Hedgehog Signaling in Cortical Development
Abstract
:1. Introduction
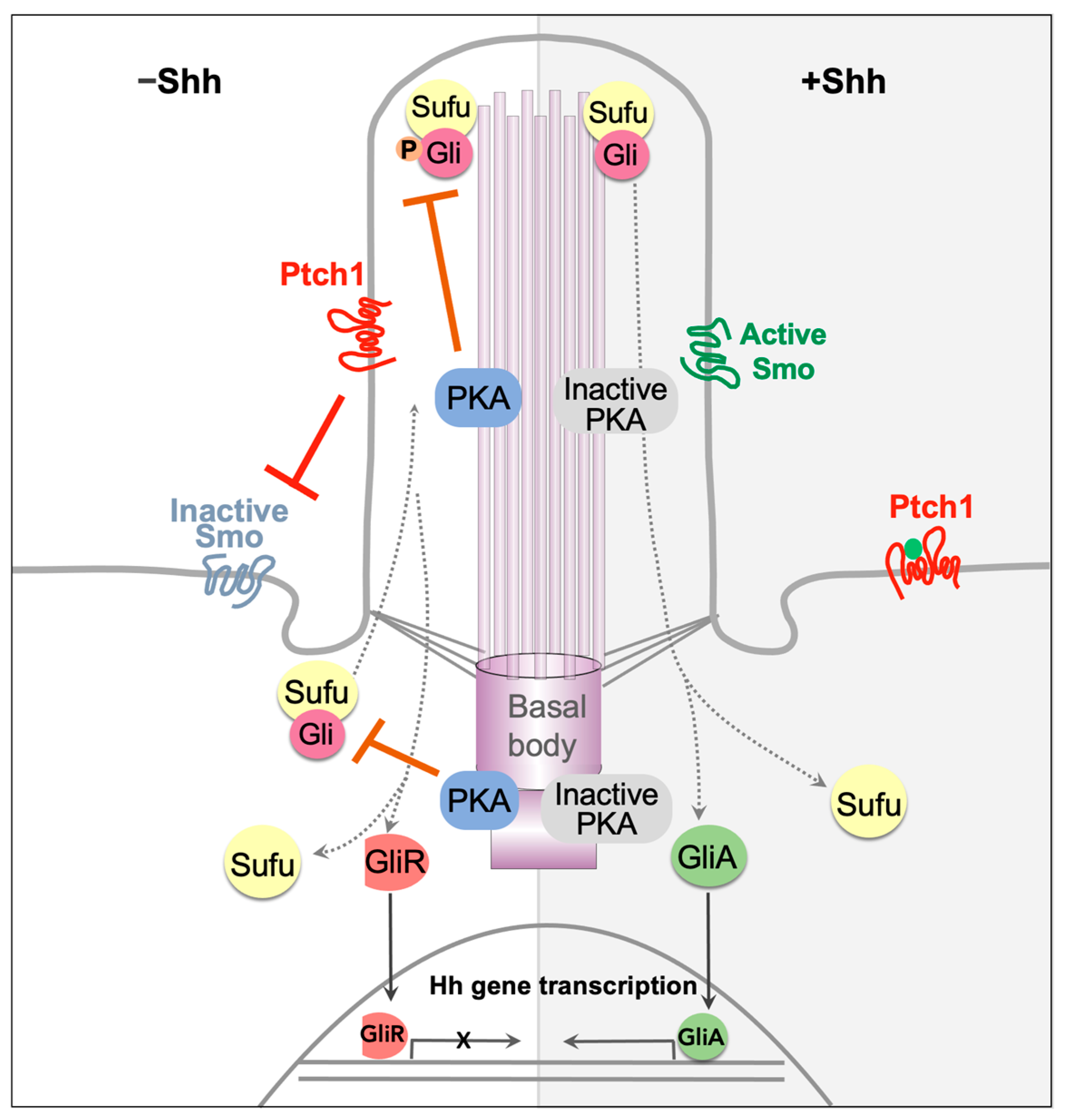
1.1. Transduction of Hh Signaling
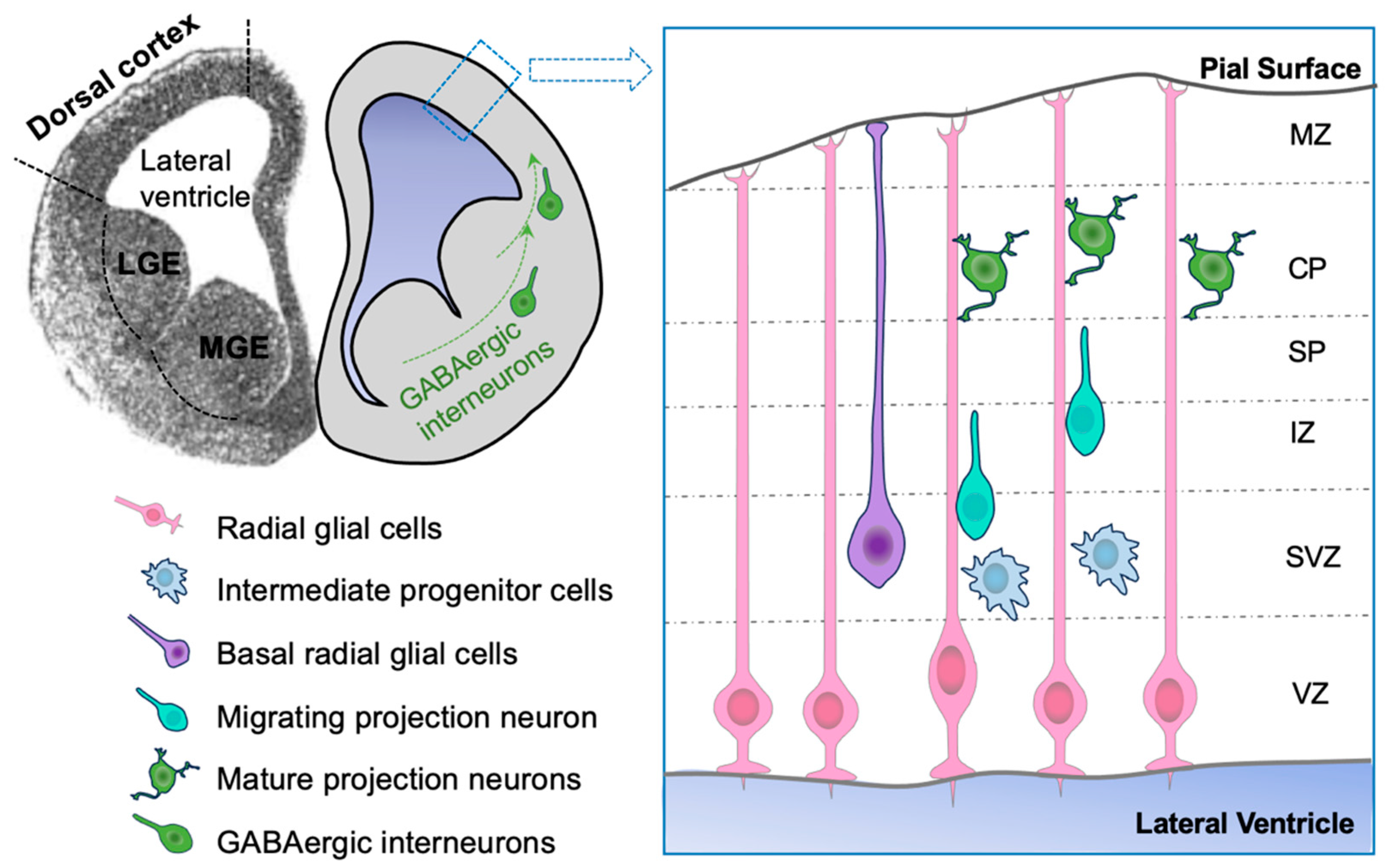
1.2. A Brief Overview of Corticogenesis
2. The Hypothesis of Expanded Hh Signaling in Gyrification of the Brain
3. Roles of Hedgehog Signaling during Cortical Development
3.1. Roles of Shh, the Ligand of Hh Pathway, in Cortical Development
3.2. The Receptor Ptch1 and Its Coreceptors in Cortical Development
3.3. The GPCR-like Signaling Transducer, Smoothened, in Cortical Development
3.4. The Negative Hh Regulator, Sufu, in Cortical Development
3.5. The Transcription Factor, Gli, in Cortical Development
3.6. Other Hh Signaling Regulators in Cortical Development
| Mutation | Onset Stage of Knockout | Hh Signaling | Phenotype | Lethality Stage | Primary Publication |
|---|---|---|---|---|---|
| Shh−/− | Global | Down | Cyclopia, probiscus-like facial features, abnormal organ formation, reduced body size | Perinatal | [51] |
| Shhlox/lox;NestinCre | E12.5 | Down | Decrease in MGE size, defective early oligodendrogenesis, fewer proliferating neural stem cells (NSCs) in postnatal SVZ and hippocampus | N/A | [72] |
| Shhlox/lox;Emx1Cre | E10.5 | Down | Smaller dorsal telencephalon, abnormal neuron position and NSC characteristics | N/A | [53] |
| Ptch−/− | Global | Up | Open and overgrown neural tube | E9-10.5 | [61] |
| Ptch+/− | Global | Up | Partially open neural tube, hindbrain defects, overgrown cerebellum, larger body size | N/A | [61] |
| Ptch1lox/lox;NestinCre | E12.5 | Up | Surface of neocortex is folded with varying thickness, distorted brain structures in neocortex, MGE, hippocampus, medial cortex | E15.5 | [62] |
| Smo−/− | Global | Down | Heart defect, cyclopia, loss of Left/Right symmetry | E9.5 | [70] |
| Smolox/lox;FoxG1Cre | E9 | Down | Dorsalization of neural tube, loss of interneurons and oligodendrocytes | Perinatal | [71] |
| Smolox/lox;Emx1Cre | E10.5 | Down | Smaller telencephalon, defects in neuronal migration, significant loss of oligodendrocytes | N/A | [53] |
| Smolox/lox;GFAPCre | E13.5 | Down | Small brain, fewer INPs, fewer bRGCs | N/A | [44] |
| SmoM2 (drive by GFAPCre) | E13.5 | Up | Folding of the cingulate cortex, higher number of bRGs and INPs | N/A | [44] |
| Sufu−/− | Global | Up | Open ventralized neural tube | E9.5 | [73] |
| Sufu−/+ | Global | Up | Normal growth, fertile, develop Gorlin-like features | N/A | [73] |
| Sufulox/lox;GFAPCre | E13.5 | Up | No obvious phenotype, survive into adulthood, expanded VZ and SVZ | N/A | [75] |
| Sufulox/lox;Emx1Cre | E10.5 | Up | Large cortical surface, no olfactory bulb, expanded lateral ventricles, thinner cortical layer | Death before weaning | [75] |
| Gli1−/− | Global | N/A | Normal, viable | N/A | [83] |
| Gli2−/− | Global | Down | Small lungs and fused lobes, no notochord regression, loss of pituitary, craniofacial defects | Perinatal | [83] |
| Gli2P1−4 (S/A mutation at four PKA sites) | Knock-in | Up | Exencephaly, partially open neural tube, extra anterior digit, enlarged facio-cranial features | Between E14.5 to birth | [5] |
| Xt (Spontaneous loss-of-function mutation in Gli3) | Global | Up | Dorsalized neural tube, reduced cortical size, absent hippocampus and choroid plexus | N/A | [88] |
| Gli3lox/lox;NestinCre | E12.5 | Up | Loss of upper layer projection neurons (PNs), defects in cortical neuron specification and positioning | N/A | [92] |
| Shh−/−;Gli3−/− | Global knockout | N/A | 91% exencephalic, relatively normal telencephalon, missing dorsal midline structure, normal pan-ventral genes such as Dlx2 Gsh2, Nkx2.1 | N/A | [52] |
| Shh−/−;Gli3+/− | Global | Down | Partial rescue of Shh-null phenotype: two discernable eyes, reduced probiscus, partial dorsal–ventral patterning rescue | N/A | [52] |
| Gli1−/−;Gli3+/− | Global | Up | Viable, polydactylyl | N/A | [83] |
| Gli1−/−;Gli2−/− | Global | Down | Decreased viability by E18.5, loss of pituitary tissue, lung lobes defects | Perinatal | [83] |
| Gli1−/−;Gli2+/− | Global | Down | Some loss of ventral spinal cord, small lungs | Perinatal | [83] |
4. Summary and Future Perspective
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Stone, D.M.; Hynes, M.; Armanini, M.; Swanson, T.A.; Gu, Q.; Johnson, R.L.; Scott, M.P.; Pennica, D.; Goddard, A.; Phillips, H.; et al. The tumour-suppressor gene patched encodes a candidate receptor for Sonic hedgehog. Nature 1996, 384, 129–134. [Google Scholar] [CrossRef] [PubMed]
- Rohatgi, R.; Milenkovic, L.; Scott, M.P. Patched1 regulates hedgehog signaling at the primary cilium. Science 2007, 317, 372–376. [Google Scholar] [CrossRef] [PubMed]
- Humke, E.W.; Dorn, K.V.; Milenkovic, L.; Scott, M.P.; Rohatgi, R. The output of Hedgehog signaling is controlled by the dynamic association between Suppressor of Fused and the Gli proteins. Genes Dev. 2010, 24, 670–682. [Google Scholar] [CrossRef] [PubMed]
- Tuson, M.; He, M.; Anderson, K.V. Protein kinase A acts at the basal body of the primary cilium to prevent Gli2 activation and ventralization of the mouse neural tube. Development 2011, 138, 4921–4930. [Google Scholar] [CrossRef]
- Pan, Y.; Wang, C.; Wang, B. Phosphorylation of Gli2 by protein kinase A is required for Gli2 processing and degradation and the Sonic Hedgehog-regulated mouse development. Dev. Biol. 2009, 326, 177–189. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.H.; Wilson, C.W.; Li, Y.J.; Law, K.K.; Lu, C.S.; Gacayan, R.; Zhang, X.; Hui, C.C.; Chuang, P.T. Cilium-independent regulation of Gli protein function by Sufu in Hedgehog signaling is evolutionarily conserved. Genes Dev. 2009, 23, 1910–1928. [Google Scholar] [CrossRef]
- Happ, J.T.; Arveseth, C.D.; Bruystens, J.; Bertinetti, D.; Nelson, I.B.; Olivieri, C.; Zhang, J.; Hedeen, D.S.; Zhu, J.F.; Capener, J.L.; et al. A PKA inhibitor motif within SMOOTHENED controls Hedgehog signal transduction. Nat. Struct. Mol. Biol. 2022, 29, 990–999. [Google Scholar] [CrossRef]
- Tukachinsky, H.; Lopez, L.V.; Salic, A. A mechanism for vertebrate Hedgehog signaling: Recruitment to cilia and dissociation of SuFu-Gli protein complexes. J. Cell Biol. 2010, 191, 415–428. [Google Scholar] [CrossRef]
- Kong, J.H.; Siebold, C.; Rohatgi, R. Biochemical mechanisms of vertebrate hedgehog signaling. Development 2019, 146, dev166892. [Google Scholar] [CrossRef]
- Briscoe, J.; Thérond, P.P. The mechanisms of Hedgehog signalling and its roles in development and disease. Nat. Rev. Mol. Cell Biol. 2013, 14, 416–429. [Google Scholar] [CrossRef]
- Bangs, F.; Anderson, K.V. Primary cilia and Mammalian Hedgehog signaling. Cold Spring Harb. Perspect. Biol. 2017, 9, a028175. [Google Scholar] [CrossRef]
- Copp, A.J.; Greene, N.D.E.; Murdoch, J.N. The genetic basis of mammalian neurulation. Nat. Rev. Genet. 2003, 4, 784–793. [Google Scholar] [CrossRef]
- Aaku-Saraste, E.; Hellwig, A.; Huttner, W.B. Loss of Occludin and Functional Tight Junctions, but Not ZO-1, during Neural Tube Closure—Remodeling of the Neuroepithelium Prior to Neurogenesis. Dev. Biol. 1996, 180, 664–679. [Google Scholar] [CrossRef]
- Hatakeyama, J.; Bessho, Y.; Katoh, K.; Ookawara, S.; Fujioka, M.; Guillemot, F.; Kageyama, R. Hes genes regulate size, shape and histogenesis of the nervous system by control of the timing of neural stem cell differentiation. Development 2004, 131, 5539–5550. [Google Scholar] [CrossRef]
- Campbell, K.; Götz, M. Radial glia: Multi-purpose cells for vertebrate brain development. Trends Neurosci. 2002, 25, 235–238. [Google Scholar] [CrossRef]
- Kriegstein, A.; Alvarez-Buylla, A. The Glial Nature of Embryonic and Adult Neural Stem Cells. Annu. Rev. Neurosci. 2009, 32, 149–184. [Google Scholar] [CrossRef]
- Kriegstein, A.R.; Götz, M. Radial glia diversity: A matter of cell fate. Glia 2003, 43, 37–43. [Google Scholar] [CrossRef]
- Paridaen, J.T.; Huttner, W.B. Neurogenesis during development of the vertebrate central nervous system. EMBO Rep. 2014, 15, 351–364. [Google Scholar] [CrossRef]
- Götz, M.; Huttner, W.B. The cell biology of neurogenesis. Nat. Rev. Mol. Cell Biol. 2005, 6, 777–788. [Google Scholar] [CrossRef]
- Noctor, S.C.; Martinez-Cerdeño, V.; Ivic, L.; Kriegstein, A.R. Cortical neurons arise in symmetric and asymmetric division zones and migrate through specific phases. Nat. Neurosci. 2004, 7, 136–144. [Google Scholar] [CrossRef]
- Rakic, P. A small step for the cell, a giant leap for mankind: A hypothesis of neocortical expansion during evolution. Trends Neurosci. 1995, 18, 383–388. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Cerdeno, V.; Noctor, S.C.; Kriegstein, A.R. The role of intermediate progenitor cells in the evolutionary expansion of the cerebral cortex. Cereb. Cortex 2006, 16 (Suppl. S1), i152–i161. [Google Scholar] [CrossRef] [PubMed]
- Farkas, L.M.; Huttner, W.B. The cell biology of neural stem and progenitor cells and its significance for their proliferation versus differentiation during mammalian brain development. Curr. Opin. Cell Biol. 2008, 20, 707–715. [Google Scholar] [CrossRef] [PubMed]
- Greig, L.C.; Woodworth, M.B.; Galazo, M.J.; Padmanabhan, H.; Macklis, J.D. Molecular logic of neocortical projection neuron specification, development and diversity. Nat. Rev. Neurosci. 2013, 14, 755–769. [Google Scholar] [CrossRef] [PubMed]
- Gleeson, J.G.; Walsh, C.A. Neuronal migration disorders: From genetic diseases to developmental mechanisms. Trends Neurosci. 2000, 23, 352–359. [Google Scholar] [CrossRef]
- Olson, E.C.; Walsh, C.A. Smooth, rough and upside-down neocortical development. Curr. Opin. Genet. Dev. 2002, 12, 320–327. [Google Scholar] [CrossRef]
- Lewitus, E.; Kelava, I.; Huttner, W.B. Conical expansion of the outer subventricular zone and the role of neocortical folding in evolution and development. Front. Hum. Neurosci. 2013, 7, 424. [Google Scholar] [CrossRef]
- Hansen, D.V.; Lui, J.H.; Parker, P.R.L.; Kriegstein, A.R. Neurogenic radial glia in the outer subventricular zone of human neocortex. Nature 2010, 464, 554–561. [Google Scholar] [CrossRef]
- Borrell, V. How Cells Fold the Cerebral Cortex. J. Neurosci. 2018, 38, 776–783. [Google Scholar] [CrossRef]
- Wang, X.; Tsai, J.W.; Lamonica, B.; Kriegstein, A.R. A new subtype of progenitor cell in the mouse embryonic neocortex. Nat. Neurosci. 2011, 14, 555–562. [Google Scholar] [CrossRef]
- Lui, J.H.; Hansen, D.V.; Kriegstein, A.R. Development and Evolution of the Human Neocortex. Cell 2011, 146, 18–36. [Google Scholar] [CrossRef] [PubMed]
- Kriegstein, A.; Noctor, S.; Martínez-Cerdeño, V. Patterns of neural stem and progenitor cell division may underlie evolutionary cortical expansion. Nat. Rev. Neurosci. 2006, 7, 883–890. [Google Scholar] [CrossRef] [PubMed]
- Florio, M.; Borrell, V.; Huttner, W.B. Human-specific genomic signatures of neocortical expansion. Curr. Opin. Neurobiol. 2017, 42, 33–44. [Google Scholar] [CrossRef] [PubMed]
- Taverna, E.; Götz, M.; Huttner, W.B. The Cell Biology of Neurogenesis: Toward an Understanding of the Development and Evolution of the Neocortex. Annu. Rev. Cell Dev. Biol. 2014, 30, 465–502. [Google Scholar] [CrossRef] [PubMed]
- Wechsler-Reya, R.J.; Scott, M.P. Control of Neuronal Precursor Proliferation in the Cerebellum by Sonic Hedgehog. Neuron 1999, 22, 103–114. [Google Scholar] [CrossRef] [PubMed]
- Wallace, V.A. Purkinje-cell-derived Sonic hedgehog regulates granule neuron precursor cell proliferation in the developing mouse cerebellum. Curr. Biol. 1999, 9, 445–448. [Google Scholar] [CrossRef]
- Dahmane, N.; Altaba, A.R.I. Sonic hedgehog regulates the growth and patterning of the cerebellum. Development 1999, 126, 3089–3100. [Google Scholar] [CrossRef]
- Charron, F.; Stein, E.; Jeong, J.; McMahon, A.P.; Tessier-Lavigne, M. The Morphogen Sonic Hedgehog Is an Axonal Chemoattractant that Collaborates with Netrin-1 in Midline Axon Guidance. Cell 2003, 113, 11–23. [Google Scholar] [CrossRef]
- Okada, A.; Charron, F.; Morin, S.; Shin, D.S.; Wong, K.; Fabre, P.J.; Tessier-Lavigne, M.; McConnell, S.K. Boc is a receptor for sonic hedgehog in the guidance of commissural axons. Nature 2006, 444, 369–373. [Google Scholar] [CrossRef]
- Yam, P.T.; Charron, F. Signaling mechanisms of non-conventional axon guidance cues: The Shh, BMP and Wnt morphogens. Curr. Opin. Neurobiol. 2013, 23, 965–973. [Google Scholar] [CrossRef]
- Zilles, K.; Palomero-Gallagher, N.; Amunts, K. Development of cortical folding during evolution and ontogeny. Trends Neurosci. 2013, 36, 275–284. [Google Scholar] [CrossRef] [PubMed]
- Molnár, Z.; Clowry, G. Cerebral cortical development in rodents and primates. In Progress in Brain Research; Elsevier B.V.: Amsterdam, The Netherlands, 2012; pp. 45–70. [Google Scholar]
- Zecevic, N.; Chen, Y.; Filipovic, R. Contributions of cortical subventricular zone to the development of the human cerebral cortex. J. Comp. Neurol. 2005, 491, 109–122. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Hou, S.; Han, Y.G. Hedgehog signaling promotes basal progenitor expansion and the growth and folding of the neocortex. Nat. Neurosci. 2016, 19, 888–896. [Google Scholar] [CrossRef] [PubMed]
- Ohtsuka, T.; Kageyama, R. Dual activation of Shh and Notch signaling induces dramatic enlargement of neocortical surface area. Neurosci. Res. 2022, 176, 18–30. [Google Scholar] [CrossRef] [PubMed]
- Hou, S.; Ho, W.-L.; Wang, L.; Kuo, B.; Park, J.Y.; Han, Y.-G. Biphasic Roles of Hedgehog Signaling in the Production and Self-Renewal of Outer Radial Glia in the Ferret Cerebral Cortex. Cereb. Cortex 2021, 31, 4730–4741. [Google Scholar] [PubMed]
- Matsumoto, N.; Tanaka, S.; Horiike, T.; Shinmyo, Y.; Kawasaki, H. A discrete subtype of neural progenitor crucial for cortical folding in the gyrencephalic mammalian brain. eLife 2020, 9, e54873. [Google Scholar] [CrossRef] [PubMed]
- Heussler, H.S.; Suri, M.; Young, I.D.; Muenke, M. Extreme variability of expression of a Sonic Hedgehog mutation: Attention difficulties and holoprosencephaly. Arch. Dis. Child. 2002, 86, 293–296. [Google Scholar] [CrossRef]
- Roessler, E.; Belloni, E.; Gaudenz, K.; Jay, P.; Berta, P.; Scherer, S.W.; Tsui, L.C.; Muenke, M. Mutations in the human Sonic Hedgehog gene cause holoprosencephaly. Nat. Genet. 1996, 14, 357–360. [Google Scholar] [CrossRef]
- Odent, S.; Atti-Bitach, T.; Blayau, M.; Mathieu, M.; Aug, J.; Delezo de, A.L.; Gall, J.Y.; Le Marec, B.; Munnich, A.; David, V.; et al. Expression of the Sonic hedgehog (SHH) Gene during Early Human Development and Phenotypic Expression of New Mutations Causing Holoprosencephaly. Hum. Mol. Genet. 1999, 8, 1683–1689. [Google Scholar] [CrossRef]
- Chiang, C.; Litingtung, Y.; Lee, E.; Young, K.E.; Corden, J.L.; Westphal, H.; Beachy, P.A. Cyclopia and defective axial patterning in mice lacking Sonic hedgehog gene function. Nature 1996, 383, 407–413. [Google Scholar] [CrossRef]
- Rallu, M.; Machold, R.; Gaiano, N.; Corbin, J.G.; McMahon, A.P.; Fishell, G. Dorsoventral patterning is established in the telencephalon of mutants lacking both Gli3 and Hedgehog signaling. Development 2002, 129, 4963–4974. [Google Scholar] [CrossRef] [PubMed]
- Komada, M.; Saitsu, H.; Kinboshi, M.; Miura, T.; Shiota, K.; Ishibashi, M. Hedgehog signaling is involved in development of the neocortex. Development 2008, 135, 2717–2727. [Google Scholar] [CrossRef] [PubMed]
- Dahmane, N.; Sánchez, P.; Gitton, Y.; Palma, V.; Sun, T.; Beyna, M.; Weiner, H.; Ruiz i Altaba, A. The Sonic Hedgehog-Gli pathway regulates dorsal brain growth and tumorigenesis. Development 2001, 128, 5201–5212. [Google Scholar] [CrossRef] [PubMed]
- Izumi, K.; Hahn, A.; Christ, L.; Curtis, C.; Neilson, D.E. Familial 9q22.3 microduplication spanning PTCH1 causes short stature syndrome with mild intellectual disability and dysmorphic features. Am. J. Med. Genet. A 2011, 155, 1384–1389. [Google Scholar] [CrossRef] [PubMed]
- Derwińska, K.; Smyk, M.; Cooper, M.L.; Bader, P.; Cheung, S.W.; Stankiewicz, P. PTCH1 duplication in a family with microcephaly and mild developmental delay. Eur. J. Hum. Genet. 2009, 17, 267–271. [Google Scholar] [CrossRef] [PubMed]
- Ming, J.E.; Kaupas, M.E.; Roessler, E.; Brunner, H.G.; Golabi, M.; Tekin, M.; Stratton, R.F.; Sujansky, E.; Bale, S.J.; Muenke, M. Mutations in PATCHED-1, the receptor for SONIC HEDGEHOG, are associated with holoprosencephaly. Hum. Genet. 2002, 110, 297–301. [Google Scholar] [CrossRef]
- Motoyama, J.; Takabatake, T.; Takeshima, K.; Hui, C. Ptch2, a second mouse Patched gene is co-expressed with Sonic hedgehog. Nat. Genet. 1998, 18, 104–106. [Google Scholar] [CrossRef]
- Lee, Y.; Miller, H.L.; Russell, H.R.; Boyd, K.; Curran, T.; McKinnon, P.J. Patched2 modulates tumorigenesis in Patched1 heterozygous mice. Cancer Res. 2006, 66, 6964–6971. [Google Scholar] [CrossRef]
- Goodrich, L.V.; Jung, D.; Higgins, K.M.; Scott, M.P. Overexpression of ptc1 Inhibits Induction of Shh Target Genes and Prevents Normal Patterning in the Neural Tube. Dev. Biol. 1999, 211, 323–334. [Google Scholar] [CrossRef]
- Goodrich, L.V.; Milenković, L.; Higgins, K.M.; Scott, M.P. Altered neural cell fates and medulloblastoma in mouse patched mutants. Science 1997, 277, 1109–1113. [Google Scholar] [CrossRef]
- Dave, R.K.; Ellis, T.; Toumpas, M.C.; Robson, J.P.; Julian, E.; Adolphe, C.; Bartlett, P.F.; Cooper, H.M.; Reynolds, B.A.; Wainwright, B.J. Sonic Hedgehog and Notch Signaling Can Cooperate to Regulate Neurogenic Divisions of Neocortical Progenitors. PLoS ONE 2011, 6, e14680. [Google Scholar] [CrossRef] [PubMed]
- Cole, F.; Krauss, R.S. Microform holoprosencephaly in mice that lack the Ig superfamily member Cdon. Curr. Biol. 2003, 13, 411–415. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Kang, J.S.; Cole, F.; Yi, M.J.; Krauss, R.S. Cdo Functions at Multiple Points in the Sonic Hedgehog Pathway, and Cdo-Deficient Mice Accurately Model Human Holoprosencephaly. Dev. Cell 2006, 10, 657–665. [Google Scholar] [CrossRef] [PubMed]
- Bae, G.U.; Domené, S.; Roessler, E.; Schachter, K.; Kang, J.S.; Muenke, M.; Krauss, R.S. Mutations in CDON, encoding a hedgehog receptor, result in holoprosencephaly and defective interactions with other hedgehog receptors. Am. J. Hum. Genet. 2011, 89, 231–240. [Google Scholar] [CrossRef] [PubMed]
- Allen, B.L.; Tenzen, T.; McMahon, A.P. The Hedgehog-binding proteins Gas1 and Cdo cooperate to positively regulate Shh signaling during mouse development. Genes Dev. 2007, 21, 1244–1257. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Hong, M.; Bae, G.; Kang, J.-S.; Krauss, R.S. Boc modifies the holoprosencephaly spectrum of Cdo mutant mice. Dis. Model. Mech. 2011, 4, 368–380. [Google Scholar] [CrossRef]
- Tenzen, T.; Allen, B.L.; Cole, F.; Kang, J.S.; Krauss, R.S.; McMahon, A.P. The Cell Surface Membrane Proteins Cdo and Boc Are Components and Targets of the Hedgehog Signaling Pathway and Feedback Network in Mice. Dev. Cell 2006, 10, 647–656. [Google Scholar] [CrossRef]
- Quezada-Ramírez, M.A.; Castañeda-Arellano, R.; Pérez-Sánchez, G.; Hernández-Soto, J.; Segovia, J. The Growth arrest specific 1 (GAS1) gene is transcriptionally regulated by NeuroD1 via two distal E-boxes. Exp. Cell Res. 2018, 363, 332–341. [Google Scholar] [CrossRef]
- Zhang, X.M.; Ramalho-Santos, M.; McMahon, A.P. Smoothened mutants reveal redundant roles for Shh and Ihh signaling including regulation of L/R asymmetry by the mouse node. Cell 2001, 105, 781–792. [Google Scholar] [CrossRef]
- Fuccillo, M.; Rallu, M.; McMahon, A.P.; Fishell, G. Temporal requirement for hedgehog signaling in ventral telencephalic patterning. Development 2004, 131, 5031–5040. [Google Scholar] [CrossRef]
- Machold, R.; Hayashi, S.; Rutlin, M.; Muzumdar, M.D.; Nery, S.; Corbin, J.G.; Gritli-Linde, A.; Dellovade, T.; Porter, J.A.; Rubin, L.L.; et al. Sonic Hedgehog Is Required for Progenitor Cell Maintenance in Telencephalic Stem Cell Niches. Neuron 2003, 39, 937–950. [Google Scholar] [CrossRef] [PubMed]
- Svärd, J.; Heby-Henricson, K.; Persson-Lek, M.; Rozell, B.; Lauth, M.; Bergström, A.; Ericson, J.; Toftgård, R.; Teglund, S. Genetic elimination of suppressor of fused reveals an essential repressor function in the mammalian hedgehog signaling pathway. Dev. Cell 2006, 10, 187–197. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Heydeck, W.; Zeng, H.; Liu, A. Dual function of suppressor of fused in Hh pathway activation and mouse spinal cord patterning. Dev. Biol. 2012, 362, 141–153. [Google Scholar] [CrossRef] [PubMed]
- Yabut, O.R.; Fernandez, G.; Huynh, T.; Yoon, K.; Pleasure, S.J. Suppressor of Fused Is Critical for Maintenance of Neuronal Progenitor Identity during Corticogenesis. Cell Rep. 2015, 12, 2021–2034. [Google Scholar] [CrossRef] [PubMed]
- Radonjić, N.V.; Memi, F.; Ortega, J.A.; Glidden, N.; Zhan, H.; Zecevic, N. The Role of Sonic Hedgehog in the Specification of Human Cortical Progenitors In Vitro. Cereb. Cortex 2016, 26, 131–143. [Google Scholar] [CrossRef] [PubMed]
- Jakovcevski, I.; Mayer, N.; Zecevic, N. Multiple origins of human neocortical interneurons are supported by distinct expression of transcription factors. Cereb. Cortex 2011, 21, 1771–1782. [Google Scholar] [CrossRef] [PubMed]
- Zecevic, N.; Hu, F.; Jakovcevski, I. Interneurons in the developing human neocortex. Dev. Neurobiol. 2011, 71, 18–33. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Zecevic, N. Dorsal radial glial cells have the potential to generate cortical interneurons in human but not in mouse brain. J. Neurosci. 2011, 31, 2413–2420. [Google Scholar] [CrossRef]
- Al-Jaberi, N.; Lindsay, S.; Sarma, S.; Bayatti, N.; Clowry, G.J. The early fetal development of human neocortical GABAergic interneurons. Cereb. Cortex 2015, 25, 631–645. [Google Scholar] [CrossRef]
- Yabut, O.; Ng, H.; Fernandez, G.; Yoon, K.; Kuhn, J.; Pleasure, S. Loss of Suppressor of Fused in Mid-Corticogenesis Leads to the Expansion of Intermediate Progenitors. J. Dev. Biol. 2016, 4, 29. [Google Scholar] [CrossRef]
- Bai, C.B.; Auerbach, W.; Lee, J.S.; Stephen, D.; Joyner, A.L. Gli2, but not Gli1, is required for initial Shh signaling and ectopic activation of the Shh pathway. Development 2002, 129, 4753–4761. [Google Scholar] [CrossRef] [PubMed]
- Park, H.L.; Bai, C.; Platt, K.A.; Matise, M.P.; Beeghly, A.; Hui, C.C.; Nakashima, M.; Joyner, A.L. Mouse Gli1 mutants are viable but have defects in SHH signaling in combination with a Gli2 mutation. Development 2000, 127, 1593–1605. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Platt, K.A.; Censullo, P.; Altaba, A.R. Gli1 is a target of Sonic hedgehog that induces ventral neural tube development. Development 1997, 124, 2537–2552. [Google Scholar] [CrossRef] [PubMed]
- Ruiz i Altaba, A.; Jessell, T.M.; Roelink, H. Restrictions to floor plate induction by hedgehog and winged-helix genes in the neural tube of frog embryos. Mol. Cell Neurosci. 1995, 6, 106–121. [Google Scholar] [CrossRef] [PubMed]
- Bai, C.B.; Stephen, D.; Joyner, A.L. All Mouse Ventral Spinal Cord Patterning by Hedgehog Is Gli Dependent and Involves an Activator Function of Gli3. Dev. Cell. 2004, 6, 103–115. [Google Scholar] [CrossRef] [PubMed]
- Persson, M.; Stamataki, D.; te Welscher, P.; Andersson, E.; Böse, J.; Rüther, U.; Ericson, J.; Briscoe, J. Dorsal-ventral patterning of the spinal cord requires Gli3 transcriptional repressor activity. Genes Dev. 2002, 16, 2865–2878. [Google Scholar] [CrossRef] [PubMed]
- Hui, C.; Joyner, A.L. A mouse model of Greig cephalo–polysyndactyly syndrome: The extra–toesJ mutation contains an intragenic deletion of the Gli3 gene. Nat. Genet. 1993, 3, 241–246. [Google Scholar] [CrossRef] [PubMed]
- Theil, T.; Alvarez-Bolado, G.; Walter, A.; Rüther, U. Gli3 is required for Emx gene expression during dorsal telencephalon development. Development 1999, 126, 3561–3571. [Google Scholar] [CrossRef]
- Tole, S.; Ragsdale, C.W.; Grove, E.A. Dorsoventral patterning of the telencephalon is disrupted in the mouse mutant extra-toes. Dev. Biol. 2000, 217, 254–265. [Google Scholar] [CrossRef]
- Huang, Y.; Roelink, H.; McKnight, G.S. Protein kinase A deficiency causes axially localized neural tube defects in mice. J. Biol. Chem. 2002, 277, 19889–19896. [Google Scholar] [CrossRef]
- Wang, H.; Ge, G.; Uchida, Y.; Luu, B.; Ahn, S. Gli3 is required for maintenance and fate specification of cortical progenitors. J. Neurosci. 2011, 31, 6440–6448. [Google Scholar] [CrossRef] [PubMed]
- Palma, V.; Ruiz i Altaba, A. Hedgehog-GLI signaling regulates the behavior of cells with stem cell properties in the developing neocortex. Development 2004, 131, 337–345. [Google Scholar] [CrossRef] [PubMed]
- Motoyama, J.; Milenkovic, L.; Iwama, M.; Shikata, Y.; Scott, M.P.; Hui, C. Differential requirement for Gli2 and Gli3 in ventral neural cell fate specification. Dev. Biol. 2003, 259, 150–161. [Google Scholar] [CrossRef] [PubMed]
- Cai, E.; Zhang, J.; Ge, X. Control of the Hedgehog pathway by compartmentalized PKA in the primary cilium. Sci. China Life Sci. 2022, 65, 500–514. [Google Scholar] [CrossRef] [PubMed]
- Truong, M.E.; Bilekova, S.; Choksi, S.P.; Li, W.; Bugaj, L.J.; Xu, K.; Reiter, J.F. Vertebrate cells differentially interpret ciliary and extraciliary cAMP. Cell 2021, 184, 2911–2926.e18. [Google Scholar] [CrossRef] [PubMed]
- Mukhopadhyay, S.; Wen, X.; Ratti, N.; Loktev, A.; Rangell, L.; Scales, S.J.; Jackson, P.K. The ciliary G-protein-coupled receptor Gpr161 negatively regulates the sonic hedgehog pathway via cAMP signaling. Cell 2013, 152, 210–223. [Google Scholar] [CrossRef] [PubMed]
- Somatilaka, B.N.; Hwang, S.H.; Palicharla, V.R.; White, K.A.; Badgandi, H.; Shelton, J.M.; Mukhopadhyay, S. Ankmy2 Prevents Smoothened-Independent Hyperactivation of the Hedgehog Pathway via Cilia-Regulated Adenylyl Cyclase Signaling. Dev. Cell 2020, 54, 710–726.e8. [Google Scholar] [CrossRef]
- Ge, X.; Milenkovic, L.; Suyama, K.; Hartl, T.; Purzner, T.; Winans, A.; Meyer, T.; Scott, M.P. Phosphodiesterase 4D acts downstream of Neuropilin to control Hedgehog signal transduction and the growth of medulloblastoma. eLife 2015, 4, e07068. [Google Scholar] [CrossRef]
- Peng, H.; Zhang, J.; Ya, A.; Ma, W.; Villa, S.; Sukenik, S.; Ge, X. Myomegalin regulates Hedgehog pathway by controlling PDE4D at the centrosome. Mol. Biol. Cell 2021, 32, 1807–1817. [Google Scholar] [CrossRef]
- Yamamoto, M.; Dräger, U.C.; Ong, D.E.; McCaffery, P. Retinoid-binding proteins in the cerebellum and choroid plexus and their relationship to regionalized retinoic acid synthesis and degradation. Eur. J. Biochem. 1998, 257, 344–350. [Google Scholar] [CrossRef]
- Chang, J.T.; Lehtinen, M.K.; Sive, H. Zebrafish cerebrospinal fluid mediates cell survival through a retinoid signaling pathway. Dev. Neurobiol. 2016, 76, 75–92. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Liu, J.; Ketova, T.; Fleming, J.T.; Grover, V.K.; Cooper, M.K.; Litingtung, Y.; Chiang, C. Transventricular delivery of sonic hedgehog is essential to cerebellar ventricular zone development. Proc. Natl. Acad. Sci. USA 2010, 107, 8422–8427. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, C.M.; Dymecki, S.M. Sonic hedgehog is required for vascular outgrowth in the hindbrain choroid plexus. Dev. Biol. 2010, 340, 430–437. [Google Scholar] [CrossRef] [PubMed]
- Lamus, F.; Martín, C.; Carnicero, E.; Moro, J.A.; Fernández, J.M.F.; Mano, A.; Gato, Á.; Alonso, M.I. FGF2/EGF contributes to brain neuroepithelial precursor proliferation and neurogenesis in rat embryos: The involvement of embryonic cerebrospinal fluid. Dev. Dyn. 2020, 249, 141–153. [Google Scholar] [CrossRef] [PubMed]
- Kaiser, K.; Gyllborg, D.; Procházka, J.; Salašová, A.; Kompaníková, P.; Molina, F.L.; Laguna-Goya, R.; Radaszkiewicz, T.; Harnoš, J.; Procházková, M.; et al. WNT5A is transported via lipoprotein particles in the cerebrospinal fluid to regulate hindbrain morphogenesis. Nat. Commun. 2019, 10, 1498. [Google Scholar] [CrossRef] [PubMed]
- Toresson, H.; Urquiza AM de Fagerström, C.; Perlmann, T.; Campbell, K. Retinoids are produced by glia in the lateral ganglionic eminence and regulate striatal neuron differentiation. Development 1999, 126, 1317–1326. [Google Scholar] [CrossRef] [PubMed]
- Lehtinen, M.K.; Walsh, C.A. Neurogenesis at the Brain–Cerebrospinal Fluid Interface. Annu. Rev. Cell Dev. Biol. 2011, 27, 653–679. [Google Scholar] [CrossRef]
- Chau, K.F.; Springel, M.W.; Broadbelt, K.G.; Park, H.Y.; Topal, S.; Lun, M.P.; Mullan, H.; Maynard, T.; Steen, H.; LaMantia, A.S.; et al. Progressive Differentiation and Instructive Capacities of Amniotic Fluid and Cerebrospinal Fluid Proteomes following Neural Tube Closure. Dev. Cell 2015, 35, 789–802. [Google Scholar] [CrossRef]
- Valente, E.M.; Rosti, R.O.; Gibbs, E.; Gleeson, J.G. Primary cilia in neurodevelopmental disorders. Nat. Rev. Neurol. 2014, 10, 27–36. [Google Scholar] [CrossRef]
- Nachury, M.V.; Loktev, A.V.; Zhang, Q.; Westlake, C.J.; Peränen, J.; Merdes, A.; Slusarski, D.C.; Scheller, R.H.; Bazan, J.F.; Sheffield, V.C.; et al. A Core Complex of BBS Proteins Cooperates with the GTPase Rab8 to Promote Ciliary Membrane Biogenesis. Cell 2007, 129, 1201–1213. [Google Scholar] [CrossRef]
- Tian, X.; Zhao, H.; Zhou, J. Organization, functions, and mechanisms of the BBSome in development, ciliopathies, and beyond. eLife 2023, 12, e87623. [Google Scholar] [CrossRef] [PubMed]
- Eggenschwiler, J.T.; Anderson, K.V. Cilia and Developmental Signaling. Annu. Rev. Cell Dev. Biol. 2007, 23, 345–373. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.-G.; Alvarez-Buylla, A. Role of primary cilia in brain development and cancer. Curr. Opin. Neurobiol. 2010, 20, 58–67. [Google Scholar] [CrossRef] [PubMed]
- Goetz, S.C.; Anderson, K.V. The primary cilium: A signalling centre during vertebrate development. Nat. Rev. Genet. 2010, 11, 331–344. [Google Scholar] [CrossRef]
- Wu, F.; Zhang, Y.; Sun, B.; McMahon, A.P.; Wang, Y. Hedgehog Signaling: From Basic Biology to Cancer Therapy. Cell Chem. Biol. 2017, 24, 252–280. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cai, E.; Barba, M.G.; Ge, X. Hedgehog Signaling in Cortical Development. Cells 2024, 13, 21. https://doi.org/10.3390/cells13010021
Cai E, Barba MG, Ge X. Hedgehog Signaling in Cortical Development. Cells. 2024; 13(1):21. https://doi.org/10.3390/cells13010021
Chicago/Turabian StyleCai, Eva, Maximiliano Gonzalez Barba, and Xuecai Ge. 2024. "Hedgehog Signaling in Cortical Development" Cells 13, no. 1: 21. https://doi.org/10.3390/cells13010021
APA StyleCai, E., Barba, M. G., & Ge, X. (2024). Hedgehog Signaling in Cortical Development. Cells, 13(1), 21. https://doi.org/10.3390/cells13010021






