M.; Zeng, S.; Zou, Y.; Shi, M.; Qiu, Q.; Xiao, Y.; Chen, G.; Yang, X.; Liang, L.; Xu, H. The suppression of bromodomain and extra-terminal domain inhibits vascular inflammation by blocking NF-kappaB and MAPK activation. Br. J. Pharmacol. 2017, 174, 101–115. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Y.; Liang, L.; Huang, M.; Qiu, Q.; Zeng, S.; Shi, M.; Zou, Y.; Ye, Y.; Yang, X.; Xu, H. Bromodomain and extra-terminal domain bromodomain inhibition prevents synovial inflammation via blocking IkappaB kinase-dependent NF-kappaB activation in rheumatoid fibroblast-like synoviocytes. Rheumatology 2016, 55, 173–184. [Google Scholar] [CrossRef]
- Devaiah, B.N.; Case-Borden, C.; Gegonne, A.; Hsu, C.H.; Chen, Q.; Meerzaman, D.; Dey, A.; Ozato, K.; Singer, D.S. BRD4 is a histone acetyltransferase that evicts nucleosomes from chromatin. Nat. Struct. Mol. Biol. 2016, 23, 540–548. [Google Scholar] [CrossRef]
- Mishima, Y.; Miyagi, S.; Saraya, A.; Negishi, M.; Endoh, M.; Endo, T.A.; Toyoda, T.; Shinga, J.; Katsumoto, T.; Chiba, T.; et al. The Hbo1-Brd1/Brpf2 complex is responsible for global acetylation of H3K14 and required for fetal liver erythropoiesis. Blood 2011, 118, 2443–2453. [Google Scholar] [CrossRef] [PubMed]
- Peña-Hernández, R.; Aprigliano, R.; Frommel, S.C.; Pietrzak, K.; Steiger, S.; Roganowicz, M.; Lerra, L.; Bizzarro, J.; Santoro, R. BAZ2A-mediated repression via H3K14ac-marked enhancers promotes prostate cancer stem cells. EMBO Rep. 2021, 22, e53014. [Google Scholar] [CrossRef]
- Liao, L.; Alicea-Velázquez, N.L.; Langbein, L.; Niu, X.; Cai, W.; Cho, E.A.; Zhang, M.; Greer, C.B.; Yan, Q.; Cosgrove, M.S.; et al. High affinity binding of H3K14ac through collaboration of bromodomains 2, 4 and 5 is critical for the molecular and tumor suppressor functions of PBRM1. Mol. Oncol. 2019, 13, 811–828. [Google Scholar] [CrossRef]
- Wu, T.; Kamikawa, Y.F.; Donohoe, M.E. Brd4’s Bromodomains Mediate Histone H3 Acetylation and Chromatin Remodeling in Pluripotent Cells through P300 and Brg1. Cell Rep. 2018, 25, 1756–1771. [Google Scholar] [CrossRef]
- Klein, K.; Gay, R.E.; Kolling, C.; Gay, S.; Ospelt, C. A Dual Role of Bromodomain Containing 1 Protein in Rheumatoid Arthritis Synovial Fibroblasts. Arthritis Rheumatol. 2016, 68 (Suppl. 10). Available online: https://acrabstracts.org/abstract/a-dual-role-of-bromodomain-containing-1-protein-in-rheumatoid-arthritis-synovial-fibroblasts/ (accessed on 28 March 2023).
- Mizzen, C.A.; Allis, C.D. Linking histone acetylation to transcriptional regulation. Cell Mol. Life Sci. 1998, 54, 6–20. [Google Scholar] [CrossRef] [PubMed]
- Dominguez-Andres, J.; Ferreira, A.V.; Jansen, T.; Smithers, N.; Prinjha, R.K.; Furze, R.C.; Netea, M.G. Bromodomain inhibitor I-BET151 suppresses immune responses during fungal-immune interaction. Eur. J. Immunol. 2019, 49, 2044–2050. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Krošel, M.; Moser, L.; Houtman, M.; Friščić, J.; Tomšič, M.; Distler, O.; Hoffmann, M.H.; Ospelt, C.; Klein, K. Bromodomain Protein Inhibitors Reorganize the Chromatin of Synovial Fibroblasts. Cells 2023, 12, 1149. https://doi.org/10.3390/cells12081149
Krošel M, Moser L, Houtman M, Friščić J, Tomšič M, Distler O, Hoffmann MH, Ospelt C, Klein K. Bromodomain Protein Inhibitors Reorganize the Chromatin of Synovial Fibroblasts. Cells. 2023; 12(8):1149. https://doi.org/10.3390/cells12081149
Chicago/Turabian StyleKrošel, Monika, Larissa Moser, Miranda Houtman, Jasna Friščić, Matija Tomšič, Oliver Distler, Markus H. Hoffmann, Caroline Ospelt, and Kerstin Klein. 2023. "Bromodomain Protein Inhibitors Reorganize the Chromatin of Synovial Fibroblasts" Cells 12, no. 8: 1149. https://doi.org/10.3390/cells12081149
APA StyleKrošel, M., Moser, L., Houtman, M., Friščić, J., Tomšič, M., Distler, O., Hoffmann, M. H., Ospelt, C., & Klein, K. (2023). Bromodomain Protein Inhibitors Reorganize the Chromatin of Synovial Fibroblasts. Cells, 12(8), 1149. https://doi.org/10.3390/cells12081149





