The Emerging Role of SIRT7 in Glucose and Lipid Metabolism
Abstract
:1. Introduction
2. The Role of Nuclear Sirtuins in Lipid Metabolism
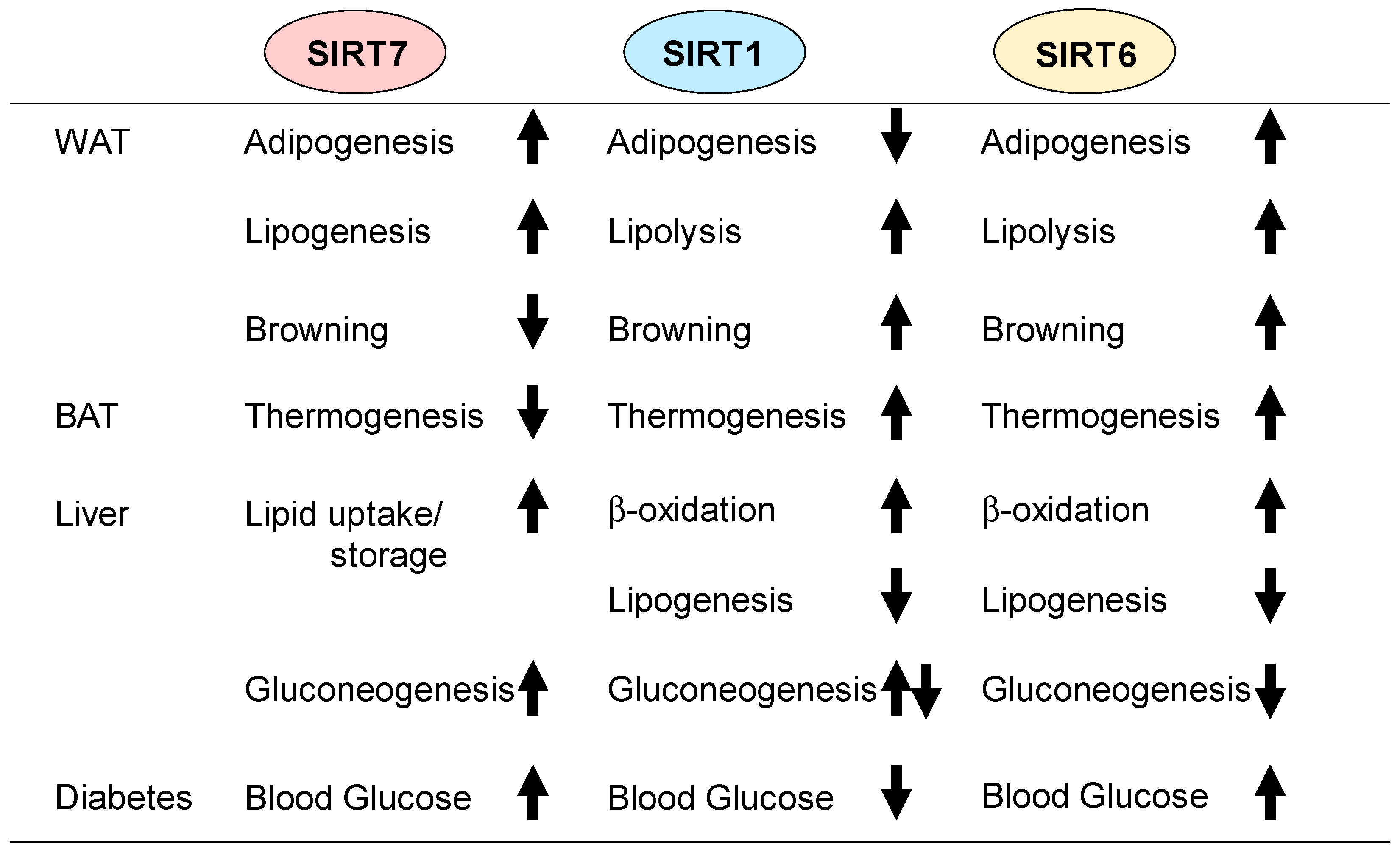
2.1. Adipogenesis
2.2. Lipolysis and Lipogenesis
2.3. Thermogenesis
2.4. Adipose Tissue Inflammation
2.5. Hepatic Lipid Metabolism
3. Nuclear Sirtuins in Glucose Metabolism
3.1. Gluconeogenesis
3.2. Glycolysis and Mitochondrial Function
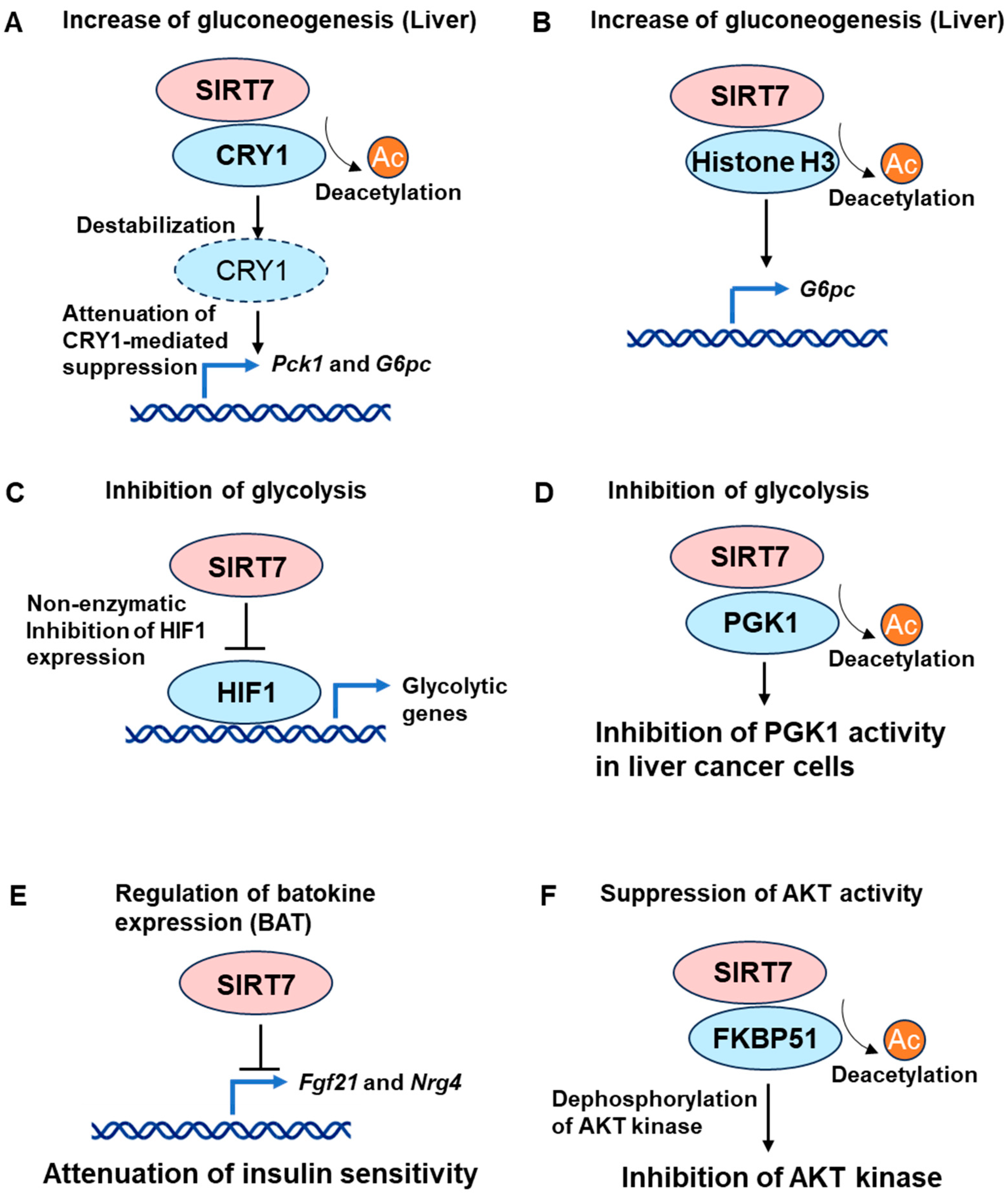
3.3. Insulin Action and Glucose Tolerance
4. Conclusions and Future Perspectives
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Giblin, W.; Skinner, M.E.; Lombard, D.B. Sirtuins: Guardians of Mammalian Healthspan. Trends Genet. 2014, 30, 271–286. [Google Scholar] [CrossRef]
- Houtkooper, R.H.; Pirinen, E.; Auwerx, J. Sirtuins as Regulators of Metabolism and Healthspan. Nat. Rev. Mol. Cell Biol. 2012, 13, 225–238. [Google Scholar] [CrossRef]
- Sebastiań, C.; Satterstrom, F.K.; Haigis, M.C.; Mostoslavsky, R. From Sirtuin Biology to Human Diseases: An Update. J. Biol. Chem. 2012, 287, 42444–42452. [Google Scholar] [CrossRef]
- Finkel, T.; Deng, C.X.; Mostoslavsky, R. Recent Progress in the Biology and Physiology of Sirtuins. Nature 2009, 460, 587–591. [Google Scholar] [CrossRef]
- Haigis, M.C.; Mostoslavsky, R.; Haigis, K.M.; Fahie, K.; Christodoulou, D.C.; Murphy, A.J.J.; Valenzuela, D.M.; Yancopoulos, G.D.; Karow, M.; Blander, G.; et al. SIRT4 Inhibits Glutamate Dehydrogenase and Opposes the Effects of Calorie Restriction in Pancreatic β Cells. Cell 2006, 126, 941–954. [Google Scholar] [CrossRef]
- Du, J.; Zhou, Y.; Su, X.; Yu, J.J.; Khan, S.; Jiang, H.; Kim, J.; Woo, J.; Kim, J.H.; Choi, B.H.; et al. Sirt5 Is a NAD-Dependent Protein Lysine Demalonylase and Desuccinylase. Science 2011, 334, 806–809. [Google Scholar] [CrossRef]
- Jiang, H.; Khan, S.; Wang, Y.; Charron, G.; He, B.; Sebastian, C.; Du, J.; Kim, R.; Ge, E.; Mostoslavsky, R.; et al. SIRT6 Regulates TNF-α Secretion through Hydrolysis of Long-Chain Fatty Acyl Lysine. Nature 2013, 496, 110–113. [Google Scholar] [CrossRef]
- Feldman, J.L.; Baeza, J.; Denu, J.M. Activation of the Protein Deacetylase SIRT6 by Long-Chain Fatty Acids and Widespread Deacylation by Mammalian Sirtuins. J. Biol. Chem. 2013, 288, 31350–31356. [Google Scholar] [CrossRef]
- Ford, E.; Voit, R.; Liszt, G.; Magin, C.; Grummt, I.; Guarente, L. Mammalian Sir2 Homolog SIRT7 Is an Activator of RNA Polymerase I Transcription. Genes. Dev. 2006, 20, 1075–1080. [Google Scholar] [CrossRef]
- Barber, M.F.; Michishita-Kioi, E.; Xi, Y.; Tasselli, L.; Kioi, M.; Moqtaderi, Z.; Tennen, R.I.; Paredes, S.; Young, N.L.; Chen, K.; et al. SIRT7 Links H3K18 Deacetylation to Maintenance of Oncogenic Transformation. Nature 2012, 487, 114–118. [Google Scholar] [CrossRef]
- Vazquez, B.N.; Thackray, J.K.; Simonet, N.G.; Kane-Goldsmith, N.; Martinez-Redondo, P.; Nguyen, T.; Bunting, S.; Vaquero, A.; Tischfield, J.A.; Serrano, L. SIRT 7 Promotes Genome Integrity and Modulates Non-homologous End Joining DNA Repair. EMBO J. 2016, 35, 1488–1503. [Google Scholar] [CrossRef]
- Wang, W.W.; Angulo-Ibanez, M.; Lyu, J.; Kurra, Y.; Tong, Z.; Wu, B.; Zhang, L.; Sharma, V.; Zhou, J.; Lin, H.; et al. A Click Chemistry Approach Reveals the Chromatin-Dependent Histone H3K36 Deacylase Nature of SIRT7. J. Am. Chem. Soc. 2019, 141, 2462–2473. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Seiler, J.; Santiago-Reichelt, M.; Felbel, K.; Grummt, I.; Voit, R. Repression of RNA Polymerase I upon Stress Is Caused by Inhibition of RNA-Dependent Deacetylation of PAF53 by SIRT7. Mol. Cell 2013, 52, 303–313. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Blank, M.F.; Iyer, A.; Huang, B.; Wang, L.; Grummt, I.; Voit, R. SIRT7-Dependent Deacetylation of the U3-55k Protein Controls Pre-RRNA Processing. Nat. Commun. 2016, 7, 10734. [Google Scholar] [CrossRef] [PubMed]
- Iyer-Bierhoff, A.; Krogh, N.; Tessarz, P.; Ruppert, T.; Nielsen, H.; Grummt, I. SIRT7-Dependent Deacetylation of Fibrillarin Controls Histone H2A Methylation and RRNA Synthesis during the Cell Cycle. Cell Rep. 2018, 25, 2946–2954.e5. [Google Scholar] [CrossRef] [PubMed]
- Blank, M.F.; Chen, S.; Poetz, F.; Schnölzer, M.; Voit, R.; Grummt, I. SIRT7-Dependent Deacetylation of CDK9 Activates RNA Polymerase II Transcription. Nucleic Acids Res. 2017, 45, 2675–2686. [Google Scholar] [CrossRef] [PubMed]
- Song, C.; Hotz-Wagenblatt, A.; Voit, R.; Grummt, I. SIRT7 and the DEAD-Box Helicase DDX21 Cooperate to Resolve Genomic R Loops and Safeguard Genome Stability. Genes. Dev. 2017, 31, 1370–1381. [Google Scholar] [CrossRef]
- Tang, M.; Li, Z.; Zhang, C.; Lu, X.; Tu, B.; Cao, Z.; Li, Y.; Chen, Y.; Jiang, L.; Wang, H.; et al. SIRT7-Mediated ATM Deacetylation Is Essential for Its Deactivation and DNA Damage Repair. Sci. Adv. 2019, 5, eaav1118. [Google Scholar] [CrossRef]
- Karim, M.F.; Yoshizawa, T.; Sobuz, S.U.; Sato, Y.; Yamagata, K. Sirtuin 7-Dependent Deacetylation of DDB1 Regulates the Expression of Nuclear Receptor TR4. Biochem. Biophys. Res. Commun. 2017, 490, 423–428. [Google Scholar] [CrossRef]
- Akter, F.; Tsuyama, T.; Yoshizawa, T.; Sobuz, S.U.; Yamagata, K. SIRT7 Regulates Lipogenesis in Adipocytes through Deacetylation of PPARγ2. J. Diabetes Investig. 2021, 12, 1765–1774. [Google Scholar] [CrossRef]
- Yoshizawa, T.; Sato, Y.; Sobuz, S.U.; Mizumoto, T.; Tsuyama, T.; Karim, M.F.; Miyata, K.; Tasaki, M.; Yamazaki, M.; Kariba, Y.; et al. SIRT7 Suppresses Energy Expenditure and Thermogenesis by Regulating Brown Adipose Tissue Functions in Mice. Nat. Commun. 2022, 13, 7439. [Google Scholar] [CrossRef] [PubMed]
- Ryu, D.; Jo, Y.S.; Lo Sasso, G.; Stein, S.; Zhang, H.; Perino, A.; Lee, J.U.; Zeviani, M.; Romand, R.; Hottiger, M.O.; et al. A SIRT7-Dependent Acetylation Switch of GABPβ1 Controls Mitochondrial Function. Cell Metab. 2014, 20, 856–869. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Qian, M.; Tang, X.; Hu, W.; Sun, S.; Li, G.; Zhang, S.; Meng, F.; Cao, X.; Sun, J.; et al. SIRT7 Couples Light-Driven Body Temperature Cues to Hepatic Circadian Phase Coherence and Gluconeogenesis. Nat. Metab. 2019, 1, 1141–1156. [Google Scholar] [CrossRef] [PubMed]
- Qi, H.; Shi, X.; Yu, M.; Liu, B.; Liu, M.; Song, S.; Chen, S.; Zou, J.; Zhu, W.G.; Luo, J. Sirtuin 7–Mediated Deacetylation of WD Repeat Domain 77 (WDR77) Suppresses Cancer Cell Growth by Reducing WDR77/ PRMT5 Transmethylase Complex Activity. J. Biol. Chem. 2018, 293, 17769–17779. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.; Zhu, W.; Qin, J.; Chen, M.; Gong, L.; Li, L.; Liu, X.; Tao, Y.; Yin, H.; Zhou, H.; et al. Acetylation of PGK1 Promotes Liver Cancer Cell Proliferation and Tumorigenesis. Hepatology 2017, 65, 515–528. [Google Scholar] [CrossRef]
- Yu, J.; Qin, B.; Wu, F.; Qin, S.; Nowsheen, S.; Shan, S.; Zayas, J.; Pei, H.; Lou, Z.; Wang, L. Regulation of Serine-Threonine Kinase Akt Activation by NAD+-Dependent Deacetylase SIRT7. Cell Rep. 2017, 18, 1229–1240. [Google Scholar] [CrossRef] [PubMed]
- Tang, X.; Shi, L.; Xie, N.; Liu, Z.; Qian, M.; Meng, F.; Xu, Q.; Zhou, M.; Cao, X.; Zhu, W.G.; et al. SIRT7 Antagonizes TGF-β Signaling and Inhibits Breast Cancer Metastasis. Nat. Commun. 2017, 8, 318. [Google Scholar] [CrossRef]
- Dong, L.; Yu, L.; Li, H.; Shi, L.; Luo, Z.; Zhao, H.; Liu, Z.; Yin, G.; Yan, X.; Lin, Z. An NAD+-Dependent Deacetylase SIRT7 Promotes HCC Development Through Deacetylation of USP39. iScience 2020, 23, 101351. [Google Scholar] [CrossRef]
- Liu, X.; Li, C.; Li, Q.; Chang, H.C.; Tang, Y.C. SIRT7 Facilitates CENP-A Nucleosome Assembly and Suppresses Intestinal Tumorigenesis. iScience 2020, 23, 101461. [Google Scholar] [CrossRef]
- Yamamura, S.; Izumiya, Y.; Araki, S.; Nakamura, T.; Kimura, Y.; Hanatani, S.; Yamada, T.; Ishida, T.; Yamamoto, M.; Onoue, Y.; et al. Cardiomyocyte Sirt (Sirtuin) 7 Ameliorates Stress-Induced Cardiac Hypertrophy by Interacting With and Deacetylating GATA4. Hypertension 2020, 75, 98–108. [Google Scholar] [CrossRef]
- Noriega, L.G.; Melo, Z.; Rajaram, R.D.; Mercado, A.; Tovar, A.R.; Velazquez-Villegas, L.A.; Castañeda-Bueno, M.; Reyes-López, Y.; Ryu, D.; Rojas-Vega, L.; et al. SIRT7 Modulates the Stability and Activity of the Renal K-Cl Cotransporter KCC4 through Deacetylation. EMBO Rep. 2021, 22, e50766. [Google Scholar] [CrossRef] [PubMed]
- Sobuz, S.U.; Sato, Y.; Yoshizawa, T.; Karim, F.; Ono, K.; Sawa, T.; Miyamoto, Y.; Oka, M.; Yamagata, K. SIRT7 Regulates the Nuclear Export of NF-κB P65 by Deacetylating Ran. Biochim. Biophys. Acta Mol. Cell Res. 2019, 1866, 1355–1367. [Google Scholar] [CrossRef] [PubMed]
- Lee, N.; Kim, D.K.; Kim, E.S.; Park, S.J.; Kwon, J.H.; Shin, J.; Park, S.M.; Moon, Y.H.; Wang, H.J.; Gho, Y.S.; et al. Comparative Interactomes of SIRT6 and SIRT7: Implication of Functional Links to Aging. Proteomics 2014, 14, 1610–1622. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Bridges, B.; Olson, J.; Weinman, S.A. The Interaction between Acetylation and Serine-574 Phosphorylation Regulates the Apoptotic Function of FOXO3. Oncogene 2017, 36, 1887–1898. [Google Scholar] [CrossRef] [PubMed]
- Vakhrusheva, O.; Smolka, C.; Gajawada, P.; Kostin, S.; Boettger, T.; Kubin, T.; Braun, T.; Bober, E. Sirt7 Increases Stress Resistance of Cardiomyocytes and Prevents Apoptosis and Inflammatory Cardiomyopathy in Mice. Circ. Res. 2008, 102, 703–710. [Google Scholar] [CrossRef] [PubMed]
- Yu, M.; Shi, X.; Ren, M.; Liu, L.; Qi, H.; Zhang, C.; Zou, J.; Qiu, X.; Zhu, W.G.; Zhang, Y.E.; et al. SIRT7 Deacetylates Strap to Regulate P53 Activity and Stability. Int. J. Mol. Sci. 2020, 21, 4122. [Google Scholar] [CrossRef] [PubMed]
- Mohrin, M.; Shin, J.; Liu, Y.; Brown, K.; Luo, H.; Xi, Y.; Haynes, C.M.; Chen, D. Stem cell aging. A mitochondrial UPR-mediated metabolic checkpoint regulates hematopoietic stem cell aging. Science 2015, 347, 1374–1377. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Tang, X.; Zhang, S.; Jin, M.; Wang, M.; Deng, Z.; Liu, Z.; Qian, M.; Shi, W.; Wang, Z.; et al. SIRT 7 Activates Quiescent Hair Follicle Stem Cells to Ensure Hair Growth in Mice. EMBO J. 2020, 39, e104365. [Google Scholar] [CrossRef]
- Li, L.; Shi, L.; Yang, S.; Yan, R.; Zhang, D.; Yang, J.; He, L.; Li, W.; Yi, X.; Sun, L.; et al. SIRT7 Is a Histone Desuccinylase That Functionally Links to Chromatin Compaction and Genome Stability. Nat. Commun. 2016, 7, 12235. [Google Scholar] [CrossRef]
- Yuan, H.F.; Zhao, M.; Zhao, L.N.; Yun, H.L.; Yang, G.; Geng, Y.; Wang, Y.F.; Zheng, W.; Yuan, Y.; Song, T.Q.; et al. PRMT5 Confers Lipid Metabolism Reprogramming, Tumour Growth and Metastasis Depending on the SIRT7-Mediated Desuccinylation of PRMT5 K387 in Tumours. Acta Pharmacol. Sin. 2022, 43, 2373–2385. [Google Scholar] [CrossRef]
- Bao, X.; Liu, Z.; Zhang, W.; Gladysz, K.; Fung, Y.M.E.; Tian, G.; Xiong, Y.; Wong, J.W.H.; Yuen, K.W.Y.; Li, X.D. Glutarylation of Histone H4 Lysine 91 Regulates Chromatin Dynamics. Mol. Cell 2019, 76, 660–675. [Google Scholar] [CrossRef] [PubMed]
- Yu, A.Q.; Wang, J.; Jiang, S.T.; Yuan, L.Q.; Ma, H.Y.; Hu, Y.M.; Han, X.M.; Tan, L.M.; Wang, Z.X. SIRT7-Induced PHF5A Decrotonylation Regulates Aging Progress Through Alternative Splicing-Mediated Downregulation of CDK2. Front. Cell Dev. Biol. 2021, 9, 710479. [Google Scholar] [CrossRef] [PubMed]
- Fukuda, M.; Yoshizawa, T.; Karim, M.F.; Sobuz, S.U.; Korogi, W.; Kobayasi, D.; Okanishi, H.; Tasaki, M.; Ono, K.; Sawa, T.; et al. SIRT7 Has a Critical Role in Bone Formation by Regulating Lysine Acylation of SP7/Osterix. Nat. Commun. 2018, 9, 2833. [Google Scholar] [CrossRef] [PubMed]
- Simonet, N.G.; Thackray, J.K.; Vazquez, B.N.; Ianni, A.; Espinosa-Alcantud, M.; Morales-Sanfrutos, J.; Hurtado-Bagès, S.; Sabidó, E.; Buschbeck, M.; Tischfield, J.; et al. SirT7 Auto-ADP-Ribosylation Regulates Glucose Starvation Response through MH2A1. Sci. Adv. 2020, 6, eaaz2590. [Google Scholar] [CrossRef] [PubMed]
- Chang, A.R.; Ferrer, C.M.; Mostoslavsky, R. SIRT6, a Mammalian Deacylase with Multitasking Abilities. Physiol. Rev. 2020, 100, 145–169. [Google Scholar] [CrossRef] [PubMed]
- Guo, Z.; Li, P.; Ge, J.; Li, H. SIRT6 in Aging, Metabolism, Inflammation and Cardiovascular Diseases. Aging Dis. 2022, 13, 1787–1822. [Google Scholar] [CrossRef] [PubMed]
- Yamagata, K.; Yoshizawa, T. Transcriptional Regulation of Metabolism by SIRT1 and SIRT7. Int. Rev. Cell Mol. Biol. 2018, 335, 143–166. [Google Scholar] [CrossRef] [PubMed]
- Tang, M.; Tang, H.; Tu, B.; Zhu, W.G. SIRT7: A Sentinel of Genome Stability. Open Biol. 2021, 11, 210047. [Google Scholar] [CrossRef]
- Lagunas-Rangel, F.A. SIRT7 in the Aging Process. Cell Mol. Life Sci. 2022, 79, 297. [Google Scholar] [CrossRef]
- Wu, S.; Jia, S. Functional Diversity of SIRT7 Across Cellular Compartments: Insights and Perspectives. Cell Biochem. Biophys. 2023, 81, 409–419. [Google Scholar] [CrossRef]
- Raza, U.; Tang, X.; Liu, Z.; Liu, B. SIRT7: The Seventh Key to Unlocking the Mystery of Aging. Physiol. Rev. 2024, 104, 253–280. [Google Scholar] [CrossRef] [PubMed]
- Lefterova, M.I.; Lazar, M.A. New Developments in Adipogenesis. Trends Endocrinol. Metab. 2009, 20, 107–114. [Google Scholar] [CrossRef] [PubMed]
- Ali, A.T.; Hochfeld, W.E.; Myburgh, R.; Pepper, M.S. Adipocyte and Adipogenesis. Eur. J. Cell Biol. 2013, 92, 229–236. [Google Scholar] [CrossRef] [PubMed]
- Ahmadian, M.; Suh, J.M.; Hah, N.; Liddle, C.; Atkins, A.R.; Downes, M.; Evans, R.M. Pparγ Signaling and Metabolism: The Good, the Bad and the Future. Nat. Med. 2013, 19, 557–566. [Google Scholar] [CrossRef] [PubMed]
- Picard, F.; Kurtev, M.; Chung, N.; Topark-Ngarm, A.; Senawong, T.; De Oliveira, R.M.; Leid, M.; McBurney, M.W.; Guarente, L. Sirt1 Promotes Fat Mobilization in White Adipocytes by Repressing PPAR-Gamma. Nature 2004, 429, 771–776. [Google Scholar] [CrossRef]
- Chen, Q.; Hao, W.; Xiao, C.; Wang, R.; Xu, X.; Lu, H.; Chen, W.; Deng, C.X. SIRT6 Is Essential for Adipocyte Differentiation by Regulating Mitotic Clonal Expansion. Cell Rep. 2017, 18, 3155–3166. [Google Scholar] [CrossRef]
- Hong, J.; Mei, C.; Abbas Raza, S.H.; Khan, R.; Cheng, G.; Zan, L. SIRT6 Cooperates with SIRT5 to Regulate Bovine Preadipocyte Differentiation and Lipid Metabolism via the AMPKα Signaling Pathway. Arch. Biochem. Biophys. 2020, 681, 108260. [Google Scholar] [CrossRef]
- Cioffi, M.; Vallespinos-Serrano, M.; Trabulo, S.M.; Fernandez-Marcos, P.J.; Firment, A.N.; Vazquez, B.N.; Vieira, C.R.; Mulero, F.; Camara, J.A.; Cronin, U.P.; et al. MiR-93 Controls Adiposity via Inhibition of Sirt7 and Tbx3. Cell Rep. 2015, 12, 1594–1605. [Google Scholar] [CrossRef]
- Fang, J.; Ianni, A.; Smolka, C.; Vakhrusheva, O.; Nolte, H.; Krüger, M.; Wietelmann, A.; Simonet, N.G.; Adrian-Segarra, J.M.; Vaquero, A.; et al. Sirt7 Promotes Adipogenesis in the Mouse by Inhibiting Autocatalytic Activation of Sirt1. Proc. Natl. Acad. Sci. USA 2017, 114, E8352–E8361. [Google Scholar] [CrossRef]
- Rangwala, S.M.; Lazar, M.A. Peroxisome Proliferator-Activated Receptor γ in Diabetes and Metabolism. Trends Pharmacol. Sci. 2004, 25, 331–336. [Google Scholar] [CrossRef]
- Zimmermann, R.; Strauss, J.G.; Haemmerle, G.; Schoiswohl, G.; Birner-Gruenberger, R.; Riederer, M.; Lass, A.; Neuberger, G.; Eisenhaber, F.; Hermetter, A.; et al. Fat Mobilization in Adipose Tissue Is Promoted by Adipose Triglyceride Lipase. Science 2004, 306, 1383–1386. [Google Scholar] [CrossRef] [PubMed]
- Haemmerle, G.; Lass, A.; Zimmermann, R.; Gorkiewicz, G.; Meyer, C.; Rozman, J.; Heldmaier, G.; Maier, R.; Theussl, C.; Eder, S.; et al. Defective Lipolysis and Altered Energy Metabolism in Mice Lacking Adipose Triglyceride Lipase. Science 2006, 312, 734–737. [Google Scholar] [CrossRef] [PubMed]
- Chakrabarti, P.; Kandror, K.V. FoxO1 Controls Insulin-Dependent Adipose Triglyceride Lipase (ATGL) Expression and Lipolysis in Adipocytes. J. Biol. Chem. 2009, 284, 13296–13300. [Google Scholar] [CrossRef] [PubMed]
- Kuang, J.; Zhang, Y.; Liu, Q.; Shen, J.; Pu, S.; Cheng, S.; Chen, L.; Li, H.; Wu, T.; Li, R.; et al. Fat-Specific Sirt6 Ablation Sensitizes Mice to High-Fat Diet-Induced Obesity and Insulin Resistance by Inhibiting Lipolysis. Diabetes 2017, 66, 1159–1171. [Google Scholar] [CrossRef] [PubMed]
- Rosen, E.D.; Spiegelman, B.M. What We Talk about When We Talk about Fat. Cell 2014, 156, 20–44. [Google Scholar] [CrossRef] [PubMed]
- Ikeda, K.; Maretich, P.; Kajimura, S. The Common and Distinct Features of Brown and Beige Adipocytes. Trends Endocrinol. Metab. 2018, 29, 191–200. [Google Scholar] [CrossRef] [PubMed]
- Lowell, B.B.; Spiegelman, B.M. Towards a Molecular Understanding of Adaptive Thermogenesis. Nature 2000, 404, 652–660. [Google Scholar] [CrossRef] [PubMed]
- Richard, D.; Carpentier, A.C.; Doré, G.; Ouellet, V.; Picard, F. Determinants of Brown Adipocyte Development and Thermogenesis. Int. J. Obes. 2010, 34 (Suppl. S2), S59–S66. [Google Scholar] [CrossRef]
- Boutant, M.; Joffraud, M.; Kulkarni, S.S.; García-Casarrubios, E.; García-Roves, P.M.; Ratajczak, J.; Fernández-Marcos, P.J.; Valverde, A.M.; Serrano, M.; Cantó, C. SIRT1 Enhances Glucose Tolerance by Potentiating Brown Adipose Tissue Function. Mol. Metab. 2015, 4, 118–131. [Google Scholar] [CrossRef]
- Xu, F.; Zheng, X.; Lin, B.; Liang, H.; Cai, M.; Cao, H.; Ye, J.; Weng, J. Diet-Induced Obesity and Insulin Resistance Are Associated with Brown Fat Degeneration in SIRT1-Deficient Mice. Obesity 2016, 24, 634–642. [Google Scholar] [CrossRef]
- Qiang, L.; Wang, L.; Kon, N.; Zhao, W.; Lee, S.; Zhang, Y.; Rosenbaum, M.; Zhao, Y.; Gu, W.; Farmer, S.R.; et al. Brown Remodeling of White Adipose Tissue by SirT1-Dependent Deacetylation of Pparγ. Cell 2012, 150, 620–632. [Google Scholar] [CrossRef]
- Yao, L.; Cui, X.; Chen, Q.; Yang, X.; Fang, F.; Zhang, J.; Liu, G.; Jin, W.; Chang, Y. Cold-Inducible SIRT6 Regulates Thermogenesis of Brown and Beige Fat. Cell Rep. 2017, 20, 641–654. [Google Scholar] [CrossRef]
- Jung, S.M.; Hung, C.M.; Hildebrand, S.R.; Sanchez-Gurmaches, J.; Martinez-Pastor, B.; Gengatharan, J.M.; Wallace, M.; Mukhopadhyay, D.; Martinez Calejman, C.; Luciano, A.K.; et al. Non-Canonical MTORC2 Signaling Regulates Brown Adipocyte Lipid Catabolism through SIRT6-FoxO1. Mol. Cell 2019, 75, 807–822. [Google Scholar] [CrossRef]
- Dai, N.; Zhao, L.; Wrighting, D.; Krämer, D.; Majithia, A.; Wang, Y.; Cracan, V.; Borges-Rivera, D.; Mootha, V.K.; Nahrendorf, M.; et al. IGF2BP2/IMP2-Deficient Mice Resist Obesity through Enhanced Translation of Ucp1 MRNA and Other MRNAs Encoding Mitochondrial Proteins. Cell Metab. 2015, 21, 609–621. [Google Scholar] [CrossRef]
- Shoelson, S.E.; Herrero, L.; Naaz, A. Obesity, Inflammation, and Insulin Resistance. Gastroenterology 2007, 132, 2169–2180. [Google Scholar] [CrossRef]
- Lumeng, C.N.; Saltiel, A.R. Inflammatory Links between Obesity and Metabolic Disease. J. Clin. Investig. 2011, 121, 2111–2117. [Google Scholar] [CrossRef]
- Yeung, F.; Hoberg, J.E.; Ramsey, C.S.; Keller, M.D.; Jones, D.R.; Frye, R.A.; Mayo, M.W. Modulation of NF-κB-Dependent Transcription and Cell Survival by the SIRT1 Deacetylase. EMBO J. 2004, 23, 2369–2380. [Google Scholar] [CrossRef]
- Yoshizaki, T.; Milne, J.C.; Imamura, T.; Schenk, S.; Sonoda, N.; Babendure, J.L.; Lu, J.-C.; Smith, J.J.; Jirousek, M.R.; Olefsky, J.M. SIRT1 Exerts Anti-Inflammatory Effects and Improves Insulin Sensitivity in Adipocytes. Mol. Cell Biol. 2009, 29, 1363–1374. [Google Scholar] [CrossRef]
- Gillum, M.P.; Kotas, M.E.; Erion, D.M.; Kursawe, R.; Chatterjee, P.; Nead, K.T.; Muise, E.S.; Hsiao, J.J.; Frederick, D.W.; Yonemitsu, S.; et al. SirT1 Regulates Adipose Tissue Inflammation. Diabetes 2011, 60, 3235–3245. [Google Scholar] [CrossRef]
- Kawahara, T.L.A.; Michishita, E.; Adler, A.S.; Damian, M.; Berber, E.; Lin, M.; McCord, R.A.; Ongaigui, K.C.L.; Boxer, L.D.; Chang, H.Y.; et al. SIRT6 Links Histone H3 Lysine 9 Deacetylation to NF-κB-Dependent Gene Expression and Organismal Life Span. Cell 2009, 136, 62–74. [Google Scholar] [CrossRef]
- Xiong, X.; Zhang, C.; Zhang, Y.; Fan, R.; Qian, X.; Dong, X.C. Fabp4-Cre-Mediated Sirt6 Deletion Impairs Adipose Tissue Function and Metabolic Homeostasis in Mice. J. Endocrinol. 2017, 233, 307–314. [Google Scholar] [CrossRef] [PubMed]
- Miyasato, Y.; Yoshizawa, T.; Sato, Y.; Nakagawa, T.; Miyasato, Y.; Kakizoe, Y.; Kuwabara, T.; Adachi, M.; Ianni, A.; Braun, T.; et al. Sirtuin 7 Deficiency Ameliorates Cisplatin-Induced Acute Kidney Injury Through Regulation of the Inflammatory Response. Sci. Rep. 2018, 8, 5927. [Google Scholar] [CrossRef] [PubMed]
- Araki, S.; Izumiya, Y.; Rokutanda, T.; Ianni, A.; Hanatani, S.; Kimura, Y.; Onoue, Y.; Senokuchi, T.; Yoshizawa, T.; Yasuda, O.; et al. Sirt7 Contributes to Myocardial Tissue Repair by Maintaining Transforming Growth Factor-β Signaling Pathway. Circulation 2015, 132, 1081–1093. [Google Scholar] [CrossRef] [PubMed]
- Wyman, A.E.; Nguyen, T.T.T.; Karki, P.; Tulapurkar, M.E.; Zhang, C.O.; Kim, J.; Feng, T.G.; Dabo, A.J.; Todd, N.W.; Luzina, I.G.; et al. SIRT7 Deficiency Suppresses Inflammation, Induces EndoMT, and Increases Vascular Permeability in Primary Pulmonary Endothelial Cells. Sci. Rep. 2020, 10, 12497. [Google Scholar] [CrossRef] [PubMed]
- Yoshizawa, T.; Karim, M.F.; Sato, Y.; Senokuchi, T.; Miyata, K.; Fukuda, T.; Go, C.; Tasaki, M.; Uchimura, K.; Kadomatsu, T.; et al. SIRT7 Controls Hepatic Lipid Metabolism by Regulating the Ubiquitin-Proteasome Pathway. Cell Metab. 2014, 19, 712–721. [Google Scholar] [CrossRef] [PubMed]
- Vega, R.B.; Huss, J.M.; Kelly, D.P. The Coactivator PGC-1 Cooperates with Peroxisome Proliferator-Activated Receptor α in Transcriptional Control of Nuclear Genes Encoding Mitochondrial Fatty Acid Oxidation Enzymes. Mol. Cell Biol. 2000, 20, 1868–1876. [Google Scholar] [CrossRef] [PubMed]
- Purushotham, A.; Schug, T.T.; Xu, Q.; Surapureddi, S.; Guo, X.; Li, X. Hepatocyte-Specific Deletion of SIRT1 Alters Fatty Acid Metabolism and Results in Hepatic Steatosis and Inflammation. Cell Metab. 2009, 9, 327–338. [Google Scholar] [CrossRef]
- Pfluger, P.T.; Herranz, D.; Velasco-Miguel, S.; Serrano, M.; Tschöp, M.H. Sirt1 Protects against High-Fat Diet-Induced Metabolic Damage. Proc. Natl. Acad. Sci. USA 2008, 105, 9793–9798. [Google Scholar] [CrossRef]
- Ponugoti, B.; Kim, D.H.; Xiao, Z.; Smith, Z.; Miao, J.; Zang, M.; Wu, S.Y.; Chiang, C.M.; Veenstra, T.D.; Kemper, J.K. SIRT1 Deacetylates and Inhibits SREBP-1C Activity in Regulation of Hepatic Lipid Metabolism. J. Biol. Chem. 2010, 285, 33959–33970. [Google Scholar] [CrossRef]
- Kim, H.S.; Xiao, C.; Wang, R.H.; Lahusen, T.; Xu, X.; Vassilopoulos, A.; Vazquez-Ortiz, G.; Jeong, W.I.; Park, O.; Ki, S.H.; et al. Hepatic-Specific Disruption of SIRT6 in Mice Results in Fatty Liver Formation Due to Enhanced Glycolysis and Triglyceride Synthesis. Cell Metab. 2010, 12, 224–236. [Google Scholar] [CrossRef]
- Esau, C.; Davis, S.; Murray, S.F.; Yu, X.X.; Pandey, S.K.; Pear, M.; Watts, L.; Booten, S.L.; Graham, M.; McKay, R.; et al. MiR-122 Regulation of Lipid Metabolism Revealed by in Vivo Antisense Targeting. Cell Metab. 2006, 3, 87–98. [Google Scholar] [CrossRef] [PubMed]
- Elhanati, S.; Ben-Hamo, R.; Kanfi, Y.; Varvak, A.; Glazz, R.; Lerrer, B.; Efroni, S.; Cohen, H.Y. Reciprocal Regulation between SIRT6 and MiR-122 Controls Liver Metabolism and Predicts Hepatocarcinoma Prognosis. Cell Rep. 2016, 14, 234–242. [Google Scholar] [CrossRef] [PubMed]
- Elhanati, S.; Kanfi, Y.; Varvak, A.; Roichman, A.; Carmel-Gross, I.; Barth, S.; Gibor, G.; Cohen, H.Y. Multiple Regulatory Layers of SREBP1/2 by SIRT6. Cell Rep. 2013, 4, 905–912. [Google Scholar] [CrossRef] [PubMed]
- Shin, J.; He, M.; Liu, Y.; Paredes, S.; Villanova, L.; Brown, K.; Qiu, X.; Nabavi, N.; Mohrin, M.; Wojnoonski, K.; et al. SIRT7 Represses Myc Activity to Suppress Er Stress and Prevent Fatty Liver Disease. Cell Rep. 2013, 5, 654–665. [Google Scholar] [CrossRef] [PubMed]
- Rodgers, J.T.; Lerin, C.; Haas, W.; Gygi, S.P.; Spiegelman, B.M.; Puigserver, P. Nutrient Control of Glucose Homeostasis through a Complex of PGC-1alpha and SIRT1. Nature 2005, 434, 113–118. [Google Scholar] [CrossRef] [PubMed]
- Frescas, D.; Valenti, L.; Accili, D. Nuclear Trapping of the Forkhead Transcription Factor FoxO1 via Sirt-Dependent Deacetylation Promotes Expression of Glucogenetic Genes. J. Biol. Chem. 2005, 280, 20589–20595. [Google Scholar] [CrossRef]
- Rodgers, J.T.; Lerin, C.; Gerhart-Hines, Z.; Puigserver, P. Metabolic Adaptations through the PGC-1α and SIRT1 Pathways. FEBS Lett. 2008, 582, 46–53. [Google Scholar] [CrossRef]
- Liu, Y.; Dentin, R.; Chen, D.; Hedrick, S.; Ravnskjaer, K.; Schenk, S.; Milne, J.; Meyers, D.J.; Cole, P.; Yates, J.; et al. A Fasting Inducible Switch Modulates Gluconeogenesis via Activator/Coactivator Exchange. Nature 2008, 456, 269–273. [Google Scholar] [CrossRef]
- Dominy, J.E.; Lee, Y.; Jedrychowski, M.P.; Chim, H.; Jurczak, M.J.; Camporez, J.P.; Ruan, H.B.; Feldman, J.; Pierce, K.; Mostoslavsky, R.; et al. The Deacetylase Sirt6 Activates the Acetyltransferase GCN5 and Suppresses Hepatic Gluconeogenesis. Mol. Cell 2012, 48, 900–913. [Google Scholar] [CrossRef]
- Zhang, P.; Tu, B.; Wang, H.; Cao, Z.; Tang, M.; Zhang, C.; Gu, B.; Li, Z.; Wang, L.; Yang, Y.; et al. Tumor Suppressor P53 Cooperates with SIRT6 to Regulate Gluconeogenesis by Promoting FoxO1 Nuclear Exclusion. Proc. Natl. Acad. Sci. USA 2014, 111, 10684–10689. [Google Scholar] [CrossRef]
- Roichman, A.; Elhanati, S.; Aon, M.A.; Abramovich, I.; Di Francesco, A.; Shahar, Y.; Avivi, M.Y.; Shurgi, M.; Rubinstein, A.; Wiesner, Y.; et al. Restoration of Energy Homeostasis by SIRT6 Extends Healthy Lifespan. Nat. Commun. 2021, 12, 3208. [Google Scholar] [CrossRef]
- Jang, H.; Lee, G.Y.; Selby, C.P.; Lee, G.; Jeon, Y.G.; Lee, J.H.; Cheng, K.K.Y.; Titchenell, P.; Birnbaum, M.J.; Xu, A.; et al. SREBP1c-CRY1 Signalling Represses Hepatic Glucose Production by Promoting FOXO1 Degradation during Refeeding. Nat. Commun. 2016, 7, 12180. [Google Scholar] [CrossRef]
- Tong, X.; Zhang, D.; Charney, N.; Jin, E.; Vandommelon, K.; Stamper, K.; Gupta, N.; Saldate, J.; Yin, L. DDB1-Mediated CRY1 Degradation Promotes FOXO1-Driven Gluconeogenesis in Liver. Diabetes 2017, 66, 2571–2582. [Google Scholar] [CrossRef]
- Jiang, L.; Xiong, J.; Zhan, J.; Yuan, F.; Tang, M.; Zhang, C.; Cao, Z.; Chen, Y.; Lu, X.; Li, Y.; et al. Ubiquitin-Specific Peptidase 7 (USP7)-Mediated Deubiquitination of the Histone Deacetylase SIRT7 Regulates Gluconeogenesis. J. Biol. Chem. 2017, 292, 13296–13311. [Google Scholar] [CrossRef]
- Hallows, W.C.; Yu, W.; Denu, J.M. Regulation of Glycolytic Enzyme Phosphoglycerate Mutase-1 by Sirt1 Protein-Mediated Deacetylation. J. Biol. Chem. 2012, 287, 3850–3858. [Google Scholar] [CrossRef]
- Aragonés, J.; Fraisl, P.; Baes, M.; Carmeliet, P. Oxygen Sensors at the Crossroad of Metabolism. Cell Metab. 2009, 9, 11–22. [Google Scholar] [CrossRef]
- Lim, J.H.; Lee, Y.M.; Chun, Y.S.; Chen, J.; Kim, J.E.; Park, J.W. Sirtuin 1 Modulates Cellular Responses to Hypoxia by Deacetylating Hypoxia-Inducible Factor 1α. Mol. Cell 2010, 38, 864–878. [Google Scholar] [CrossRef]
- Zhong, L.; D’Urso, A.; Toiber, D.; Sebastian, C.; Henry, R.E.; Vadysirisack, D.D.; Guimaraes, A.; Marinelli, B.; Wikstrom, J.D.; Nir, T.; et al. The Histone Deacetylase Sirt6 Regulates Glucose Homeostasis via Hif1α. Cell 2010, 140, 280–293. [Google Scholar] [CrossRef]
- Hubbi, M.E.; Hu, H.; Kshitiz; Gilkes, D.M.; Semenza, G.L. Sirtuin-7 Inhibits the Activity of Hypoxia-Inducible Factors. J. Biol. Chem. 2013, 288, 20768–20775. [Google Scholar] [CrossRef]
- Yan, W.; Liang, Y.; Zhang, Q.; Wang, D.; Lei, M.; Qu, J.; He, X.; Lei, Q.; Wang, Y. Arginine Methylation of SIRT 7 Couples Glucose Sensing with Mitochondria Biogenesis. EMBO Rep. 2018, 19, e46377. [Google Scholar] [CrossRef]
- Roden, M.; Shulman, G.I. The Integrative Biology of Type 2 Diabetes. Nature 2019, 576, 51–60. [Google Scholar] [CrossRef] [PubMed]
- Czech, M.P. Mechanisms of Insulin Resistance Related to White, Beige, and Brown Adipocytes. Mol. Metab. 2020, 34, 27–42. [Google Scholar] [CrossRef] [PubMed]
- Shulman, G.I. Ectopic Fat in Insulin Resistance, Dyslipidemia, and Cardiometabolic Disease. N. Engl. J. Med. 2014, 371, 1131–1141. [Google Scholar] [CrossRef] [PubMed]
- Qatanani, M.; Lazar, M.A. Mechanisms of Obesity-Associated Insulin Resistance: Many Choices on the Menu. Genes Dev. 2007, 21, 1443–1455. [Google Scholar] [CrossRef] [PubMed]
- Feige, J.N.; Lagouge, M.; Canto, C.; Strehle, A.; Houten, S.M.; Milne, J.C.; Lambert, P.D.; Mataki, C.; Elliott, P.J.; Auwerx, J. Specific SIRT1 Activation Mimics Low Energy Levels and Protects against Diet-Induced Metabolic Disorders by Enhancing Fat Oxidation. Cell Metab. 2008, 8, 347–358. [Google Scholar] [CrossRef] [PubMed]
- Milne, J.C.; Lambert, P.D.; Schenk, S.; Carney, D.P.; Smith, J.J.; Gagne, D.J.; Jin, L.; Boss, O.; Perni, R.B.; Vu, C.B.; et al. Small Molecule Activators of SIRT1 as Therapeutics for the Treatment of Type 2 Diabetes. Nature 2007, 450, 712–716. [Google Scholar] [CrossRef] [PubMed]
- Xiao, C.; Kim, H.S.; Lahusen, T.; Wang, R.H.; Xu, X.; Gavrilova, O.; Jou, W.; Gius, D.; Deng, C.X. SIRT6 Deficiency Results in Severe Hypoglycemia by Enhancing Both Basal and Insulin-Stimulated Glucose Uptake in Mice. J. Biol. Chem. 2010, 285, 36776–36784. [Google Scholar] [CrossRef]
- Sociali, G.; Magnone, M.; Ravera, S.; Damonte, P.; Vigliarolo, T.; von Holtey, M.; Vellone, V.G.; Millo, E.; Caffa, I.; Cea, M.; et al. Pharmacological Sirt6 Inhibition Improves Glucose Tolerance in a Type 2 Diabetes Mouse Model. FASEB J. 2017, 31, 3138–3149. [Google Scholar] [CrossRef]
- Cui, X.; Yao, L.; Yang, X.; Gao, Y.; Fang, F.; Zhang, J.; Wang, Q.; Chang, Y. SIRT6 Regulates Metabolic Homeostasis in Skeletal Muscle through Activation of AMPK. Am. J. Physiol. Endocrinol. Metab. 2017, 313, 493–505. [Google Scholar] [CrossRef]
- Kanfi, Y.; Peshti, V.; Gil, R.; Naiman, S.; Nahum, L.; Levin, E.; Kronfeld-Schor, N.; Cohen, H.Y. SIRT6 Protects against Pathological Damage Caused by Diet-Induced Obesity. Aging Cell 2010, 9, 162–173. [Google Scholar] [CrossRef]
- Kanfi, Y.; Naiman, S.; Amir, G.; Peshti, V.; Zinman, G.; Nahum, L.; Bar-Joseph, Z.; Cohen, H.Y. The Sirtuin SIRT6 Regulates Lifespan in Male Mice. Nature 2012, 483, 218–221. [Google Scholar] [CrossRef] [PubMed]
- BonDurant, L.D.; Potthoff, M.J. Fibroblast Growth Factor 21: A Versatile Regulator of Metabolic Homeostasis. Annu. Rev. Nutr. 2018, 38, 173–196. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Wang, G.X.; Ma, S.L.; Jung, D.Y.; Ha, H.; Altamimi, T.; Zhao, X.Y.; Guo, L.; Zhang, P.; Hu, C.R.; et al. Nrg4 Promotes Fuel Oxidation and a Healthy Adipokine Profile to Ameliorate Diet-Induced Metabolic Disorders. Mol. Metab. 2017, 6, 863–872. [Google Scholar] [CrossRef] [PubMed]
- Mizumoto, T.; Yoshizawa, T.; Sato, Y.; Ito, T.; Tsuyama, T.; Satoh, A.; Araki, S.; Tsujita, K.; Tamura, M.; Oike, Y.; et al. SIRT7 Deficiency Protects against Aging-Associated Glucose Intolerance and Extends Lifespan in Male Mice. Cells 2022, 11, 3609. [Google Scholar] [CrossRef] [PubMed]
- Moynihan, K.A.; Grimm, A.A.; Plueger, M.M.; Bernal-Mizrachi, E.; Ford, E.; Cras-Méneur, C.; Permutt, M.A.; Imai, S.I. Increased Dosage of Mammalian Sir2 in Pancreatic β Cells Enhances Glucose-Stimulated Insulin Secretion in Mice. Cell Metab. 2005, 2, 105–117. [Google Scholar] [CrossRef] [PubMed]
- Qin, K.; Zhang, N.; Zhang, Z.; Nipper, M.; Zhu, Z.; Leighton, J.; Xu, K.; Musi, N.; Wang, P. SIRT6-Mediated Transcriptional Suppression of Txnip Is Critical for Pancreatic Beta Cell Function and Survival in Mice. Diabetologia 2018, 61, 906–918. [Google Scholar] [CrossRef] [PubMed]
- Ramadori, G.; Fujikawa, T.; Fukuda, M.; Anderson, J.; Morgan, D.A.; Mostoslavsky, R.; Stuart, R.C.; Perello, M.; Vianna, C.R.; Nillni, E.A.; et al. SIRT1 Deacetylase in POMC Neurons Is Required for Homeostatic Defenses against Diet-Induced Obesity. Cell Metab. 2010, 12, 78–87. [Google Scholar] [CrossRef] [PubMed]
- Tang, Q.; Gao, Y.; Liu, Q.; Yang, X.; Wu, T.; Huang, C.; Huang, Y.; Zhang, J.; Zhang, Z.; Li, R.; et al. Sirt6 in Pro-Opiomelanocortin Neurons Controls Energy Metabolism by Modulating Leptin Signaling. Mol. Metab. 2020, 37, 100994. [Google Scholar] [CrossRef]
- Li, S.; Wu, B.; Zheng, W. Cyclic Tripeptide-Based Potent Human SIRT7 Inhibitors. Bioorg Med. Chem. Lett. 2019, 29, 461–465. [Google Scholar] [CrossRef]
- Kim, J.H.; Kim, D.; Cho, S.J.; Jung, K.Y.; Kim, J.H.; Lee, J.M.; Jung, H.J.; Kim, K.R. Identification of a Novel SIRT7 Inhibitor as Anticancer Drug Candidate. Biochem. Biophys. Res. Commun. 2019, 508, 451–457. [Google Scholar] [CrossRef]
- Zhang, C.; Li, Y.; Liu, B.; Ning, C.; Li, Y.; Wang, Y.; Li, Z. Discovery of SIRT7 Inhibitor as New Therapeutic Options Against Liver Cancer. Front. Cell Dev. Biol. 2022, 9, 813233. [Google Scholar] [CrossRef] [PubMed]

| Activity | Target Protein | Function | |
|---|---|---|---|
| Deacetylation | Gene expression ☐ ☐ ☐ ☐ ☐ | Histone H3K18 | Tumorigenesis and DNA repair [10,11] |
| Histone H3K36/K37 | Heterochromatin silencing [12] | ||
| PAF53 | Synthesis of pre-rRNA [13] | ||
| U3-55k | Processing of pre-rRNA [14] | ||
| Fibrillarin | rRNA synthesis [15] | ||
| CDK9 | RNA polymerase II transcription [16] | ||
| DNA stability ☐ | DDX21 | Genome stability [17] | |
| ATM | DNA repair [18] | ||
| Metabolism ☐ ☐ ☐ ☐ | DDB1 | Lipid metabolism [19] | |
| PPARγ2 | Lipogenesis [20] | ||
| IMP2/IGF2BP2 | Thermogenesis [21] | ||
| GABPβ1 | Mitochondrial homeostasis [22] | ||
| CRY1 | Circadian phase [23] | ||
| Cancer ☐ ☐ ☐ ☐ ☐ | WDR77 | Transmethylase activity [24] | |
| PGK1 | Glycolysis [25] | ||
| FKBP51 | Akt inactivation [26] | ||
| SMAD4 | Cancer metastasis [27] | ||
| USP39 | Cancer growth [28] | ||
| HAT1 | Tumorigenesis [29] | ||
| Cardiorenal disease ☐ | GATA4 | Regulation of cardiac hypertrophy [30] | |
| KCC4 | Regulation of ion flux [31] | ||
| Immunity | RAN | Regulation of inflammation [32] | |
| Stress response ☐ ☐ ☐ | NPM1 | Aging and p53 stability [33] | |
| FOXO3 | Regulation of apoptosis [34] | ||
| p53 | Apoptosis [35] | ||
| STRAP | p53 activity and stability [36] | ||
| Stem cell | NRF1 | Mitochondrial homeostasis [37] | |
| NFATc1 | Hair follicle initiation [38] | ||
| Desuccinylation | Histone H3K122 | Chromatin compaction [39] | |
| PRMT5 | Lipid metabolism [40] | ||
| Deglutarylation | Histone H4K91 | Chromatin structure [41] | |
| Decrotonylation | PHF5A | Aging [42] | |
| Deacylation | Osterix | Bone formation [43] | |
| Mono-ADP ribosylation | SIRT7 | Stress response [44] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yamagata, K.; Mizumoto, T.; Yoshizawa, T. The Emerging Role of SIRT7 in Glucose and Lipid Metabolism. Cells 2024, 13, 48. https://doi.org/10.3390/cells13010048
Yamagata K, Mizumoto T, Yoshizawa T. The Emerging Role of SIRT7 in Glucose and Lipid Metabolism. Cells. 2024; 13(1):48. https://doi.org/10.3390/cells13010048
Chicago/Turabian StyleYamagata, Kazuya, Tomoya Mizumoto, and Tatsuya Yoshizawa. 2024. "The Emerging Role of SIRT7 in Glucose and Lipid Metabolism" Cells 13, no. 1: 48. https://doi.org/10.3390/cells13010048
APA StyleYamagata, K., Mizumoto, T., & Yoshizawa, T. (2024). The Emerging Role of SIRT7 in Glucose and Lipid Metabolism. Cells, 13(1), 48. https://doi.org/10.3390/cells13010048






