Experimental Validation and Prediction of Super-Enhancers: Advances and Challenges
Abstract
1. Introduction
2. Constitutive Super-Enhancers in Different Cell Types
2.1. Defining Constitutive Super-Enhancers: Current View
- Enhancers were considered to be the sites bound by all three master regulators, Oct4, Sox2, and Nanog, according to ChIP-seq;
- Enhancers within 12.5 kb of each other were stitched together to form a single entity;
- The stitched SE entities and the remaining TEs were then ranked by the total background-normalized level of Med1 signaling within the locus.
2.2. Techniques Used and Proposed for SE Discovery and Research
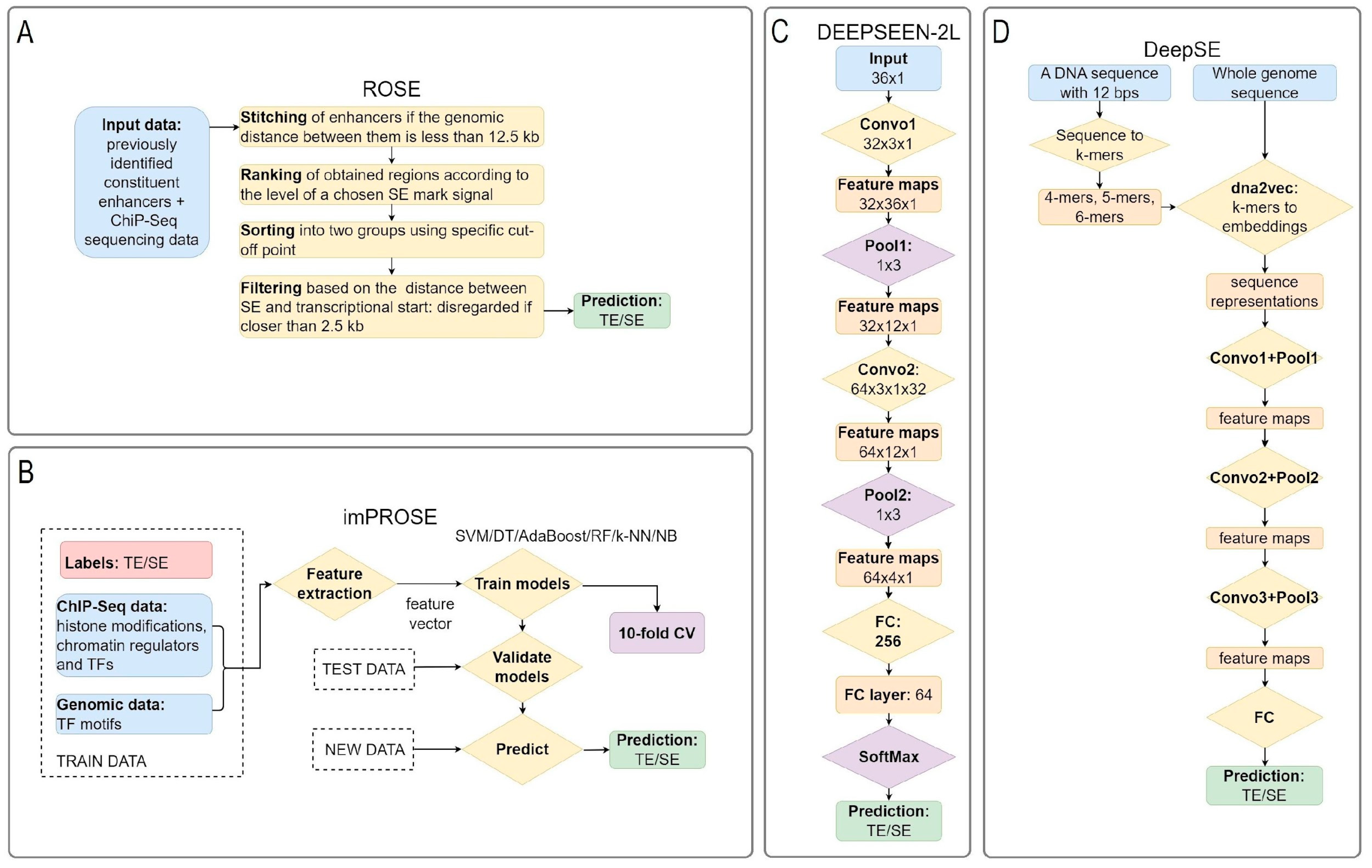
2.2.1. Computational Methods for Searching and Predicting Super-Enhancers
2.2.2. Experimental Validation and Characterization of Super-Enhancers
2.3. Three-Dimensional Organization of Super-Enhancers
2.4. SEs and Transcription Regulation in Different Cell Types
2.5. SE Conservation in Humans, Mice, and Other Placental Organisms
3. Biological Role of Super-Enhancers in the Development of Diseases and Healthy Tissues
3.1. The Role of Super-Enhancers in Non-Pathological Processes
3.2. Super-Enhancers and the Development of Diseases
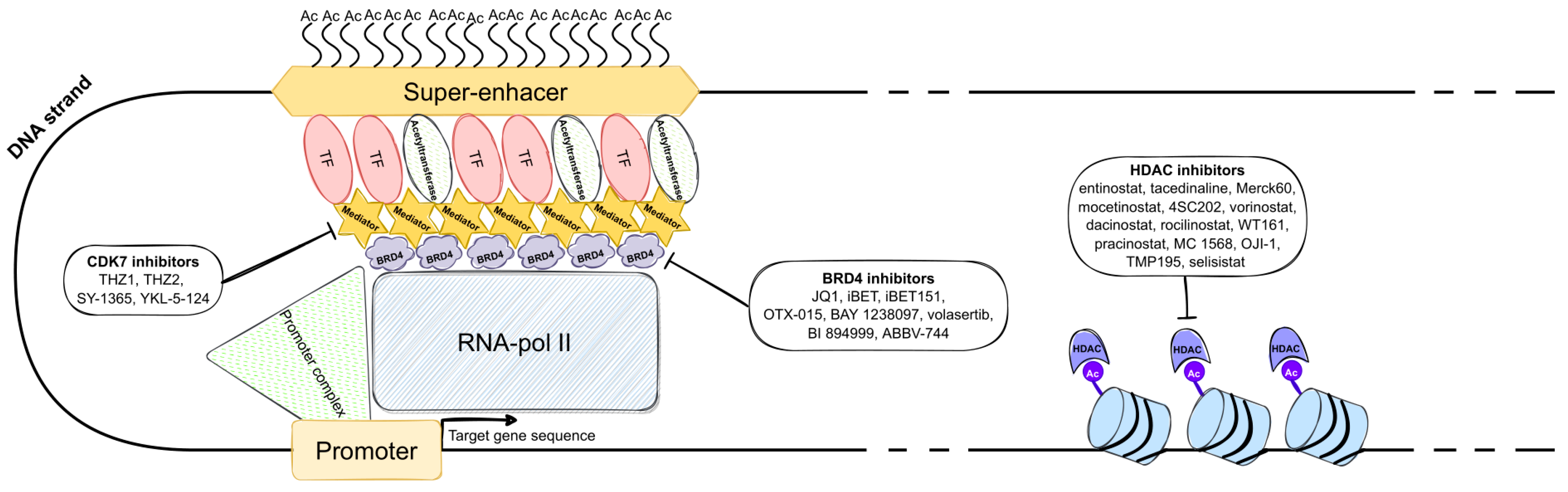
4. Inhibitors of the SE-Mediated Transcription Positive Regulators
4.1. Selective Inhibitors of BET Bromodomains
4.2. Selective CDK7 Inhibitors
4.3. Histone Deacetylase and Demethylase Inhibitors
4.4. Other Potential Inhibitors and Their Targets
4.5. Combined Treatment Strategies
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Pennacchio, L.A.; Bickmore, W.; Dean, A.; Nobrega, M.A.; Bejerano, G. Enhancers: Five Essential Questions. Nat. Rev. Genet. 2013, 14, 288–295. [Google Scholar] [CrossRef] [PubMed]
- Blobel, G.A.; Higgs, D.R.; Mitchell, J.A.; Notani, D.; Young, R.A. Testing the Super-Enhancer Concept. Nat. Rev. Genet. 2021, 22, 749–755. [Google Scholar] [CrossRef] [PubMed]
- Whyte, W.A.; Orlando, D.A.; Hnisz, D.; Abraham, B.J.; Lin, C.Y.; Kagey, M.H.; Rahl, P.B.; Lee, T.I.; Young, R.A. Master Transcription Factors and Mediator Establish Super-Enhancers at Key Cell Identity Genes. Cell 2013, 153, 307–319. [Google Scholar] [CrossRef] [PubMed]
- Parker, S.C.J.; Stitzel, M.L.; Taylor, D.L.; Orozco, J.M.; Erdos, M.R.; Akiyama, J.A.; van Bueren, K.L.; Chines, P.S.; Narisu, N.; NISC Comparative Sequencing Program; et al. Chromatin Stretch Enhancer States Drive Cell-Specific Gene Regulation and Harbor Human Disease Risk Variants. Proc. Natl. Acad. Sci. USA 2013, 110, 17921–17926. [Google Scholar] [CrossRef] [PubMed]
- Hnisz, D.; Abraham, B.J.; Lee, T.I.; Lau, A.; Saint-André, V.; Sigova, A.A.; Hoke, H.A.; Young, R.A. Super-Enhancers in the Control of Cell Identity and Disease. Cell 2013, 155, 934–947. [Google Scholar] [CrossRef]
- Pott, S.; Lieb, J.D. What Are Super-Enhancers? Nat. Genet. 2015, 47, 8–12. [Google Scholar] [CrossRef]
- The ENCODE Project Consortium. An Integrated Encyclopedia of DNA Elements in the Human Genome. Nature 2012, 489, 57–74. [Google Scholar] [CrossRef]
- Chen, Y.; Yao, B.; Zhu, Z.; Yi, Y.; Lin, X.; Zhang, Z.; Shen, G. A Constitutive Super-Enhancer: Homologous Region 3 of Bombyx Mori Nucleopolyhedrovirus. Biochem. Biophys. Res. Commun. 2004, 318, 1039–1044. [Google Scholar] [CrossRef]
- Lovén, J.; Hoke, H.A.; Lin, C.Y.; Lau, A.; Orlando, D.A.; Vakoc, C.R.; Bradner, J.E.; Lee, T.I.; Young, R.A. Selective Inhibition of Tumor Oncogenes by Disruption of Super-Enhancers. Cell 2013, 153, 320–334. [Google Scholar] [CrossRef]
- Sengupta, S.; George, R.E. Super-Enhancer-Driven Transcriptional Dependencies in Cancer. Trends Cancer 2017, 3, 269–281. [Google Scholar] [CrossRef]
- Hnisz, D.; Shrinivas, K.; Young, R.A.; Chakraborty, A.K.; Sharp, P.A. A Phase Separation Model for Transcriptional Control. Cell 2017, 169, 13–23. [Google Scholar] [CrossRef] [PubMed]
- Bruter, A.V.; Rodionova, M.D.; Varlamova, E.A.; Shtil, A.A. Super-Enhancers in the Regulation of Gene Transcription: General Aspects and Antitumor Targets. Acta Nat. 2021, 13, 4–15. [Google Scholar] [CrossRef] [PubMed]
- Roadmap Epigenomics Consortium; Kundaje, A.; Meuleman, W.; Ernst, J.; Bilenky, M.; Yen, A.; Heravi-Moussavi, A.; Kheradpour, P.; Zhang, Z.; Wang, J.; et al. Integrative Analysis of 111 Reference Human Epigenomes. Nature 2015, 518, 317–330. [Google Scholar] [CrossRef] [PubMed]
- Niu, D.-K.; Jiang, L. Can ENCODE Tell Us How Much Junk DNA We Carry in Our Genome? Biochem. Biophys. Res. Commun. 2013, 430, 1340–1343. [Google Scholar] [CrossRef] [PubMed]
- Doolittle, W.F. Is Junk DNA Bunk? A Critique of ENCODE. Proc. Natl. Acad. Sci. USA 2013, 110, 5294–5300. [Google Scholar] [CrossRef] [PubMed]
- Eddy, S.R. The ENCODE Project: Missteps Overshadowing a Success. Curr. Biol. 2013, 23, R259–R261. [Google Scholar] [CrossRef]
- Kellis, M.; Wold, B.; Snyder, M.P.; Bernstein, B.E.; Kundaje, A.; Marinov, G.K.; Ward, L.D.; Birney, E.; Crawford, G.E.; Dekker, J.; et al. Defining Functional DNA Elements in the Human Genome. Proc. Natl. Acad. Sci. USA 2014, 111, 6131–6138. [Google Scholar] [CrossRef]
- Zhang, T.; Zhang, Z.; Dong, Q.; Xiong, J.; Zhu, B. Histone H3K27 Acetylation Is Dispensable for Enhancer Activity in Mouse Embryonic Stem Cells. Genome Biol. 2020, 21, 45. [Google Scholar] [CrossRef]
- Hay, D.; Hughes, J.R.; Babbs, C.; Davies, J.O.J.; Graham, B.J.; Hanssen, L.L.P.; Kassouf, M.T.; Oudelaar, A.M.; Sharpe, J.A.; Suciu, M.C.; et al. Genetic Dissection of the α-Globin Super-Enhancer in Vivo. Nat. Genet. 2016, 48, 895–903. [Google Scholar] [CrossRef]
- Schulz, V.P.; Lezon-Geyda, K.; Maksimova, Y.; Gallagher, P.G. Enhancers and Super Enhancers Are Associated with Genes That Control Phenotypic Traits In Primary Human Erythroid Cells. Blood 2013, 122, 1200. [Google Scholar] [CrossRef]
- Majumder, P.; Lee, J.T.; Rahmberg, A.R.; Kumar, G.; Mi, T.; Scharer, C.D.; Boss, J.M. A Super Enhancer Controls Expression and Chromatin Architecture within the MHC Class II Locus. J. Exp. Med. 2020, 217, e20190668. [Google Scholar] [CrossRef] [PubMed]
- Quang, D.X.; Erdos, M.R.; Parker, S.C.J.; Collins, F.S. Motif Signatures in Stretch Enhancers Are Enriched for Disease-Associated Genetic Variants. Epigenetics Chromatin 2015, 8, 23. [Google Scholar] [CrossRef] [PubMed]
- Vahedi, G.; Kanno, Y.; Furumoto, Y.; Jiang, K.; Parker, S.C.J.; Erdos, M.R.; Davis, S.R.; Roychoudhuri, R.; Restifo, N.P.; Gadina, M.; et al. Super-Enhancers Delineate Disease-Associated Regulatory Nodes in T Cells. Nature 2015, 520, 558–562. [Google Scholar] [CrossRef] [PubMed]
- Khan, A.; Mathelier, A.; Zhang, X. Super-Enhancers Are Transcriptionally More Active and Cell Type-Specific than Stretch Enhancers. Epigenetics 2018, 13, 910–922. [Google Scholar] [CrossRef]
- Dukler, N.; Gulko, B.; Huang, Y.-F.; Siepel, A. Is a Super-Enhancer Greater than the Sum of Its Parts? Nat. Genet. 2017, 49, 2–3. [Google Scholar] [CrossRef] [PubMed]
- Hnisz, D.; Schuijers, J.; Lin, C.Y.; Weintraub, A.S.; Abraham, B.J.; Lee, T.I.; Bradner, J.E.; Young, R.A. Convergence of Developmental and Oncogenic Signaling Pathways at Transcriptional Super-Enhancers. Mol. Cell 2015, 58, 362–370. [Google Scholar] [CrossRef] [PubMed]
- Khan, A.; Zhang, X. DbSUPER: A Database of Super-Enhancers in Mouse and Human Genome. Nucleic Acids Res. 2016, 44, D164–D171. [Google Scholar] [CrossRef]
- Chen, C.; Zhou, D.; Gu, Y.; Wang, C.; Zhang, M.; Lin, X.; Xing, J.; Wang, H.; Zhang, Y. SEA Version 3.0: A Comprehensive Extension and Update of the Super-Enhancer Archive. Nucleic Acids Res. 2019, 48, gkz1028. [Google Scholar] [CrossRef]
- Jiang, Y.; Qian, F.; Bai, X.; Liu, Y.; Wang, Q.; Ai, B.; Han, X.; Shi, S.; Zhang, J.; Li, X.; et al. SEdb: A Comprehensive Human Super-Enhancer Database. Nucleic Acids Res. 2019, 47, D235–D243. [Google Scholar] [CrossRef]
- Wang, X.; Cairns, M.J.; Yan, J. Super-Enhancers in Transcriptional Regulation and Genome Organization. Nucleic Acids Res. 2019, 47, gkz1038. [Google Scholar] [CrossRef]
- Ryu, J.; Kim, H.; Yang, D.; Lee, A.J.; Jung, I. A New Class of Constitutively Active Super-Enhancers Is Associated with Fast Recovery of 3D Chromatin Loops. BMC Bioinform. 2019, 20, 127. [Google Scholar] [CrossRef] [PubMed]
- Khan, A.; Zhang, X. Integrative Modeling Reveals Key Chromatin and Sequence Signatures Predicting Super-Enhancers. Sci. Rep. 2019, 9, 2877. [Google Scholar] [CrossRef] [PubMed]
- Kursa, M.B.; Rudnicki, W.R. Feature Selection with the Boruta Package. J. Stat. Soft. 2010, 36, 1–13. [Google Scholar] [CrossRef]
- Bu, H.; Hao, J.; Gan, Y.; Zhou, S.; Guan, J. DEEPSEN: A Convolutional Neural Network Based Method for Super-Enhancer Prediction. BMC Bioinform. 2019, 20, 598. [Google Scholar] [CrossRef]
- Ji, Q.-Y.; Gong, X.-J.; Li, H.-M.; Du, P.-F. DeepSE: Detecting Super-Enhancers among Typical Enhancers Using Only Sequence Feature Embeddings. Genomics 2021, 113, 4052–4060. [Google Scholar] [CrossRef]
- Gross, D.S.; Garrard, W.T. Nuclease hypersensitive sites in chromatin. Annu. Rev. Biochem. 1988, 57, 159–197. [Google Scholar] [CrossRef]
- Xi, H.; Shulha, H.P.; Lin, J.M.; Vales, T.R.; Fu, Y.; Bodine, D.M.; McKay, R.D.G.; Chenoweth, J.G.; Tesar, P.J.; Furey, T.S.; et al. Identification and Characterization of Cell Type–Specific and Ubiquitous Chromatin Regulatory Structures in the Human Genome. PLoS Genet. 2007, 3, e136. [Google Scholar] [CrossRef]
- Boyle, A.P.; Davis, S.; Shulha, H.P.; Meltzer, P.; Margulies, E.H.; Weng, Z.; Furey, T.S.; Crawford, G.E. High-Resolution Mapping and Characterization of Open Chromatin across the Genome. Cell 2008, 132, 311–322. [Google Scholar] [CrossRef] [PubMed]
- Johnson, D.S.; Mortazavi, A.; Myers, R.M.; Wold, B. Genome-Wide Mapping of in Vivo Protein-DNA Interactions. Science 2007, 316, 1497–1502. [Google Scholar] [CrossRef]
- Barski, A.; Cuddapah, S.; Cui, K.; Roh, T.-Y.; Schones, D.E.; Wang, Z.; Wei, G.; Chepelev, I.; Zhao, K. High-Resolution Profiling of Histone Methylations in the Human Genome. Cell 2007, 129, 823–837. [Google Scholar] [CrossRef]
- Mikkelsen, T.S.; Ku, M.; Jaffe, D.B.; Issac, B.; Lieberman, E.; Giannoukos, G.; Alvarez, P.; Brockman, W.; Kim, T.-K.; Koche, R.P.; et al. Genome-Wide Maps of Chromatin State in Pluripotent and Lineage-Committed Cells. Nature 2007, 448, 553–560. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Liu, X.; Han, L.; Chen, X.; Wu, X.; Wu, J.; Yan, D.; Wang, Y.; Liu, S.; Shan, L.; et al. BRD4-Directed Super-Enhancer Organization of Transcription Repression Programs Links to Chemotherapeutic Efficacy in Breast Cancer. Proc. Natl. Acad. Sci. USA 2022, 119, e2109133119. [Google Scholar] [CrossRef] [PubMed]
- Visel, A.; Blow, M.J.; Li, Z.; Zhang, T.; Akiyama, J.A.; Holt, A.; Plajzer-Frick, I.; Shoukry, M.; Wright, C.; Chen, F.; et al. ChIP-Seq Accurately Predicts Tissue-Specific Activity of Enhancers. Nature 2009, 457, 854–858. [Google Scholar] [CrossRef] [PubMed]
- Creyghton, M.P.; Cheng, A.W.; Welstead, G.G.; Kooistra, T.; Carey, B.W.; Steine, E.J.; Hanna, J.; Lodato, M.A.; Frampton, G.M.; Sharp, P.A.; et al. Histone H3K27ac Separates Active from Poised Enhancers and Predicts Developmental State. Proc. Natl. Acad. Sci. USA 2010, 107, 21931–21936. [Google Scholar] [CrossRef] [PubMed]
- Buenrostro, J.D.; Giresi, P.G.; Zaba, L.C.; Chang, H.Y.; Greenleaf, W.J. Transposition of Native Chromatin for Fast and Sensitive Epigenomic Profiling of Open Chromatin, DNA-Binding Proteins and Nucleosome Position. Nat. Methods 2013, 10, 1213–1218. [Google Scholar] [CrossRef] [PubMed]
- Honnell, V.; Norrie, J.L.; Patel, A.G.; Ramirez, C.; Zhang, J.; Lai, Y.-H.; Wan, S.; Dyer, M.A. Identification of a Modular Super-Enhancer in Murine Retinal Development. Nat. Commun. 2022, 13, 253. [Google Scholar] [CrossRef]
- Krishna, V.; Yin, X.; Song, Q.; Walsh, A.; Pocalyko, D.; Bachman, K.; Anderson, I.; Madakamutil, L.; Nagpal, S. Integration of the Transcriptome and Genome-Wide Landscape of BRD2 and BRD4 Binding Motifs Identifies Key Superenhancer Genes and Reveals the Mechanism of Bet Inhibitor Action in Rheumatoid Arthritis Synovial Fibroblasts. J. Immunol. 2021, 206, 422–431. [Google Scholar] [CrossRef]
- Yates, K.B.; Tonnerre, P.; Martin, G.E.; Gerdemann, U.; Al Abosy, R.; Comstock, D.E.; Weiss, S.A.; Wolski, D.; Tully, D.C.; Chung, R.T.; et al. Epigenetic Scars of CD8+ T Cell Exhaustion Persist after Cure of Chronic Infection in Humans. Nat. Immunol. 2021, 22, 1020–1029. [Google Scholar] [CrossRef]
- Giresi, P.G.; Kim, J.; McDaniell, R.M.; Iyer, V.R.; Lieb, J.D. FAIRE (Formaldehyde-Assisted Isolation of Regulatory Elements) Isolates Active Regulatory Elements from Human Chromatin. Genome Res. 2007, 17, 877–885. [Google Scholar] [CrossRef]
- Thakurela, S.; Sahu, S.K.; Garding, A.; Tiwari, V.K. Dynamics and Function of Distal Regulatory Elements during Neurogenesis and Neuroplasticity. Genome Res. 2015, 25, 1309–1324. [Google Scholar] [CrossRef]
- Core, L.J.; Waterfall, J.J.; Lis, J.T. Nascent RNA Sequencing Reveals Widespread Pausing and Divergent Initiation at Human Promoters. Science 2008, 322, 1845–1848. [Google Scholar] [CrossRef] [PubMed]
- Lopes, R.; Reuven, A.; Gozde, K. GRO-Seq, A Tool for Identification of Transcripts Regulating Gene Expression. Methods Mol. Biol. 2017, 1543, 45–55. [Google Scholar] [CrossRef] [PubMed]
- Meng, F.-L.; Du, Z.; Federation, A.; Hu, J.; Wang, Q.; Kieffer-Kwon, K.-R.; Meyers, R.M.; Amor, C.; Wasserman, C.R.; Neuberg, D.; et al. Convergent Transcription at Intragenic Super-Enhancers Targets AID-Initiated Genomic Instability. Cell 2014, 159, 1538–1548. [Google Scholar] [CrossRef] [PubMed]
- Hah, N.; Benner, C.; Chong, L.-W.; Yu, R.T.; Downes, M.; Evans, R.M. Inflammation-Sensitive Super Enhancers Form Domains of Coordinately Regulated Enhancer RNAs. Proc. Natl. Acad. Sci. USA 2015, 112, E297–E302. [Google Scholar] [CrossRef]
- Lu, Y.; Shou, J.; Jia, Z.; Wu, Y.; Li, J.; Guo, Y.; Wu, Q. Genetic Evidence for Asymmetric Blocking of Higher-Order Chromatin Structure by CTCF/Cohesin. Protein Cell 2019, 10, 914–920. [Google Scholar] [CrossRef]
- Li, Y.; Rivera, C.M.; Ishii, H.; Jin, F.; Selvaraj, S.; Lee, A.Y.; Dixon, J.R.; Ren, B. CRISPR Reveals a Distal Super-Enhancer Required for Sox2 Expression in Mouse Embryonic Stem Cells. PLoS ONE 2014, 9, e114485. [Google Scholar] [CrossRef]
- Zhang, X.; Choi, P.S.; Francis, J.M.; Imielinski, M.; Watanabe, H.; Cherniack, A.D.; Meyerson, M. Identification of Focally Amplified Lineage-Specific Super-Enhancers in Human Epithelial Cancers. Nat. Genet. 2016, 48, 176–182. [Google Scholar] [CrossRef]
- Cui, S.; Wu, Q.; Liu, M.; Su, M.; Liu, S.; Shao, L.; Han, X.; He, H. EphA2 Super-Enhancer Promotes Tumor Progression by Recruiting FOSL2 and TCF7L2 to Activate the Target Gene EphA2. Cell Death Dis. 2021, 12, 264. [Google Scholar] [CrossRef]
- Mushimiyimana, I.; Niskanen, H.; Beter, M.; Laakkonen, J.P.; Kaikkonen, M.U.; Ylä-Herttuala, S.; Laham-Karam, N. Characterization of a Functional Endothelial Super-Enhancer That Regulates ADAMTS18 and Angiogenesis. Nucleic Acids Res. 2021, 49, 8078–8096. [Google Scholar] [CrossRef]
- Guo, Y.; Xu, Q.; Canzio, D.; Shou, J.; Li, J.; Gorkin, D.U.; Jung, I.; Wu, H.; Zhai, Y.; Tang, Y.; et al. CRISPR Inversion of CTCF Sites Alters Genome Topology and Enhancer/Promoter Function. Cell 2015, 162, 900–910. [Google Scholar] [CrossRef]
- Toyoda, S.; Kawaguchi, M.; Kobayashi, T.; Tarusawa, E.; Toyama, T.; Okano, M.; Oda, M.; Nakauchi, H.; Yoshimura, Y.; Sanbo, M.; et al. Developmental Epigenetic Modification Regulates Stochastic Expression of Clustered Protocadherin Genes, Generating Single Neuron Diversity. Neuron 2014, 82, 94–108. [Google Scholar] [CrossRef] [PubMed]
- Monahan, K.; Horta, A.; Lomvardas, S. LHX2- and LDB1-Mediated Trans Interactions Regulate Olfactory Receptor Choice. Nature 2019, 565, 448–453. [Google Scholar] [CrossRef] [PubMed]
- Mach, P.; Kos, P.I.; Zhan, Y.; Cramard, J.; Gaudin, S.; Tünnermann, J.; Marchi, E.; Eglinger, J.; Zuin, J.; Kryzhanovska, M.; et al. Cohesin and CTCF Control the Dynamics of Chromosome Folding. Nat. Genet. 2022, 54, 1907–1918. [Google Scholar] [CrossRef] [PubMed]
- Canzio, D.; Nwakeze, C.L.; Horta, A.; Rajkumar, S.M.; Coffey, E.L.; Duffy, E.E.; Duffié, R.; Monahan, K.; O’Keeffe, S.; Simon, M.D.; et al. Antisense LncRNA Transcription Mediates DNA Demethylation to Drive Stochastic Protocadherin α Promoter Choice. Cell 2019, 177, 639–653.e15. [Google Scholar] [CrossRef]
- Damaschke, N.A.; Gawdzik, J.; Avilla, M.; Yang, B.; Svaren, J.; Roopra, A.; Luo, J.-H.; Yu, Y.P.; Keles, S.; Jarrard, D.F. CTCF Loss Mediates Unique DNA Hypermethylation Landscapes in Human Cancers. Clin. Epigenet. 2020, 12, 80. [Google Scholar] [CrossRef]
- Ing-Simmons, E.; Seitan, V.C.; Faure, A.J.; Flicek, P.; Carroll, T.; Dekker, J.; Fisher, A.G.; Lenhard, B.; Merkenschlager, M. Spatial Enhancer Clustering and Regulation of Enhancer-Proximal Genes by Cohesin. Genome Res. 2015, 25, 504–513. [Google Scholar] [CrossRef]
- Willi, M.; Yoo, K.H.; Reinisch, F.; Kuhns, T.M.; Lee, H.K.; Wang, C.; Hennighausen, L. Facultative CTCF Sites Moderate Mammary Super-Enhancer Activity and Regulate Juxtaposed Gene in Non-Mammary Cells. Nat. Commun 2017, 8, 16069. [Google Scholar] [CrossRef]
- Vos, E.S.M.; Valdes-Quezada, C.; Huang, Y.; Allahyar, A.; Verstegen, M.J.A.M.; Felder, A.-K.; van der Vegt, F.; Uijttewaal, E.C.H.; Krijger, P.H.L.; de Laat, W. Interplay between CTCF Boundaries and a Super Enhancer Controls Cohesin Extrusion Trajectories and Gene Expression. Mol. Cell 2021, 81, 3082–3095.e6. [Google Scholar] [CrossRef]
- Jia, Z.; Li, J.; Ge, X.; Wu, Y.; Guo, Y.; Wu, Q. Tandem CTCF Sites Function as Insulators to Balance Spatial Chromatin Contacts and Topological Enhancer-Promoter Selection. Genome Biol. 2020, 21, 75. [Google Scholar] [CrossRef]
- Cho, W.-K.; Spille, J.-H.; Hecht, M.; Lee, C.; Li, C.; Grube, V.; Cisse, I.I. Mediator and RNA Polymerase II Clusters Associate in Transcription-Dependent Condensates. Science 2018, 361, 412–415. [Google Scholar] [CrossRef]
- Sabari, B.R.; Dall’Agnese, A.; Boija, A.; Klein, I.A.; Coffey, E.L.; Shrinivas, K.; Abraham, B.J.; Hannett, N.M.; Zamudio, A.V.; Manteiga, J.C.; et al. Coactivator Condensation at Super-Enhancers Links Phase Separation and Gene Control. Science 2018, 361, eaar3958. [Google Scholar] [CrossRef] [PubMed]
- Lee, R.; Kang, M.-K.; Kim, Y.-J.; Yang, B.; Shim, H.; Kim, S.; Kim, K.; Yang, C.M.; Min, B.; Jung, W.-J.; et al. CTCF-Mediated Chromatin Looping Provides a Topological Framework for the Formation of Phase-Separated Transcriptional Condensates. Nucleic Acids Res. 2022, 50, 207–226. [Google Scholar] [CrossRef] [PubMed]
- Khoury, A.; Achinger-Kawecka, J.; Bert, S.A.; Smith, G.C.; French, H.J.; Luu, P.-L.; Peters, T.J.; Du, Q.; Parry, A.J.; Valdes-Mora, F.; et al. Constitutively Bound CTCF Sites Maintain 3D Chromatin Architecture and Long-Range Epigenetically Regulated Domains. Nat. Commun. 2020, 11, 54. [Google Scholar] [CrossRef] [PubMed]
- Bansal, K.; Michelson, D.A.; Ramirez, R.N.; Viny, A.D.; Levine, R.L.; Benoist, C.; Mathis, D. Aire Regulates Chromatin Looping by Evicting CTCF from Domain Boundaries and Favoring Accumulation of Cohesin on Superenhancers. Proc. Natl. Acad. Sci. USA 2021, 118, e2110991118. [Google Scholar] [CrossRef]
- Osterwalder, M.; Barozzi, I.; Tissières, V.; Fukuda-Yuzawa, Y.; Mannion, B.J.; Afzal, S.Y.; Lee, E.A.; Zhu, Y.; Plajzer-Frick, I.; Pickle, C.S.; et al. Enhancer Redundancy Provides Phenotypic Robustness in Mammalian Development. Nature 2018, 554, 239–243. [Google Scholar] [CrossRef]
- Kai, Y.; Li, B.E.; Zhu, M.; Li, G.Y.; Chen, F.; Han, Y.; Cha, H.J.; Orkin, S.H.; Cai, W.; Huang, J.; et al. Mapping the Evolving Landscape of Super-Enhancers during Cell Differentiation. Genome Biol. 2021, 22, 269. [Google Scholar] [CrossRef]
- Liu, X.; Zhao, B.; Shaw, T.I.; Fridley, B.L.; Duckett, D.R.; Tan, A.C.; Teng, M. Summarizing Internal Dynamics Boosts Differential Analysis and Functional Interpretation of Super Enhancers. Nucleic Acids Res. 2022, 50, 3115–3127. [Google Scholar] [CrossRef]
- Vanhille, L.; Griffon, A.; Maqbool, M.A.; Zacarias-Cabeza, J.; Dao, L.T.M.; Fernandez, N.; Ballester, B.; Andrau, J.C.; Spicuglia, S. High-Throughput and Quantitative Assessment of Enhancer Activity in Mammals by CapStarr-Seq. Nat. Commun. 2015, 6, 6905. [Google Scholar] [CrossRef]
- Bergman, D.T.; Jones, T.R.; Liu, V.; Ray, J.; Jagoda, E.; Siraj, L.; Kang, H.Y.; Nasser, J.; Kane, M.; Rios, A.; et al. Compatibility Rules of Human Enhancer and Promoter Sequences. Nature 2022, 607, 176–184. [Google Scholar] [CrossRef]
- Kassouf, M.T.; Francis, H.S.; Gosden, M.; Suciu, M.C.; Downes, D.J.; Harrold, C.; Larke, M.; Oudelaar, M.; Cornell, L.; Blayney, J.; et al. Multipartite Super-Enhancers Function in an Orientation-Dependent Manner. bioRxiv 2022. [Google Scholar] [CrossRef]
- Blayney, J.; Francis, H.; Camellato, B.; Mitchell, L.; Stolper, R.; Boeke, J.; Higgs, D.; Kassouf, M. Super-Enhancers Require a Combination of Classical Enhancers and Novel Facilitator Elements to Drive High Levels of Gene Expression. bioRxiv 2022. [Google Scholar] [CrossRef]
- Wong, E.S.; Zheng, D.; Tan, S.Z.; Bower, N.I.; Garside, V.; Vanwalleghem, G.; Gaiti, F.; Scott, E.; Hogan, B.M.; Kikuchi, K.; et al. Deep Conservation of the Enhancer Regulatory Code in Animals. Science 2020, 370, eaax8137. [Google Scholar] [CrossRef] [PubMed]
- Villar, D.; Berthelot, C.; Aldridge, S.; Rayner, T.F.; Lukk, M.; Pignatelli, M.; Park, T.J.; Deaville, R.; Erichsen, J.T.; Jasinska, A.J.; et al. Enhancer Evolution across 20 Mammalian Species. Cell 2015, 160, 554–566. [Google Scholar] [CrossRef] [PubMed]
- Flores, M.A.; Ovcharenko, I. Enhancer Reprogramming in Mammalian Genomes. BMC Bioinform. 2018, 19, 316. [Google Scholar] [CrossRef]
- Zboril, E.; Yoo, H.; Chen, L.; Liu, Z. Dynamic Interactions of Transcription Factors and Enhancer Reprogramming in Cancer Progression. Front. Oncol. 2021, 11, 753051. [Google Scholar] [CrossRef]
- Bi, M.; Zhang, Z.; Jiang, Y.-Z.; Xue, P.; Wang, H.; Lai, Z.; Fu, X.; De Angelis, C.; Gong, Y.; Gao, Z.; et al. Enhancer Reprogramming Driven by High-Order Assemblies of Transcription Factors Promotes Phenotypic Plasticity and Breast Cancer Endocrine Resistance. Nat. Cell Biol. 2020, 22, 701–715. [Google Scholar] [CrossRef]
- Kamm, G.B.; Pisciottano, F.; Kliger, R.; Franchini, L.F. The Developmental Brain Gene NPAS3 Contains the Largest Number of Accelerated Regulatory Sequences in the Human Genome. Mol. Biol. Evol. 2013, 30, 1088–1102. [Google Scholar] [CrossRef]
- May, D.; Blow, M.J.; Kaplan, T.; McCulley, D.J.; Jensen, B.C.; Akiyama, J.A.; Holt, A.; Plajzer-Frick, I.; Shoukry, M.; Wright, C.; et al. Large-Scale Discovery of Enhancers from Human Heart Tissue. Nat. Genet. 2012, 44, 89–93. [Google Scholar] [CrossRef]
- Prabhakar, S.; Visel, A.; Akiyama, J.A.; Shoukry, M.; Lewis, K.D.; Holt, A.; Plajzer-Frick, I.; Morrison, H.; FitzPatrick, D.R.; Afzal, V.; et al. Human-Specific Gain of Function in a Developmental Enhancer. Science 2008, 321, 1346–1350. [Google Scholar] [CrossRef]
- Sumiyama, K.; Saitou, N. Loss-of-Function Mutation in a Repressor Module of Human-Specifically Activated Enhancer HACNS1. Mol. Biol. Evol. 2011, 28, 3005–3007. [Google Scholar] [CrossRef]
- Capra, J.A.; Erwin, G.D.; McKinsey, G.; Rubenstein, J.L.R.; Pollard, K.S. Many Human Accelerated Regions Are Developmental Enhancers. Phil. Trans. R. Soc. B 2013, 368, 20130025. [Google Scholar] [CrossRef] [PubMed]
- See, Y.X.; Chen, K.; Fullwood, M.J. MYC Overexpression Leads to Increased Chromatin Interactions at Super-Enhancers and MYC Binding Sites. Genome Res. 2022, 32, 629–642. [Google Scholar] [CrossRef] [PubMed]
- Oudelaar, A.M.; Davies, J.O.J.; Hanssen, L.L.P.; Telenius, J.M.; Schwessinger, R.; Liu, Y.; Brown, J.M.; Downes, D.J.; Chiariello, A.M.; Bianco, S.; et al. Single-Allele Chromatin Interactions Identify Regulatory Hubs in Dynamic Compartmentalized Domains. Nat. Genet. 2018, 50, 1744–1751. [Google Scholar] [CrossRef] [PubMed]
- Allahyar, A.; Vermeulen, C.; Bouwman, B.A.M.; Krijger, P.H.L.; Verstegen, M.J.A.M.; Geeven, G.; van Kranenburg, M.; Pieterse, M.; Straver, R.; Haarhuis, J.H.I.; et al. Enhancer Hubs and Loop Collisions Identified from Single-Allele Topologies. Nat. Genet. 2018, 50, 1151–1160. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Zhou, Y.; Yue, W.; Zhu, Z.; Wu, X.; Yu, S.; Shen, Q.; Pan, Q.; Xu, W.; Zhang, R.; et al. Deeply Conserved Super-Enhancers Maintain Stem Cell Pluripotency in Placental Mammals. bioRxiv 2022. [Google Scholar] [CrossRef]
- Peng, Y.; Kang, H.; Luo, J.; Zhang, Y. A Comparative Analysis of Super-Enhancers and Broad H3K4me3 Domains in Pig, Human, and Mouse Tissues. Front. Genet. 2021, 12, 701049. [Google Scholar] [CrossRef]
- Luan, Y.; Zhang, L.; Hu, M.; Xu, Y.; Hou, Y.; Li, X.; Zhao, S.; Zhao, Y.; Li, C. Identification and Conservation Analysis of Cis-Regulatory Elements in Pig Liver. Genes 2019, 10, 348. [Google Scholar] [CrossRef]
- Pérez-Rico, Y.A.; Boeva, V.; Mallory, A.C.; Bitetti, A.; Majello, S.; Barillot, E.; Shkumatava, A. Comparative Analyses of Super-Enhancers Reveal Conserved Elements in Vertebrate Genomes. Genome Res. 2017, 27, 259–268. [Google Scholar] [CrossRef]
- Zhao, Y.; Hou, Y.; Xu, Y.; Luan, Y.; Zhou, H.; Qi, X.; Hu, M.; Wang, D.; Wang, Z.; Fu, Y.; et al. A Compendium and Comparative Epigenomics Analysis of Cis-Regulatory Elements in the Pig Genome. Nat. Commun. 2021, 12, 2217. [Google Scholar] [CrossRef]
- Man, J.C.K.; van Duijvenboden, K.; Krijger, P.H.L.; Hooijkaas, I.B.; van der Made, I.; de Gier-de Vries, C.; Wakker, V.; Creemers, E.E.; de Laat, W.; Boukens, B.J.; et al. Genetic Dissection of a Super Enhancer Controlling the Nppa-Nppb Cluster in the Heart. Circ. Res. 2021, 128, 115–129. [Google Scholar] [CrossRef]
- Shiau, C.-K.; Huang, J.-H.; Liu, Y.-T.; Tsai, H.-K. Genome-Wide Identification of Associations between Enhancer and Alternative Splicing in Human and Mouse. BMC Genom. 2021, 22, 919. [Google Scholar] [CrossRef] [PubMed]
- Hazan, I.; Monin, J.; Bouwman, B.A.M.; Crosetto, N.; Aqeilan, R.I. Activation of Oncogenic Super-Enhancers Is Coupled with DNA Repair by RAD51. Cell Rep. 2019, 29, 560–572.e4. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Yang, Y.; Gu, H.; Tao, B.; Zhang, E.; Wei, J.; Wang, Z.; Liu, A.; Sun, R.; Chen, M.; et al. Multi-omics Analysis Reveals the Functional Transcription and Potential Translation of Enhancers. Int. J. Cancer 2020, 147, 2210–2224. [Google Scholar] [CrossRef] [PubMed]
- Rennie, S.; Dalby, M.; Lloret-Llinares, M.; Bakoulis, S.; Dalager Vaagensø, C.; Heick Jensen, T.; Andersson, R. Transcription Start Site Analysis Reveals Widespread Divergent Transcription in D. Melanogaster and Core Promoter-Encoded Enhancer Activities. Nucleic Acids Res. 2018, 46, 5455–5469. [Google Scholar] [CrossRef] [PubMed]
- Kainth, A.S.; Chowdhary, S.; Pincus, D.; Gross, D.S. Primordial Super-Enhancers: Heat Shock-Induced Chromatin Organization in Yeast. Trends Cell Biol. 2021, 31, 801–813. [Google Scholar] [CrossRef] [PubMed]
- Chowdhary, S.; Kainth, A.S.; Paracha, S.; Gross, D.S.; Pincus, D. Inducible Transcriptional Condensates Drive 3D Genome Reorganization in the Heat Shock Response. Mol. Cell 2022, 82, 4386–4399.e7. [Google Scholar] [CrossRef] [PubMed]
- Benton, M.L.; Talipineni, S.C.; Kostka, D.; Capra, J.A. Genome-Wide Enhancer Annotations Differ Significantly in Genomic Distribution, Evolution, and Function. BMC Genom. 2019, 20, 511. [Google Scholar] [CrossRef]
- Chen, L.; Fish, A.E.; Capra, J.A. Prediction of Gene Regulatory Enhancers across Species Reveals Evolutionarily Conserved Sequence Properties. PLoS Comput. Biol. 2018, 14, e1006484. [Google Scholar] [CrossRef]
- Young, R.A. Control of the Embryonic Stem Cell State. Cell 2011, 144, 940–954. [Google Scholar] [CrossRef]
- Ng, H.-H.; Surani, M.A. The Transcriptional and Signalling Networks of Pluripotency. Nat. Cell Biol. 2011, 13, 490–496. [Google Scholar] [CrossRef]
- Tsai, P.-H.; Chien, Y.; Wang, M.-L.; Hsu, C.-H.; Laurent, B.; Chou, S.-J.; Chang, W.-C.; Chien, C.-S.; Li, H.-Y.; Lee, H.-C.; et al. Ash2l Interacts with Oct4-Stemness Circuitry to Promote Super-Enhancer-Driven Pluripotency Network. Nucleic Acids Res. 2019, 47, 10115–10133. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.Y.; Katsman, Y.; Dhaliwal, N.K.; Davidson, S.; Macpherson, N.N.; Sakthidevi, M.; Collura, F.; Mitchell, J.A. A Sox2 Distal Enhancer Cluster Regulates Embryonic Stem Cell Differentiation Potential. Genes Dev. 2014, 28, 2699–2711. [Google Scholar] [CrossRef] [PubMed]
- Yue, F.; Cheng, Y.; Breschi, A.; Vierstra, J.; Wu, W.; Ryba, T.; Sandstrom, R.; Ma, Z.; Davis, C.; Pope, B.D.; et al. A Comparative Encyclopedia of DNA Elements in the Mouse Genome. Nature 2014, 515, 355–364. [Google Scholar] [CrossRef] [PubMed]
- Blinka, S.; Reimer, M.H.; Pulakanti, K.; Rao, S. Super-Enhancers at the Nanog Locus Differentially Regulate Neighboring Pluripotency-Associated Genes. Cell Rep. 2016, 17, 19–28. [Google Scholar] [CrossRef]
- Di Micco, R.; Fontanals-Cirera, B.; Low, V.; Ntziachristos, P.; Yuen, S.K.; Lovell, C.D.; Dolgalev, I.; Yonekubo, Y.; Zhang, G.; Rusinova, E.; et al. Control of Embryonic Stem Cell Identity by BRD4-Dependent Transcriptional Elongation of Super-Enhancer-Associated Pluripotency Genes. Cell Rep. 2014, 9, 234–247. [Google Scholar] [CrossRef] [PubMed]
- Bortvin, A.; Winston, F. Evidence That Spt6p Controls Chromatin Structure by a Direct Interaction with Histones. Science 1996, 272, 1473–1476. [Google Scholar] [CrossRef] [PubMed]
- Wang, A.H.; Juan, A.H.; Ko, K.D.; Tsai, P.-F.; Zare, H.; Dell’Orso, S.; Sartorelli, V. The Elongation Factor Spt6 Maintains ESC Pluripotency by Controlling Super-Enhancers and Counteracting Polycomb Proteins. Mol. Cell 2017, 68, 398–413.e6. [Google Scholar] [CrossRef]
- Zhang, J.; Zhou, Y.; Yue, W.; Zhu, Z.; Wu, X.; Yu, S.; Shen, Q.; Pan, Q.; Xu, W.; Zhang, R.; et al. Super-Enhancers Conserved within Placental Mammals Maintain Stem Cell Pluripotency. Proc. Natl. Acad. Sci. USA 2022, 119, e2204716119. [Google Scholar] [CrossRef]
- Wang, C.; Tian, W.; Hu, S.-Y.; Di, C.-X.; He, C.-Y.; Cao, Q.-L.; Hao, R.-H.; Dong, S.-S.; Liu, C.-C.; Rong, Y.; et al. Lineage-Selective Super Enhancers Mediate Core Regulatory Circuitry during Adipogenic and Osteogenic Differentiation of Human Mesenchymal Stem Cells. Cell Death Dis. 2022, 13, 866. [Google Scholar] [CrossRef]
- Huang, Z.; Jia, B.; Wang, Q.; Wang, N.; Zhao, J. The Potential Function of Super Enhancers in Human Bone Marrow Mesenchymal Stem Cells during Osteogenic Differentiation. BioMed Res. Int. 2021, 2021, 6614762. [Google Scholar] [CrossRef]
- Factor, D.C.; Corradin, O.; Zentner, G.E.; Saiakhova, A.; Song, L.; Chenoweth, J.G.; McKay, R.D.; Crawford, G.E.; Scacheri, P.C.; Tesar, P.J. Epigenomic Comparison Reveals Activation of “Seed” Enhancers during Transition from Naive to Primed Pluripotency. Cell Stem Cell 2014, 14, 854–863. [Google Scholar] [CrossRef] [PubMed]
- Lee, B.-K.; Jang, Y.; Kim, M.; LeBlanc, L.; Rhee, C.; Lee, J.; Beck, S.; Shen, W.; Kim, J. Super-Enhancer-Guided Mapping of Regulatory Networks Controlling Mouse Trophoblast Stem Cells. Nat. Commun. 2019, 10, 4749. [Google Scholar] [CrossRef] [PubMed]
- Klein, R.H.; Hu, W.; Kashgari, G.; Lin, Z.; Nguyen, T.; Doan, M.; Andersen, B. Characterization of Enhancers and the Role of the Transcription Factor KLF7 in Regulating Corneal Epithelial Differentiation. J. Biol. Chem. 2017, 292, 18937–18950. [Google Scholar] [CrossRef] [PubMed]
- Siersbæk, R.; Rabiee, A.; Nielsen, R.; Sidoli, S.; Traynor, S.; Loft, A.; Poulsen, L.L.C.; Rogowska-Wrzesinska, A.; Jensen, O.N.; Mandrup, S. Transcription Factor Cooperativity in Early Adipogenic Hotspots and Super-Enhancers. Cell Rep. 2014, 7, 1443–1455. [Google Scholar] [CrossRef] [PubMed]
- Brunmeir, R.; Wu, J.; Peng, X.; Kim, S.-Y.; Julien, S.G.; Zhang, Q.; Xie, W.; Xu, F. Comparative Transcriptomic and Epigenomic Analyses Reveal New Regulators of Murine Brown Adipogenesis. PLoS Genet. 2016, 12, e1006474. [Google Scholar] [CrossRef]
- Huang, J.; Liu, X.; Li, D.; Shao, Z.; Cao, H.; Zhang, Y.; Trompouki, E.; Bowman, T.V.; Zon, L.I.; Yuan, G.-C.; et al. Dynamic Control of Enhancer Repertoires Drives Lineage and Stage-Specific Transcription during Hematopoiesis. Dev. Cell 2016, 36, 9–23. [Google Scholar] [CrossRef]
- Aranda-Orgilles, B.; Saldaña-Meyer, R.; Wang, E.; Trompouki, E.; Fassl, A.; Lau, S.; Mullenders, J.; Rocha, P.P.; Raviram, R.; Guillamot, M.; et al. MED12 Regulates HSC-Specific Enhancers Independently of Mediator Kinase Activity to Control Hematopoiesis. Cell Stem Cell 2016, 19, 784–799. [Google Scholar] [CrossRef]
- Liu, C.-F.; Lefebvre, V. The Transcription Factors SOX9 and SOX5/SOX6 Cooperate Genome-Wide through Super-Enhancers to Drive Chondrogenesis. Nucleic Acids Res. 2015, 43, 8183–8203. [Google Scholar] [CrossRef]
- Ang, Y.-S.; Rivas, R.N.; Ribeiro, A.J.S.; Srivas, R.; Rivera, J.; Stone, N.R.; Pratt, K.; Mohamed, T.M.A.; Fu, J.-D.; Spencer, C.I.; et al. Disease Model of GATA4 Mutation Reveals Transcription Factor Cooperativity in Human Cardiogenesis. Cell 2016, 167, 1734–1749.e22. [Google Scholar] [CrossRef]
- Zhao, Y.; Zhou, J.; He, L.; Li, Y.; Yuan, J.; Sun, K.; Chen, X.; Bao, X.; Esteban, M.A.; Sun, H.; et al. MyoD Induced Enhancer RNA Interacts with HnRNPL to Activate Target Gene Transcription during Myogenic Differentiation. Nat. Commun. 2019, 10, 5787. [Google Scholar] [CrossRef]
- Caputo, V.S.; Trasanidis, N.; Xiao, X.; Robinson, M.E.; Katsarou, A.; Ponnusamy, K.; Prinjha, R.K.; Smithers, N.; Chaidos, A.; Auner, H.W.; et al. Brd2/4 and Myc Regulate Alternative Cell Lineage Programmes during Early Osteoclast Differentiation in Vitro. iScience 2021, 24, 101989. [Google Scholar] [CrossRef] [PubMed]
- Horie, M.; Miyashita, N.; Mikami, Y.; Noguchi, S.; Yamauchi, Y.; Suzukawa, M.; Fukami, T.; Ohta, K.; Asano, Y.; Sato, S.; et al. TBX4 Is Involved in the Super-Enhancer-Driven Transcriptional Programs Underlying Features Specific to Lung Fibroblasts. Am. J. Physiol.-Lung Cell. Mol. Physiol. 2018, 314, L177–L191. [Google Scholar] [CrossRef] [PubMed]
- Paraiso, K.D.; Blitz, I.L.; Coley, M.; Cheung, J.; Sudou, N.; Taira, M.; Cho, K.W.Y. Endodermal Maternal Transcription Factors Establish Super-Enhancers during Zygotic Genome Activation. Cell Rep. 2019, 27, 2962–2977.e5. [Google Scholar] [CrossRef] [PubMed]
- Kitagawa, Y.; Ohkura, N.; Kidani, Y.; Vandenbon, A.; Hirota, K.; Kawakami, R.; Yasuda, K.; Motooka, D.; Nakamura, S.; Kondo, M.; et al. Guidance of Regulatory T Cell Development by Satb1-Dependent Super-Enhancer Establishment. Nat. Immunol. 2017, 18, 173–183. [Google Scholar] [CrossRef]
- Thomas, G.D.; Hanna, R.N.; Vasudevan, N.T.; Hamers, A.A.; Romanoski, C.E.; McArdle, S.; Ross, K.D.; Blatchley, A.; Yoakum, D.; Hamilton, B.A.; et al. Deleting an Nr4a1 Super-Enhancer Subdomain Ablates Ly6C Low Monocytes While Preserving Macrophage Gene Function. Immunity 2016, 45, 975–987. [Google Scholar] [CrossRef]
- Shin, H.Y.; Willi, M.; Yoo, K.H.; Zeng, X.; Wang, C.; Metser, G.; Hennighausen, L. Hierarchy within the Mammary STAT5-Driven Wap Super-Enhancer. Nat. Genet. 2016, 48, 904–911. [Google Scholar] [CrossRef]
- Lee, H.K.; Willi, M.; Shin, H.Y.; Liu, C.; Hennighausen, L. Progressing Super-Enhancer Landscape during Mammary Differentiation Controls Tissue-Specific Gene Regulation. Nucleic Acids Res. 2018, 46, 10796–10809. [Google Scholar] [CrossRef]
- Martinez, M.F.; Medrano, S.; Brown, E.A.; Tufan, T.; Shang, S.; Bertoncello, N.; Guessoum, O.; Adli, M.; Belyea, B.C.; Sequeira-Lopez, M.L.S.; et al. Super-Enhancers Maintain Renin-Expressing Cell Identity and Memory to Preserve Multi-System Homeostasis. J. Clin. Investig. 2018, 128, 4787–4803. [Google Scholar] [CrossRef]
- Alvarez-Dominguez, J.R.; Knoll, M.; Gromatzky, A.A.; Lodish, H.F. The Super-Enhancer-Derived AlncRNA-EC7/Bloodlinc Potentiates Red Blood Cell Development in Trans. Cell Rep. 2017, 19, 2503–2514. [Google Scholar] [CrossRef]
- Dubois-Chevalier, J.; Dubois, V.; Staels, B.; Lefebvre, P.; Eeckhoute, J. Perspectives on the Use of Super-Enhancers as a Defining Feature of Cell/Tissue-Identity Genes. Epigenomics 2020, 12, 715–723. [Google Scholar] [CrossRef]
- Nott, A.; Holtman, I.R.; Coufal, N.G.; Schlachetzki, J.C.M.; Yu, M.; Hu, R.; Han, C.Z.; Pena, M.; Xiao, J.; Wu, Y.; et al. Brain Cell Type–Specific Enhancer–Promoter Interactome Maps and Disease—Risk Association. Science 2019, 366, 1134–1139. [Google Scholar] [CrossRef]
- Achour, M.; Le Gras, S.; Keime, C.; Parmentier, F.; Lejeune, F.-X.; Boutillier, A.-L.; Neri, C.; Davidson, I.; Merienne, K. Neuronal Identity Genes Regulated by Super-Enhancers Are Preferentially down-Regulated in the Striatum of Huntington’s Disease Mice. Hum. Mol. Genet. 2015, 24, 3481–3496. [Google Scholar] [CrossRef] [PubMed]
- Brown, J.D.; Lin, C.Y.; Duan, Q.; Griffin, G.; Federation, A.J.; Paranal, R.M.; Bair, S.; Newton, G.; Lichtman, A.H.; Kung, A.L.; et al. NF-ΚB Directs Dynamic Super Enhancer Formation in Inflammation and Atherogenesis. Mol. Cell 2014, 56, 219–231. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, S.F.; Larsen, B.D.; Loft, A.; Nielsen, R.; Madsen, J.G.S.; Mandrup, S. Acute TNF-Induced Repression of Cell Identity Genes Is Mediated by NFκB-Directed Redistribution of Cofactors from Super-Enhancers. Genome Res. 2015, 25, 1281–1294. [Google Scholar] [CrossRef] [PubMed]
- Higashijima, Y.; Matsui, Y.; Shimamura, T.; Nakaki, R.; Nagai, N.; Tsutsumi, S.; Abe, Y.; Link, V.M.; Osaka, M.; Yoshida, M.; et al. Coordinated Demethylation of H3K9 and H3K27 Is Required for Rapid Inflammatory Responses of Endothelial Cells. EMBO J. 2020, 39, e103949. [Google Scholar] [CrossRef]
- Xiao, X.; Fan, Y.; Li, J.; Zhang, X.; Lou, X.; Dou, Y.; Shi, X.; Lan, P.; Xiao, Y.; Minze, L.; et al. Guidance of Super-Enhancers in Regulation of IL-9 Induction and Airway Inflammation. J. Exp. Med. 2018, 215, 559–574. [Google Scholar] [CrossRef]
- Peeters, J.G.C.; Vervoort, S.J.; Tan, S.C.; Mijnheer, G.; de Roock, S.; Vastert, S.J.; Nieuwenhuis, E.E.S.; van Wijk, F.; Prakken, B.J.; Creyghton, M.P.; et al. Inhibition of Super-Enhancer Activity in Autoinflammatory Site-Derived T Cells Reduces Disease-Associated Gene Expression. Cell Rep. 2015, 12, 1986–1996. [Google Scholar] [CrossRef]
- Rakyan, V.K.; Down, T.A.; Balding, D.J.; Beck, S. Epigenome-Wide Association Studies for Common Human Diseases. Nat. Rev. Genet. 2011, 12, 529–541. [Google Scholar] [CrossRef]
- Lacey, M.; Baribault, C.; Ehrlich, K.C.; Ehrlich, M. Atherosclerosis-Associated Differentially Methylated Regions Can Reflect the Disease Phenotype and Are Often at Enhancers. Atherosclerosis 2019, 280, 183–191. [Google Scholar] [CrossRef]
- Sun, W.; Yao, S.; Tang, J.; Liu, S.; Chen, J.; Deng, D.; Zeng, C. Integrative Analysis of Super Enhancer SNPs for Type 2 Diabetes. PLoS ONE 2018, 13, e0192105. [Google Scholar] [CrossRef]
- Kubota, S.; Tokunaga, K.; Umezu, T.; Yokomizo-Nakano, T.; Sun, Y.; Oshima, M.; Tan, K.T.; Yang, H.; Kanai, A.; Iwanaga, E.; et al. Lineage-Specific RUNX2 Super-Enhancer Activates MYC and Promotes the Development of Blastic Plasmacytoid Dendritic Cell Neoplasm. Nat. Commun. 2019, 10, 1653. [Google Scholar] [CrossRef] [PubMed]
- Gröschel, S.; Sanders, M.A.; Hoogenboezem, R.; de Wit, E.; Bouwman, B.A.M.; Erpelinck, C.; van der Velden, V.H.J.; Havermans, M.; Avellino, R.; van Lom, K.; et al. A Single Oncogenic Enhancer Rearrangement Causes Concomitant EVI1 and GATA2 Deregulation in Leukemia. Cell 2014, 157, 369–381. [Google Scholar] [CrossRef]
- Affer, M.; Chesi, M.; Chen, W.D.; Keats, J.J.; Demchenko, Y.N.; Tamizhmani, K.; Garbitt, V.M.; Riggs, D.L.; Brents, L.A.; Roschke, A.V.; et al. Promiscuous MYC Locus Rearrangements Hijack Enhancers but Mostly Super-Enhancers to Dysregulate MYC Expression in Multiple Myeloma. Leukemia 2014, 28, 1725–1735. [Google Scholar] [CrossRef] [PubMed]
- Drier, Y.; Cotton, M.J.; Williamson, K.E.; Gillespie, S.M.; Ryan, R.J.H.; Kluk, M.J.; Carey, C.D.; Rodig, S.J.; Sholl, L.M.; Afrogheh, A.H.; et al. An Oncogenic MYB Feedback Loop Drives Alternate Cell Fates in Adenoid Cystic Carcinoma. Nat. Genet. 2016, 48, 265–272. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Jiang, Y.-Y.; Lin, D.-C. Super-Enhancer-Mediated Core Regulatory Circuitry in Human Cancer. Comput. Struct. Biotechnol. J. 2021, 19, 2790–2795. [Google Scholar] [CrossRef] [PubMed]
- The Cancer Genome Atlas Research Network Integrated Genomic Characterization of Oesophageal Carcinoma. Nature 2017, 541, 169–175. [CrossRef] [PubMed]
- Hao, J.-J.; Lin, D.-C.; Dinh, H.Q.; Mayakonda, A.; Jiang, Y.-Y.; Chang, C.; Jiang, Y.; Lu, C.-C.; Shi, Z.-Z.; Xu, X.; et al. Spatial Intratumoral Heterogeneity and Temporal Clonal Evolution in Esophageal Squamous Cell Carcinoma. Nat. Genet. 2016, 48, 1500–1507. [Google Scholar] [CrossRef]
- Lin, D.-C.; Wang, M.-R.; Koeffler, H.P. Genomic and Epigenomic Aberrations in Esophageal Squamous Cell Carcinoma and Implications for Patients. Gastroenterology 2018, 154, 374–389. [Google Scholar] [CrossRef]
- Cao, W.; Wu, W.; Yan, M.; Tian, F.; Ma, C.; Zhang, Q.; Li, X.; Han, P.; Liu, Z.; Gu, J.; et al. Multiple Region Whole-Exome Sequencing Reveals Dramatically Evolving Intratumor Genomic Heterogeneity in Esophageal Squamous Cell Carcinoma. Oncogenesis 2015, 4, e175. [Google Scholar] [CrossRef]
- Chapuy, B.; McKeown, M.R.; Lin, C.Y.; Monti, S.; Roemer, M.G.M.; Qi, J.; Rahl, P.B.; Sun, H.H.; Yeda, K.T.; Doench, J.G.; et al. Discovery and Characterization of Super-Enhancer-Associated Dependencies in Diffuse Large B Cell Lymphoma. Cancer Cell 2013, 24, 777–790. [Google Scholar] [CrossRef]
- Betancur, P.A.; Abraham, B.J.; Yiu, Y.Y.; Willingham, S.B.; Khameneh, F.; Zarnegar, M.; Kuo, A.H.; McKenna, K.; Kojima, Y.; Leeper, N.J.; et al. A CD47-Associated Super-Enhancer Links pro-Inflammatory Signalling to CD47 Upregulation in Breast Cancer. Nat. Commun. 2017, 8, 14802. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Liang, H. A High-Resolution Map of Human Enhancer RNA Loci Characterizes Super-Enhancer Activities in Cancer. Cancer Cell 2020, 38, 701–715.e5. [Google Scholar] [CrossRef] [PubMed]
- Kelly, M.R.; Wisniewska, K.; Regner, M.J.; Lewis, M.W.; Perreault, A.A.; Davis, E.S.; Phanstiel, D.H.; Parker, J.S.; Franco, H.L. A Multi-Omic Dissection of Super-Enhancer Driven Oncogenic Gene Expression Programs in Ovarian Cancer. Nat. Commun. 2022, 13, 4247. [Google Scholar] [CrossRef] [PubMed]
- Yokoyama, Y.; Zhu, H.; Lee, J.H.; Kossenkov, A.V.; Wu, S.Y.; Wickramasinghe, J.M.; Yin, X.; Palozola, K.C.; Gardini, A.; Showe, L.C.; et al. BET Inhibitors Suppress ALDH Activity by Targeting ALDH1A1 Super-Enhancer in Ovarian Cancer. Cancer Res. 2016, 76, 6320–6330. [Google Scholar] [CrossRef] [PubMed]
- Pfeffer, S.R.; Yang, C.H.; Pfeffer, L.M. The Role of MiR-21 in Cancer: The Role of MiR-21 In Cancer. Drug Dev. Res. 2015, 76, 270–277. [Google Scholar] [CrossRef] [PubMed]
- Kumarswamy, R.; Volkmann, I.; Thum, T. Regulation and Function of MiRNA-21 in Health and Disease. RNA Biol. 2011, 8, 706–713. [Google Scholar] [CrossRef]
- Fujita, S.; Ito, T.; Mizutani, T.; Minoguchi, S.; Yamamichi, N.; Sakurai, K.; Iba, H. MiR-21 Gene Expression Triggered by AP-1 Is Sustained through a Double-Negative Feedback Mechanism. J. Mol. Biol. 2008, 378, 492–504. [Google Scholar] [CrossRef]
- Ribas, J.; Lupold, S.E. The Transcriptional Regulation of MiR-21, Its Multiple Transcripts and Their Implication in Prostate Cancer. Cell Cycle 2010, 9, 923–929. [Google Scholar] [CrossRef]
- Wan, Y.; Hoyle, R.G.; Xie, N.; Wang, W.; Cai, H.; Zhang, M.; Ma, Z.; Xiong, G.; Xu, X.; Huang, Z.; et al. A Super-Enhancer Driven by FOSL1 Controls MiR-21-5p Expression in Head and Neck Squamous Cell Carcinoma. Front. Oncol. 2021, 11, 656628. [Google Scholar] [CrossRef]
- Jang, M.K.; Mochizuki, K.; Zhou, M.; Jeong, H.-S.; Brady, J.N.; Ozato, K. The Bromodomain Protein Brd4 Is a Positive Regulatory Component of P-TEFb and Stimulates RNA Polymerase II-Dependent Transcription. Mol. Cell 2005, 19, 523–534. [Google Scholar] [CrossRef]
- Fisher, R.P.; Morgan, D.O. A Novel Cyclin Associates with M015/CDK7 to Form the CDK-Activating Kinase. Cell 1994, 78, 713–724. [Google Scholar] [CrossRef] [PubMed]
- Dahmus, M.E. Reversible Phosphorylation of the C-Terminal Domain of RNA Polymerase II. J. Biol. Chem. 1996, 271, 19009–19012. [Google Scholar] [CrossRef] [PubMed]
- Filippakopoulos, P.; Picaud, S.; Mangos, M.; Keates, T.; Lambert, J.-P.; Barsyte-Lovejoy, D.; Felletar, I.; Volkmer, R.; Müller, S.; Pawson, T.; et al. Histone Recognition and Large-Scale Structural Analysis of the Human Bromodomain Family. Cell 2012, 149, 214–231. [Google Scholar] [CrossRef] [PubMed]
- Filippakopoulos, P.; Qi, J.; Picaud, S.; Shen, Y.; Smith, W.B.; Fedorov, O.; Morse, E.M.; Keates, T.; Hickman, T.T.; Felletar, I.; et al. Selective Inhibition of BET Bromodomains. Nature 2010, 468, 1067–1073. [Google Scholar] [CrossRef] [PubMed]
- Tolani, B.; Gopalakrishnan, R.; Punj, V.; Matta, H.; Chaudhary, P.M. Targeting Myc in KSHV-Associated Primary Effusion Lymphoma with BET Bromodomain Inhibitors. Oncogene 2014, 33, 2928–2937. [Google Scholar] [CrossRef]
- Sengupta, D.; Kannan, A.; Kern, M.; Moreno, M.A.; Vural, E.; Stack, B.; Suen, J.Y.; Tackett, A.J.; Gao, L. Disruption of BRD4 at H3K27Ac-Enriched Enhancer Region Correlates with Decreased c-Myc Expression in Merkel Cell Carcinoma. Epigenetics 2015, 10, 460–466. [Google Scholar] [CrossRef]
- Tögel, L.; Nightingale, R.; Chueh, A.C.; Jayachandran, A.; Tran, H.; Phesse, T.; Wu, R.; Sieber, O.M.; Arango, D.; Dhillon, A.S.; et al. Dual Targeting of Bromodomain and Extraterminal Domain Proteins, and WNT or MAPK Signaling, Inhibits c-MYC Expression and Proliferation of Colorectal Cancer Cells. Mol. Cancer Ther. 2016, 15, 1217–1226. [Google Scholar] [CrossRef]
- Chen, D.; Zhao, Z.; Huang, Z.; Chen, D.-C.; Zhu, X.-X.; Wang, Y.-Z.; Yan, Y.-W.; Tang, S.; Madhavan, S.; Ni, W.; et al. Super Enhancer Inhibitors Suppress MYC Driven Transcriptional Amplification and Tumor Progression in Osteosarcoma. Bone Res. 2018, 6, 11. [Google Scholar] [CrossRef]
- Murakami, S.; Li, R.; Nagari, A.; Chae, M.; Camacho, C.V.; Kraus, W.L. Distinct Roles for BET Family Members in Estrogen Receptor α Enhancer Function and Gene Regulation in Breast Cancer Cells. Mol. Cancer Res. 2019, 17, 2356–2368. [Google Scholar] [CrossRef]
- Sin-Chan, P.; Mumal, I.; Suwal, T.; Ho, B.; Fan, X.; Singh, I.; Du, Y.; Lu, M.; Patel, N.; Torchia, J.; et al. A C19MC-LIN28A-MYCN Oncogenic Circuit Driven by Hijacked Super-Enhancers Is a Distinct Therapeutic Vulnerability in ETMRs: A Lethal Brain Tumor. Cancer Cell 2019, 36, 51–67.e7. [Google Scholar] [CrossRef]
- Nagasawa, M.; Tomimatsu, K.; Terada, K.; Kondo, K.; Miyazaki, K.; Miyazaki, M.; Motooka, D.; Okuzaki, D.; Yoshida, T.; Kageyama, S.; et al. Long Non-Coding RNA MANCR Is a Target of BET Bromodomain Protein BRD4 and Plays a Critical Role in Cellular Migration and Invasion Abilities of Prostate Cancer. Biochem. Biophys. Res. Commun. 2020, 526, 128–134. [Google Scholar] [CrossRef] [PubMed]
- Nicodeme, E.; Jeffrey, K.L.; Schaefer, U.; Beinke, S.; Dewell, S.; Chung, C.; Chandwani, R.; Marazzi, I.; Wilson, P.; Coste, H.; et al. Suppression of Inflammation by a Synthetic Histone Mimic. Nature 2010, 468, 1119–1123. [Google Scholar] [CrossRef] [PubMed]
- Shu, S.; Lin, C.Y.; He, H.H.; Witwicki, R.M.; Tabassum, D.P.; Roberts, J.M.; Janiszewska, M.; Jin Huh, S.; Liang, Y.; Ryan, J.; et al. Response and Resistance to BET Bromodomain Inhibitors in Triple-Negative Breast Cancer. Nature 2016, 529, 413–417. [Google Scholar] [CrossRef] [PubMed]
- Gibbons, H.R.; Mi, D.J.; Farley, V.M.; Esmond, T.; Kaood, M.B.; Aune, T.M. Bromodomain Inhibitor JQ1 Reversibly Blocks IFN-γ Production. Sci. Rep. 2019, 9, 10280. [Google Scholar] [CrossRef] [PubMed]
- Kwiatkowski, N.; Zhang, T.; Rahl, P.B.; Abraham, B.J.; Reddy, J.; Ficarro, S.B.; Dastur, A.; Amzallag, A.; Ramaswamy, S.; Tesar, B.; et al. Targeting Transcription Regulation in Cancer with a Covalent CDK7 Inhibitor. Nature 2014, 511, 616–620. [Google Scholar] [CrossRef]
- Christensen, C.L.; Kwiatkowski, N.; Abraham, B.J.; Carretero, J.; Al-Shahrour, F.; Zhang, T.; Chipumuro, E.; Herter-Sprie, G.S.; Akbay, E.A.; Altabef, A.; et al. Targeting Transcriptional Addictions in Small Cell Lung Cancer with a Covalent CDK7 Inhibitor. Cancer Cell 2014, 26, 909–922. [Google Scholar] [CrossRef]
- Eliades, P.; Abraham, B.J.; Ji, Z.; Miller, D.M.; Christensen, C.L.; Kwiatkowski, N.; Kumar, R.; Njauw, C.N.; Taylor, M.; Miao, B.; et al. High MITF Expression Is Associated with Super-Enhancers and Suppressed by CDK7 Inhibition in Melanoma. J. Investig. Dermatol. 2018, 138, 1582–1590. [Google Scholar] [CrossRef]
- Chipumuro, E.; Marco, E.; Christensen, C.L.; Kwiatkowski, N.; Zhang, T.; Hatheway, C.M.; Abraham, B.J.; Sharma, B.; Yeung, C.; Altabef, A.; et al. CDK7 Inhibition Suppresses Super-Enhancer-Linked Oncogenic Transcription in MYCN-Driven Cancer. Cell 2014, 159, 1126–1139. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, T.; Kwiatkowski, N.; Abraham, B.J.; Lee, T.I.; Xie, S.; Yuzugullu, H.; Von, T.; Li, H.; Lin, Z.; et al. CDK7-Dependent Transcriptional Addiction in Triple-Negative Breast Cancer. Cell 2015, 163, 174–186. [Google Scholar] [CrossRef]
- Zhao, X.; Ren, Y.; Lawlor, M.; Shah, B.D.; Park, P.M.C.; Lwin, T.; Wang, X.; Liu, K.; Wang, M.; Gao, J.; et al. BCL2 Amplicon Loss and Transcriptional Remodeling Drives ABT-199 Resistance in B Cell Lymphoma Models. Cancer Cell 2019, 35, 752–766.e9. [Google Scholar] [CrossRef]
- Sanchez, G.J.; Richmond, P.A.; Bunker, E.N.; Karman, S.S.; Azofeifa, J.; Garnett, A.T.; Xu, Q.; Wheeler, G.E.; Toomey, C.M.; Zhang, Q.; et al. Genome-Wide Dose-Dependent Inhibition of Histone Deacetylases Studies Reveal Their Roles in Enhancer Remodeling and Suppression of Oncogenic Super-Enhancers. Nucleic Acids Res. 2018, 46, 1756–1776. [Google Scholar] [CrossRef] [PubMed]
- Gryder, B.E.; Pomella, S.; Sayers, C.; Wu, X.S.; Song, Y.; Chiarella, A.M.; Bagchi, S.; Chou, H.-C.; Sinniah, R.S.; Walton, A.; et al. Histone Hyperacetylation Disrupts Core Gene Regulatory Architecture in Rhabdomyosarcoma. Nat. Genet. 2019, 51, 1714–1722. [Google Scholar] [CrossRef] [PubMed]
- Caslini, C.; Hong, S.; Ban, Y.J.; Chen, X.S.; Ince, T.A. HDAC7 Regulates Histone 3 Lysine 27 Acetylation and Transcriptional Activity at Super-Enhancer-Associated Genes in Breast Cancer Stem Cells. Oncogene 2019, 38, 6599–6614. [Google Scholar] [CrossRef] [PubMed]
- Gryder, B.E.; Wu, L.; Woldemichael, G.M.; Pomella, S.; Quinn, T.R.; Park, P.M.C.; Cleveland, A.; Stanton, B.Z.; Song, Y.; Rota, R.; et al. Chemical Genomics Reveals Histone Deacetylases Are Required for Core Regulatory Transcription. Nat. Commun. 2019, 10, 3004. [Google Scholar] [CrossRef] [PubMed]
- Shi, K.; Yin, X.; Cai, M.-C.; Yan, Y.; Jia, C.; Ma, P.; Zhang, S.; Zhang, Z.; Gu, Z.; Zhang, M.; et al. PAX8 Regulon in Human Ovarian Cancer Links Lineage Dependency with Epigenetic Vulnerability to HDAC Inhibitors. eLife 2019, 8, e44306. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.T.T.; Zhang, Y.; Shang, E.; Shu, C.; Torrini, C.; Zhao, J.; Bianchetti, E.; Mela, A.; Humala, N.; Mahajan, A.; et al. HDAC Inhibitors Elicit Metabolic Reprogramming by Targeting Super-Enhancers in Glioblastoma Models. J. Clin. Investig. 2020, 130, 3699–3716. [Google Scholar] [CrossRef] [PubMed]
- Sugino, N.; Kawahara, M.; Tatsumi, G.; Kanai, A.; Matsui, H.; Yamamoto, R.; Nagai, Y.; Fujii, S.; Shimazu, Y.; Hishizawa, M.; et al. A Novel LSD1 Inhibitor NCD38 Ameliorates MDS-Related Leukemia with Complex Karyotype by Attenuating Leukemia Programs via Activating Super-Enhancers. Leukemia 2017, 31, 2303–2314. [Google Scholar] [CrossRef]
- Tatsumi, G.; Kawahara, M.; Yamamoto, R.; Hishizawa, M.; Kito, K.; Suzuki, T.; Takaori-Kondo, A.; Andoh, A. LSD1-Mediated Repression of GFI1 Super-Enhancer Plays an Essential Role in Erythroleukemia. Leukemia 2020, 34, 746–758. [Google Scholar] [CrossRef]
- Zhang, J.; Ying, Y.; Li, M.; Wang, M.; Huang, X.; Jia, M.; Zeng, J.; Ma, C.; Zhang, Y.; Li, C.; et al. Targeted Inhibition of KDM6 Histone Demethylases Eradicates Tumor-Initiating Cells via Enhancer Reprogramming in Colorectal Cancer. Theranostics 2020, 10, 10016–10030. [Google Scholar] [CrossRef]
- Meloche, S.; Pouysségur, J. The ERK1/2 Mitogen-Activated Protein Kinase Pathway as a Master Regulator of the G1- to S-Phase Transition. Oncogene 2007, 26, 3227–3239. [Google Scholar] [CrossRef]
- Hilger, R.A.; Scheulen, M.E.; Strumberg, D. The Ras-Raf-MEK-ERK Pathway in the Treatment of Cancer. Oncol. Res. Treat. 2002, 25, 511–518. [Google Scholar] [CrossRef] [PubMed]
- Yohe, M.E.; Gryder, B.E.; Shern, J.F.; Song, Y.K.; Chou, H.-C.; Sindiri, S.; Mendoza, A.; Patidar, R.; Zhang, X.; Guha, R.; et al. MEK Inhibition Induces MYOG and Remodels Super-Enhancers in RAS-Driven Rhabdomyosarcoma. Sci. Transl. Med. 2018, 10, eaan4470. [Google Scholar] [CrossRef] [PubMed]
- Pomella, S.; Sreenivas, P.; Gryder, B.E.; Wang, L.; Milewski, D.; Cassandri, M.; Baxi, K.; Hensch, N.R.; Carcarino, E.; Song, Y.; et al. Interaction between SNAI2 and MYOD Enhances Oncogenesis and Suppresses Differentiation in Fusion Negative Rhabdomyosarcoma. Nat. Commun. 2021, 12, 192. [Google Scholar] [CrossRef]
- Mack, S.C.; Singh, I.; Wang, X.; Hirsch, R.; Wu, Q.; Villagomez, R.; Bernatchez, J.A.; Zhu, Z.; Gimple, R.C.; Kim, L.J.Y.; et al. Chromatin Landscapes Reveal Developmentally Encoded Transcriptional States That Define Human Glioblastoma. J. Exp. Med. 2019, 216, 1071–1090. [Google Scholar] [CrossRef] [PubMed]
- Furqan, M.; Mukhi, N.; Lee, B.; Liu, D. Dysregulation of JAK-STAT Pathway in Hematological Malignancies and JAK Inhibitors for Clinical Application. Biomark. Res. 2013, 1, 5. [Google Scholar] [CrossRef] [PubMed]
- Pelish, H.E.; Liau, B.B.; Nitulescu, I.I.; Tangpeerachaikul, A.; Poss, Z.C.; Da Silva, D.H.; Caruso, B.T.; Arefolov, A.; Fadeyi, O.; Christie, A.L.; et al. Mediator Kinase Inhibition Further Activates Super-Enhancer-Associated Genes in AML. Nature 2015, 526, 273–276. [Google Scholar] [CrossRef]
- Call, S.G.; Duren, R.P.; Panigrahi, A.K.; Nguyen, L.; Freire, P.R.; Grimm, S.L.; Coarfa, C.; Conneely, O.M. Targeting Oncogenic Super Enhancers in MYC-Dependent AML Using a Small Molecule Activator of NR4A Nuclear Receptors. Sci. Rep. 2020, 10, 2851. [Google Scholar] [CrossRef]
- Bolin, S.; Borgenvik, A.; Persson, C.U.; Sundström, A.; Qi, J.; Bradner, J.E.; Weiss, W.A.; Cho, Y.-J.; Weishaupt, H.; Swartling, F.J. Combined BET Bromodomain and CDK2 Inhibition in MYC-Driven Medulloblastoma. Oncogene 2018, 37, 2850–2862. [Google Scholar] [CrossRef]
- Wiese, M.; Hamdan, F.H.; Kubiak, K.; Diederichs, C.; Gielen, G.H.; Nussbaumer, G.; Carcaboso, A.M.; Hulleman, E.; Johnsen, S.A.; Kramm, C.M. Combined Treatment with CBP and BET Inhibitors Reverses Inadvertent Activation of Detrimental Super Enhancer Programs in DIPG Cells. Cell Death Dis. 2020, 11, 673. [Google Scholar] [CrossRef]
- Chen, Z.; Cao, Z.; Wang, Y.; Zhang, X.; Xu, L.; Wang, Y.; Chen, Y.; Yang, C.; Ding, J.; Meng, L. Repressing MYC by Targeting BET Synergizes with Selective Inhibition of PI3Kα against B Cell Lymphoma. Cancer Lett. 2022, 524, 206–218. [Google Scholar] [CrossRef]
- Tee, A.E.; Ciampa, O.C.; Wong, M.; Fletcher, J.I.; Kamili, A.; Chen, J.; Ho, N.; Sun, Y.; Carter, D.R.; Cheung, B.B.; et al. Combination Therapy with the CDK7 Inhibitor and the Tyrosine Kinase Inhibitor Exerts Synergistic Anticancer Effects against MYCN -amplified Neuroblastoma. Int. J. Cancer 2020, 147, 1928–1938. [Google Scholar] [CrossRef] [PubMed]
- Shang, E.; Nguyen, T.T.T.; Shu, C.; Westhoff, M.-A.; Karpel-Massler, G.; Siegelin, M.D. Epigenetic Targeting of Mcl-1 Is Synthetically Lethal with Bcl-XL/Bcl-2 Inhibition in Model Systems of Glioblastoma. Cancers 2020, 12, 2137. [Google Scholar] [CrossRef] [PubMed]
- Nagaraja, S.; Vitanza, N.A.; Woo, P.J.; Taylor, K.R.; Liu, F.; Zhang, L.; Li, M.; Meng, W.; Ponnuswami, A.; Sun, W.; et al. Transcriptional Dependencies in Diffuse Intrinsic Pontine Glioma. Cancer Cell 2017, 31, 635–652.e6. [Google Scholar] [CrossRef]
- Wong, M.; Sun, Y.; Xi, Z.; Milazzo, G.; Poulos, R.C.; Bartenhagen, C.; Bell, J.L.; Mayoh, C.; Ho, N.; Tee, A.E.; et al. JMJD6 Is a Tumorigenic Factor and Therapeutic Target in Neuroblastoma. Nat. Commun. 2019, 10, 3319. [Google Scholar] [CrossRef] [PubMed]
- Shen, Z.; Bao, W.; Huang, D.-S. Recurrent Neural Network for Predicting Transcription Factor Binding Sites. Sci. Rep. 2018, 8, 15270. [Google Scholar] [CrossRef] [PubMed]
- Ji, Y.; Zhou, Z.; Liu, H.; Davuluri, R.V. DNABERT: Pre-Trained Bidirectional Encoder Representations from Transformers Model for DNA-Language in Genome. Bioinformatics 2021, 37, 2112–2120. [Google Scholar] [CrossRef]
- Spirin, P.V.; Lebedev, T.D.; Orlova, N.N.; Gornostaeva, A.S.; Prokofjeva, M.M.; Nikitenko, N.A.; Dmitriev, S.E.; Buzdin, A.A.; Borisov, N.M.; Aliper, A.M.; et al. Silencing AML1-ETO Gene Expression Leads to Simultaneous Activation of Both pro-Apoptotic and Proliferation Signaling. Leukemia 2014, 28, 2222–2228. [Google Scholar] [CrossRef]
- Lebedev, T.D.; Spirin, P.V.; Orlova, N.N.; Prokofjeva, M.M.; Prassolov, V.S. Comparative Analysis of Gene Expression: Targeted Antitumor Therapy in Neuroblastoma Cell Lines. Mol. Biol. 2015, 49, 939–942. [Google Scholar] [CrossRef]
- Mitkevich, V.A.; Orlova, N.N.; Petrushanko, I.Y.; Simonenko, O.V.; Spirin, P.V.; Prokof’eva, M.M.; Gornostaeva, A.S.; Stocking, C.; Makarov, A.A.; Prassolov, V.S. Expression of the FLT3-ITD Oncogene Sensitizes Murine Progenitor B-Cell Line BAF3 to Cytotoxic Action of Binase. Mol. Biol. 2013, 47, 248–251. [Google Scholar] [CrossRef]
- Orlova, N.N.; Lebedev, T.D.; Spirin, P.V.; Prassolov, V.S. Key Molecular Mechanisms Associated with Cell Malignant Transformation in Acute Myeloid Leukemia. Mol. Biol. 2016, 50, 344–352. [Google Scholar] [CrossRef]
- Ivanov, A.E.; Pushkarev, A.V.; Orlov, A.V.; Nikitin, M.P.; Nikitin, P.I. Interferometric Detection of Chloramphenicol via Its Immunochemical Recognition at Polymer-Coated Nano-Corrugated Surfaces. Sens. Actuators B Chem. 2019, 282, 984–991. [Google Scholar] [CrossRef]
- Novichikhin, D.O.; Orlov, A.V.; Antopolsky, M.L.; Znoyko, S.L.; Nikitin, P.I. Specific and Sensitive Determination of Folic Acid by Label-Free Chemosensors with Microscope Glass Slips as Single-Use Consumables. Chemosensors 2022, 11, 17. [Google Scholar] [CrossRef]
- Orlov, A.V.; Novichikhin, D.O.; Pushkarev, A.V.; Malkerov, Y.A.; Znoiko, S.L.; Guteneva, N.V.; Orlova, N.N.; Gorshkov, B.G.; Nikitin, P.I. Registering the Kinetics of Intermolecular Interactions by Low-Coherence Interferometry for the Development of Biomarker Immunoassays for Cardiovascular Diseases. Dokl. Phys. 2022, 67, 193–196. [Google Scholar] [CrossRef]
- Bragina, V.A.; Orlov, A.V.; Znoyko, S.L.; Pushkarev, A.V.; Novichikhin, D.O.; Guteneva, N.V.; Nikitin, M.P.; Gorshkov, B.G.; Nikitin, P.I. Nanobiosensing Based on Optically Selected Antibodies and Superparamagnetic Labels for Rapid and Highly Sensitive Quantification of Polyvalent Hepatitis B Surface Antigen. Anal. Methods 2021, 13, 2424–2433. [Google Scholar] [CrossRef]
- Pushkarev, A.V.; Orlov, A.V.; Znoyko, S.L.; Bragina, V.A.; Nikitin, P.I. Rapid and Easy-to-Use Method for Accurate Characterization of Target Binding and Kinetics of Magnetic Particle Bioconjugates for Biosensing. Sensors 2021, 21, 2802. [Google Scholar] [CrossRef]
- Orlov, A.V.; Malkerov, J.A.; Novichikhin, D.O.; Znoyko, S.L.; Nikitin, P.I. Express High-Sensitive Detection of Ochratoxin A in Food by a Lateral Flow Immunoassay Based on Magnetic Biolabels. Food Chem. 2022, 383, 132427. [Google Scholar] [CrossRef]
- Orlov, A.V.; Malkerov, J.A.; Novichikhin, D.O.; Znoyko, S.L.; Nikitin, P.I. Multiplex Label-Free Kinetic Characterization of Antibodies for Rapid Sensitive Cardiac Troponin I Detection Based on Functionalized Magnetic Nanotags. Int. J. Mol. Sci. 2022, 23, 4474. [Google Scholar] [CrossRef]
- Burenin, A.G.; Nikitin, M.P.; Orlov, A.V.; Ksenevich, T.I.; Nikitin, P.I. Detection of Pyrethroids by Spectral Correlation Interferometry. Appl. Biochem. Microbiol. 2013, 49, 306. [Google Scholar] [CrossRef]
- Nekrasov, N.; Kudriavtseva, A.; Orlov, A.V.; Gadjanski, I.; Nikitin, P.I.; Bobrinetskiy, I.; Knežević, N.Ž. One-Step Photochemical Immobilization of Aptamer on Graphene for Label-Free Detection of NT-ProBNP. Biosensors 2022, 12, 1071. [Google Scholar] [CrossRef]
- Nekrasov, N.; Yakunina, N.; Pushkarev, A.V.; Orlov, A.V.; Gadjanski, I.; Pesquera, A.; Centeno, A.; Zurutuza, A.; Nikitin, P.I.; Bobrinetskiy, I. Spectral-Phase Interferometry Detection of Ochratoxin a via Aptamer-Functionalized Graphene Coated Glass. Nanomaterials 2021, 11, 226. [Google Scholar] [CrossRef]
- Orlova, N.N.; Bogatova, O.V.; Orlov, A.V. High-Performance Method for Identification of Super Enhancers from ChIP-Seq Data with Configurable Cloud Virtual Machines. MethodsX 2020, 7, 101165. [Google Scholar] [CrossRef] [PubMed]
- Orlova, N.N.; Nikitina, A.S.; Orlov, A.V. Identification and Analysis of Super-Enhancers as Novel Biomarkers and Potential Therapeutic Targets for Age-Associated Diseases. FEBS Open Bio. 2019, 9, 394–395. [Google Scholar] [CrossRef]
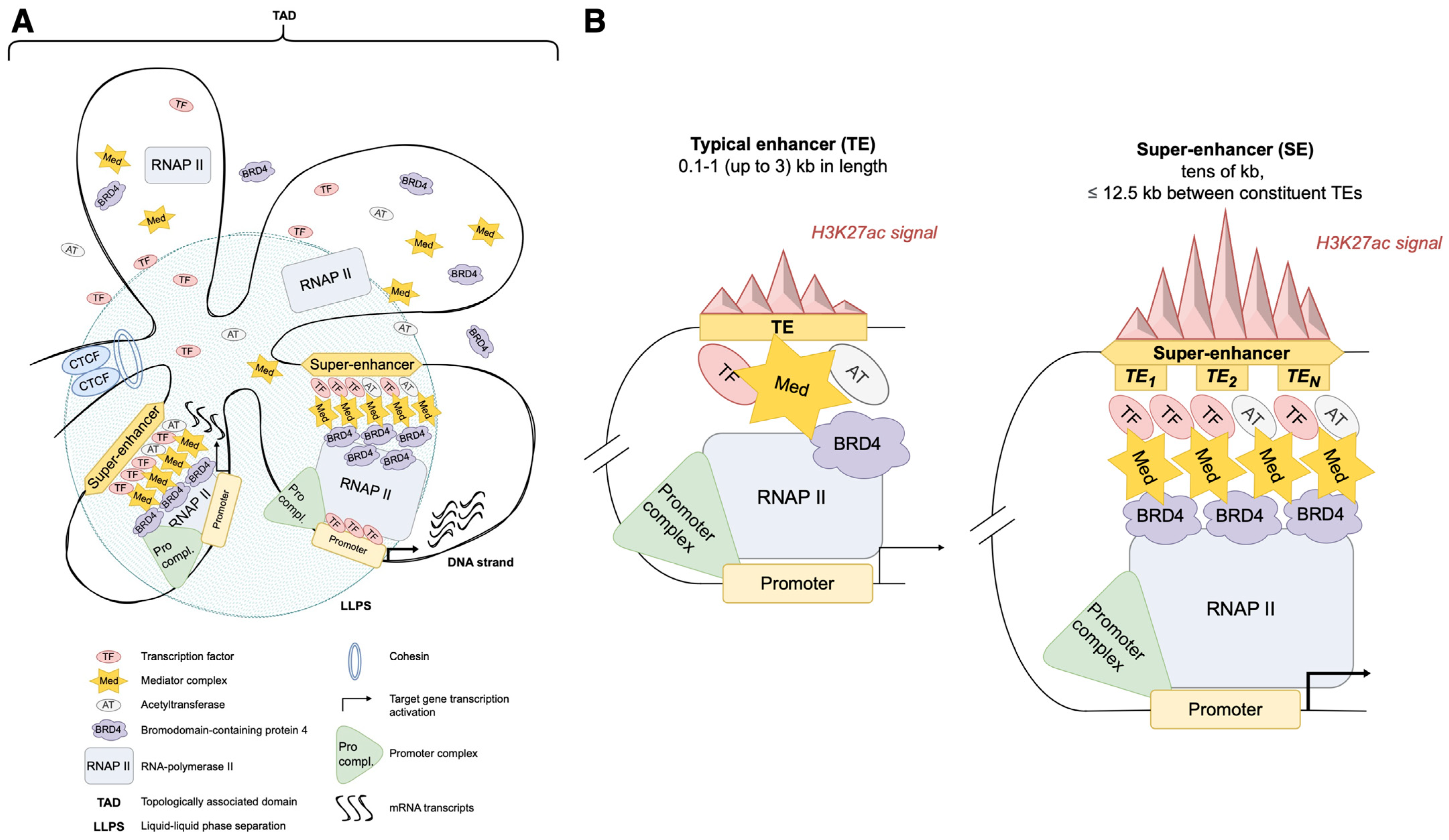
| Parameter | Typical Enhancer (TE) | Super-Enhancer (SE) |
|---|---|---|
| Length (Size) | ~1500 bp on average | ≤12.5 kb between constituent TEs, ten of kilobases long |
| Constitution | Detached region | Multiple enhancer regions—constituent TEs |
| Histone marks | H3K4me1—● * H3K4me3—∅ * H3K27ac—● * | H3K4me1—● * H3K4me3—? * H3K27ac—●●● * |
| Transcription complexes | Mostly bound by small amounts (usually—one) of TFs and Mediator | Reacher in TFs (BRD4, CDK7, p300, CBP) and Mediators, also bound by numerous co-regulators, might involve several RNAP II complexes |
| Relative transcriptional output | Mostly acting in cis Moderate transcription levels—●/●● * | Might be acting in trans as well as in cis Higher transcription levels—●●●● * |
| Bioinformatical identification approaches | ChIP-seq data analysis Gene expression annotation—GREAT, ChromHMM | ChIP-seq data analysis Gene expression annotation—GREAT, ChromHMM Identification by stitching algorithms—ROSE or its analogues, including DNABERT and other ML-alternatives (CNN, RNN) |
| SEs | Tissue/Cell Type | Key TFs | Target Genes | Refs. |
|---|---|---|---|---|
| 340 SEs | Adipocytes in early adipogenesis | C/EBPβ, KLF4, KLF5, TIF-1β, GR, C/EBPδ, STAT1, STAT5A, JunB, FOSL2, ATF7, PBX1 | Genes related with early adipogenesis (proliferation, extracellular matrix–receptor interactions, and cell growth) | [124] |
| BA SEs | Brown adipocytes | PPARγ, RXR, C/EBP, SIX1 (late-stage) | Cebpa, Fabp4, Scd, late stages: Ucp1, Cidea, Fgf21, Ppara, etc. | [125] |
| SLC25A37 SE | Erythroid cells | GATA1, TAL1 | SLC25A37 (encodes Mitoferrin 1) | [126] |
| 543 SEs | Hematopoietic stem/progenitor cells | RUNX1, GATA2, FLI1 | ETV6, ERG, KIT, LMO2, MEIS1, etc. | [127] |
| 724 SEs | Chondrocytes | Sox6, Sox9 | Chst11, Col9a1, Wwp2, Acan, Sox9, Sox6, Runx2, and Fgfr3 | [128] |
| Cardiac SEs | Cardiac tissues | GATA4 and TBX5 | Genes involved in cardiac development | [129] |
| 2 SEs producing seRNA-1 and seRNA-2 | Myocytes | MyoD, MyoG, TCF12, TCF3, MEF2D, PBX1, FoxO3 | seRNA-1: myoglobin (Mb) and apolipoprotein L6 (Apol6); seRNA-2: Atp1a1 and Igsf3 | [130] |
| 728 SEs | Osteoclasts | Irf8, Fli1, Mafb, Fosl1/2, Junb, Myc, Nfkb2, Spi1, Relb | TFs such as Irf8 and Fos | [131] |
| 784 SEs | Lung fibroblasts | TBX2, TBX4, TBX5, HOXA5, FOXL1, FOXP1, MEIS1, TGIF1 | Genes associated with response to hypoxia and extracellular matrix organization | [132] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kravchuk, E.V.; Ashniev, G.A.; Gladkova, M.G.; Orlov, A.V.; Vasileva, A.V.; Boldyreva, A.V.; Burenin, A.G.; Skirda, A.M.; Nikitin, P.I.; Orlova, N.N. Experimental Validation and Prediction of Super-Enhancers: Advances and Challenges. Cells 2023, 12, 1191. https://doi.org/10.3390/cells12081191
Kravchuk EV, Ashniev GA, Gladkova MG, Orlov AV, Vasileva AV, Boldyreva AV, Burenin AG, Skirda AM, Nikitin PI, Orlova NN. Experimental Validation and Prediction of Super-Enhancers: Advances and Challenges. Cells. 2023; 12(8):1191. https://doi.org/10.3390/cells12081191
Chicago/Turabian StyleKravchuk, Ekaterina V., German A. Ashniev, Marina G. Gladkova, Alexey V. Orlov, Anastasiia V. Vasileva, Anna V. Boldyreva, Alexandr G. Burenin, Artemiy M. Skirda, Petr I. Nikitin, and Natalia N. Orlova. 2023. "Experimental Validation and Prediction of Super-Enhancers: Advances and Challenges" Cells 12, no. 8: 1191. https://doi.org/10.3390/cells12081191
APA StyleKravchuk, E. V., Ashniev, G. A., Gladkova, M. G., Orlov, A. V., Vasileva, A. V., Boldyreva, A. V., Burenin, A. G., Skirda, A. M., Nikitin, P. I., & Orlova, N. N. (2023). Experimental Validation and Prediction of Super-Enhancers: Advances and Challenges. Cells, 12(8), 1191. https://doi.org/10.3390/cells12081191





