Deciphering Acute Myeloid Leukemia Associated Transcription Factors in Human Primary CD34+ Hematopoietic Stem/Progenitor Cells
Abstract
1. Introduction: AML and Driver Fusion Genes
2. Defined Chimeric Transcription Factors Expand Human CD34+ Progenitors in Ex Vivo Cultures as Single Factors
2.1. KMT2A Rearrangements (KMT2A-r)
2.2. CBF Rearrangements
2.3. NUP98::HOXA9 Rearrangement
2.4. PML::RARA Rearrangement
3. “Two-Hit” Models for Human CD34+ Ex Vivo Progenitor Cell Expansion
4. Synopsis
Author Contributions
Funding
Conflicts of Interest
References
- Enciso, J.; Mendoza, L.; Pelayo, R. Normal vs. Malignant hematopoiesis: The complexity of acute leukemia through systems biology. Front. Genet. 2015, 6, 290. [Google Scholar] [CrossRef] [PubMed]
- Shallis, R.M.; Wang, R.; Davidoff, A.; Ma, X.; Zeidan, A.M. Epidemiology of acute myeloid leukemia: Recent progress and enduring challenges. Blood Rev. 2019, 36, 70–87. [Google Scholar] [CrossRef] [PubMed]
- Hou, H.A.; Tien, H.F. Genomic landscape in acute myeloid leukemia and its implications in risk classification and targeted therapies. J. Biomed. Sci. 2020, 27, 81. [Google Scholar] [CrossRef] [PubMed]
- Kelly, L.M.; Gilliland, D.G. Genetics of myeloid leukemias. Annu. Rev. Genom. Hum. Genet. 2002, 3, 179–198. [Google Scholar] [CrossRef]
- Chen, S.J.; Shen, Y.; Chen, Z. A panoramic view of acute myeloid leukemia. Nat. Genet. 2013, 45, 586–587. [Google Scholar] [CrossRef] [PubMed]
- Padmakumar, D.; Chandraprabha, V.R.; Gopinath, P.; Vimala Devi, A.R.T.; Anitha, G.R.J.; Sreelatha, M.M.; Padmakumar, A.; Sreedharan, H. A concise review on the molecular genetics of acute myeloid leukemia. Leuk. Res. 2021, 111, 106727. [Google Scholar] [CrossRef] [PubMed]
- Cancer Genome Atlas Research Network; Ley, T.J.; Miller, C.; Ding, L.; Raphael, B.J.; Mungall, A.J.; Robertson, A.; Hoadley, K.; Triche, T.J., Jr.; Laird, P.W.; et al. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N. Engl. J. Med. 2013, 368, 2059–2074. [Google Scholar] [CrossRef]
- Rowley, J.D. Letter: A new consistent chromosomal abnormality in chronic myelogenous leukaemia identified by quinacrine fluorescence and Giemsa staining. Nature 1973, 243, 290–293. [Google Scholar] [CrossRef]
- Khoury, J.D.; Solary, E.; Abla, O.; Akkari, Y.; Alaggio, R.; Apperley, J.F.; Bejar, R.; Berti, E.; Busque, L.; Chan, J.K.C.; et al. The 5th edition of the World Health Organization Classification of Haematolymphoid Tumours: Myeloid and Histiocytic/Dendritic Neoplasms. Leukemia 2022, 36, 1703–1719. [Google Scholar] [CrossRef]
- Arber, D.A.; Orazi, A.; Hasserjian, R.P.; Borowitz, M.J.; Calvo, K.R.; Kvasnicka, H.M.; Wang, S.A.; Bagg, A.; Barbui, T.; Branford, S.; et al. International Consensus Classification of Myeloid Neoplasms and Acute Leukemias: Integrating morphologic, clinical, and genomic data. Blood 2022, 140, 1200–1228. [Google Scholar] [CrossRef]
- Huber, S.; Baer, C.; Hutter, S.; Dicker, F.; Meggendorfer, M.; Pohlkamp, C.; Kern, W.; Haferlach, T.; Haferlach, C.; Hoermann, G. AML classification in the year 2023: How to avoid a Babylonian confusion of languages. Leukemia 2023, 37, 1413–1420. [Google Scholar] [CrossRef] [PubMed]
- Mulloy, J.C.; Cammenga, J.; Berguido, F.J.; Wu, K.; Zhou, P.; Comenzo, R.L.; Jhanwar, S.; Moore, M.A.; Nimer, S.D. Maintaining the self-renewal and differentiation potential of human CD34+ hematopoietic cells using a single genetic element. Blood 2003, 102, 4369–4376. [Google Scholar] [CrossRef] [PubMed]
- Mulloy, J.C.; Cammenga, J.; MacKenzie, K.L.; Berguido, F.J.; Moore, M.A.; Nimer, S.D. The AML1-ETO fusion protein promotes the expansion of human hematopoietic stem cells. Blood 2002, 99, 15–23. [Google Scholar] [CrossRef] [PubMed]
- Wunderlich, M.; Krejci, O.; Wei, J.; Mulloy, J.C. Human CD34+ cells expressing the inv(16) fusion protein exhibit a myelomonocytic phenotype with greatly enhanced proliferative ability. Blood 2006, 108, 1690–1697. [Google Scholar] [CrossRef] [PubMed]
- Montes, R.; Ayllon, V.; Gutierrez-Aranda, I.; Prat, I.; Hernandez-Lamas, M.C.; Ponce, L.; Bresolin, S.; Te Kronnie, G.; Greaves, M.; Bueno, C.; et al. Enforced expression of MLL-AF4 fusion in cord blood CD34+ cells enhances the hematopoietic repopulating cell function and clonogenic potential but is not sufficient to initiate leukemia. Blood 2011, 117, 4746–4758. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Rizo, A.; Horton, S.J.; Olthof, S.; Dontje, B.; Ausema, A.; van Os, R.; van den Boom, V.; Vellenga, E.; de Haan, G.; Schuringa, J.J. BMI1 collaborates with BCR-ABL in leukemic transformation of human CD34+ cells. Blood 2010, 116, 4621–4630. [Google Scholar] [CrossRef]
- Wei, J.; Wunderlich, M.; Fox, C.; Alvarez, S.; Cigudosa, J.C.; Wilhelm, J.S.; Zheng, Y.; Cancelas, J.A.; Gu, Y.; Jansen, M.; et al. Microenvironment determines lineage fate in a human model of MLL-AF9 leukemia. Cancer Cell 2008, 13, 483–495. [Google Scholar] [CrossRef]
- Chung, K.Y.; Morrone, G.; Schuringa, J.J.; Plasilova, M.; Shieh, J.H.; Zhang, Y.; Zhou, P.; Moore, M.A. Enforced expression of NUP98-HOXA9 in human CD34(+) cells enhances stem cell proliferation. Cancer Res. 2006, 66, 11781–11791. [Google Scholar] [CrossRef]
- Gerritsen, M.; Yi, G.; Tijchon, E.; Kuster, J.; Schuringa, J.J.; Martens, J.H.A.; Vellenga, E. RUNX1 mutations enhance self-renewal and block granulocytic differentiation in human in vitro models and primary AMLs. Blood Adv. 2019, 3, 320–332. [Google Scholar] [CrossRef]
- Barabe, F.; Kennedy, J.A.; Hope, K.J.; Dick, J.E. Modeling the initiation and progression of human acute leukemia in mice. Science 2007, 316, 600–604. [Google Scholar] [CrossRef]
- Hess, J.L. MLL: A histone methyltransferase disrupted in leukemia. Trends Mol. Med. 2004, 10, 500–507. [Google Scholar] [CrossRef] [PubMed]
- Daser, A.; Rabbitts, T.H. The versatile mixed lineage leukaemia gene MLL and its many associations in leukaemogenesis. Semin. Cancer Biol. 2005, 15, 175–188. [Google Scholar] [CrossRef] [PubMed]
- Creutzig, U.; van den Heuvel-Eibrink, M.M.; Gibson, B.; Dworzak, M.N.; Adachi, S.; de Bont, E.; Harbott, J.; Hasle, H.; Johnston, D.; Kinoshita, A.; et al. Diagnosis and management of acute myeloid leukemia in children and adolescents: Recommendations from an international expert panel. Blood 2012, 120, 3187–3205. [Google Scholar] [CrossRef] [PubMed]
- Yu, B.D.; Hess, J.L.; Horning, S.E.; Brown, G.A.; Korsmeyer, S.J. Altered Hox expression and segmental identity in Mll-mutant mice. Nature 1995, 378, 505–508. [Google Scholar] [CrossRef] [PubMed]
- Yu, B.D.; Hanson, R.D.; Hess, J.L.; Horning, S.E.; Korsmeyer, S.J. MLL, a mammalian trithorax-group gene, functions as a transcriptional maintenance factor in morphogenesis. Proc. Natl. Acad. Sci. USA 1998, 95, 10632–10636. [Google Scholar] [CrossRef] [PubMed]
- Ernst, P.; Mabon, M.; Davidson, A.J.; Zon, L.I.; Korsmeyer, S.J. An Mll-dependent Hox program drives hematopoietic progenitor expansion. Curr. Biol. 2004, 14, 2063–2069. [Google Scholar] [CrossRef] [PubMed]
- Artinger, E.L.; Mishra, B.P.; Zaffuto, K.M.; Li, B.E.; Chung, E.K.; Moore, A.W.; Chen, Y.; Cheng, C.; Ernst, P. An MLL-dependent network sustains hematopoiesis. Proc. Natl. Acad. Sci. USA 2013, 110, 12000–12005. [Google Scholar] [CrossRef]
- Nakamura, T.; Mori, T.; Tada, S.; Krajewski, W.; Rozovskaia, T.; Wassell, R.; Dubois, G.; Mazo, A.; Croce, C.M.; Canaani, E. ALL-1 is a histone methyltransferase that assembles a supercomplex of proteins involved in transcriptional regulation. Mol. Cell 2002, 10, 1119–1128. [Google Scholar] [CrossRef]
- Milne, T.A.; Briggs, S.D.; Brock, H.W.; Martin, M.E.; Gibbs, D.; Allis, C.D.; Hess, J.L. MLL targets SET domain methyltransferase activity to Hox gene promoters. Mol. Cell 2002, 10, 1107–1117. [Google Scholar] [CrossRef]
- Meyer, C.; Larghero, P.; Almeida Lopes, B.; Burmeister, T.; Groger, D.; Sutton, R.; Venn, N.C.; Cazzaniga, G.; Corral Abascal, L.; Tsaur, G.; et al. The KMT2A recombinome of acute leukemias in 2023. Leukemia 2023, 37, 988–1005. [Google Scholar] [CrossRef]
- Corral, J.; Lavenir, I.; Impey, H.; Warren, A.J.; Forster, A.; Larson, T.A.; Bell, S.; McKenzie, A.N.; King, G.; Rabbitts, T.H. An Mll-AF9 fusion gene made by homologous recombination causes acute leukemia in chimeric mice: A method to create fusion oncogenes. Cell 1996, 85, 853–861. [Google Scholar] [CrossRef] [PubMed]
- Kohnken, R.; Porcu, P.; Mishra, A. Overview of the Use of Murine Models in Leukemia and Lymphoma Research. Front. Oncol. 2017, 7, 22. [Google Scholar] [CrossRef] [PubMed]
- Doulatov, S.; Notta, F.; Laurenti, E.; Dick, J.E. Hematopoiesis: A human perspective. Cell Stem Cell 2012, 10, 120–136. [Google Scholar] [CrossRef] [PubMed]
- Sitnicka, E.; Buza-Vidas, N.; Larsson, S.; Nygren, J.M.; Liuba, K.; Jacobsen, S.E. Human CD34+ hematopoietic stem cells capable of multilineage engrafting NOD/SCID mice express flt3: Distinct flt3 and c-kit expression and response patterns on mouse and candidate human hematopoietic stem cells. Blood 2003, 102, 881–886. [Google Scholar] [CrossRef] [PubMed]
- Mestas, J.; Hughes, C.C. Of mice and not men: Differences between mouse and human immunology. J. Immunol. 2004, 172, 2731–2738. [Google Scholar] [CrossRef] [PubMed]
- Bitoun, E.; Oliver, P.L.; Davies, K.E. The mixed-lineage leukemia fusion partner AF4 stimulates RNA polymerase II transcriptional elongation and mediates coordinated chromatin remodeling. Hum. Mol. Genet. 2007, 16, 92–106. [Google Scholar] [CrossRef] [PubMed]
- Yokoyama, A.; Lin, M.; Naresh, A.; Kitabayashi, I.; Cleary, M.L. A higher-order complex containing AF4 and ENL family proteins with P-TEFb facilitates oncogenic and physiologic MLL-dependent transcription. Cancer Cell 2010, 17, 198–212. [Google Scholar] [CrossRef]
- Peterlin, B.M.; Price, D.H. Controlling the elongation phase of transcription with P-TEFb. Mol. Cell 2006, 23, 297–305. [Google Scholar] [CrossRef]
- Carroll, D. Genome engineering with targetable nucleases. Annu. Rev. Biochem. 2014, 83, 409–439. [Google Scholar] [CrossRef]
- Ferrari, S.; Vavassori, V.; Canarutto, D.; Jacob, A.; Castiello, M.C.; Javed, A.O.; Genovese, P. Gene Editing of Hematopoietic Stem Cells: Hopes and Hurdles toward Clinical Translation. Front. Genome Ed. 2021, 3, 618378. [Google Scholar] [CrossRef]
- Barrangou, R.; Doudna, J.A. Applications of CRISPR technologies in research and beyond. Nat. Biotechnol. 2016, 34, 933–941. [Google Scholar] [CrossRef] [PubMed]
- Rice, S.; Jackson, T.; Crump, N.T.; Fordham, N.; Elliott, N.; O’Byrne, S.; Fanego, M.; Addy, D.; Crabb, T.; Dryden, C.; et al. A human fetal liver-derived infant MLL-AF4 acute lymphoblastic leukemia model reveals a distinct fetal gene expression program. Nat. Commun. 2021, 12, 6905. [Google Scholar] [CrossRef] [PubMed]
- Marschalek, R. The reciprocal world of MLL fusions: A personal view. Biochim. Biophys. Acta Gene Regul. Mech. 2020, 1863, 194547. [Google Scholar] [CrossRef] [PubMed]
- Wilhelm, A.; Marschalek, R. The role of reciprocal fusions in MLL-r acute leukemia: Studying the chromosomal translocation t(4;11). Oncogene 2021, 40, 6093–6102. [Google Scholar] [CrossRef] [PubMed]
- Collins, E.C.; Appert, A.; Ariza-McNaughton, L.; Pannell, R.; Yamada, Y.; Rabbitts, T.H. Mouse Af9 is a controller of embryo patterning, like Mll, whose human homologue fuses with Af9 after chromosomal translocation in leukemia. Mol. Cell. Biol. 2002, 22, 7313–7324. [Google Scholar] [CrossRef] [PubMed]
- Calvanese, V.; Nguyen, A.T.; Bolan, T.J.; Vavilina, A.; Su, T.; Lee, L.K.; Wang, Y.; Lay, F.D.; Magnusson, M.; Crooks, G.M.; et al. MLLT3 governs human haematopoietic stem-cell self-renewal and engraftment. Nature 2019, 576, 281–286. [Google Scholar] [CrossRef]
- Schneidawind, C.; Jeong, J.; Schneidawind, D.; Kim, I.S.; Duque-Afonso, J.; Wong, S.H.K.; Iwasaki, M.; Breese, E.H.; Zehnder, J.L.; Porteus, M.; et al. MLL leukemia induction by t(9;11) chromosomal translocation in human hematopoietic stem cells using genome editing. Blood Adv. 2018, 2, 832–845. [Google Scholar] [CrossRef]
- Zhou, J.; Ng, Y.; Chng, W.J. ENL: Structure, function, and roles in hematopoiesis and acute myeloid leukemia. Cell. Mol. Life Sci. 2018, 75, 3931–3941. [Google Scholar] [CrossRef]
- Mueller, D.; Bach, C.; Zeisig, D.; Garcia-Cuellar, M.P.; Monroe, S.; Sreekumar, A.; Zhou, R.; Nesvizhskii, A.; Chinnaiyan, A.; Hess, J.L.; et al. A role for the MLL fusion partner ENL in transcriptional elongation and chromatin modification. Blood 2007, 110, 4445–4454. [Google Scholar] [CrossRef]
- Chen, J.; Liao, S.; Xiao, Z.; Pan, Q.; Wang, X.; Shen, K.; Wang, S.; Yang, L.; Guo, F.; Liu, H.F.; et al. The development and improvement of immunodeficient mice and humanized immune system mouse models. Front. Immunol. 2022, 13, 1007579. [Google Scholar] [CrossRef]
- Rossi, J.G.; Bernasconi, A.R.; Alonso, C.N.; Rubio, P.L.; Gallego, M.S.; Carrara, C.A.; Guitter, M.R.; Eberle, S.E.; Cocce, M.; Zubizarreta, P.A.; et al. Lineage switch in childhood acute leukemia: An unusual event with poor outcome. Am. J. Hematol. 2012, 87, 890–897. [Google Scholar] [CrossRef] [PubMed]
- Windisch, R.; Kreissig, S.; Wichmann, C. Defined Human Leukemic CD34+ Liquid Cultures to Study HDAC/Transcriptional Repressor Complexes. Methods Mol. Biol. 2023, 2589, 27–49. [Google Scholar] [CrossRef] [PubMed]
- Buechele, C.; Breese, E.H.; Schneidawind, D.; Lin, C.H.; Jeong, J.; Duque-Afonso, J.; Wong, S.H.; Smith, K.S.; Negrin, R.S.; Porteus, M.; et al. MLL leukemia induction by genome editing of human CD34+ hematopoietic cells. Blood 2015, 126, 1683–1694. [Google Scholar] [CrossRef] [PubMed]
- Reimer, J.; Knoss, S.; Labuhn, M.; Charpentier, E.M.; Gohring, G.; Schlegelberger, B.; Klusmann, J.H.; Heckl, D. CRISPR-Cas9-induced t(11;19)/MLL-ENL translocations initiate leukemia in human hematopoietic progenitor cells in vivo. Haematologica 2017, 102, 1558–1566. [Google Scholar] [CrossRef] [PubMed]
- de Bruijn, M.F.; Speck, N.A. Core-binding factors in hematopoiesis and immune function. Oncogene 2004, 23, 4238–4248. [Google Scholar] [CrossRef] [PubMed]
- Miyoshi, H.; Shimizu, K.; Kozu, T.; Maseki, N.; Kaneko, Y.; Ohki, M. t(8;21) breakpoints on chromosome 21 in acute myeloid leukemia are clustered within a limited region of a single gene, AML1. Proc. Natl. Acad. Sci. USA 1991, 88, 10431–10434. [Google Scholar] [CrossRef] [PubMed]
- Lam, K.; Zhang, D.E. RUNX1 and RUNX1-ETO: Roles in hematopoiesis and leukemogenesis. Front. Biosci. 2012, 17, 1120–1139. [Google Scholar] [CrossRef]
- Lin, S.; Mulloy, J.C.; Goyama, S. RUNX1-ETO Leukemia. Adv. Exp. Med. Biol. 2017, 962, 151–173. [Google Scholar] [CrossRef]
- Wang, J.; Hoshino, T.; Redner, R.L.; Kajigaya, S.; Liu, J.M. ETO, fusion partner in t(8;21) acute myeloid leukemia, represses transcription by interaction with the human N-CoR/mSin3/HDAC1 complex. Proc. Natl. Acad. Sci. USA 1998, 95, 10860–10865. [Google Scholar] [CrossRef]
- Wang, L.; Gural, A.; Sun, X.J.; Zhao, X.; Perna, F.; Huang, G.; Hatlen, M.A.; Vu, L.; Liu, F.; Xu, H.; et al. The leukemogenicity of AML1-ETO is dependent on site-specific lysine acetylation. Science 2011, 333, 765–769. [Google Scholar] [CrossRef]
- Chen, G.; Liu, A.; Xu, Y.; Gao, L.; Jiang, M.; Li, Y.; Lv, N.; Zhou, L.; Wang, L.; Yu, L.; et al. The RUNX1-ETO fusion protein trans-activates c-KIT expression by recruiting histone acetyltransferase P300 on its promoter. FEBS J. 2019, 286, 901–912. [Google Scholar] [CrossRef] [PubMed]
- Chen-Wichmann, L.; Shvartsman, M.; Preiss, C.; Hockings, C.; Windisch, R.; Redondo Monte, E.; Leubolt, G.; Spiekermann, K.; Lausen, J.; Brendel, C.; et al. Compatibility of RUNX1/ETO fusion protein modules driving CD34+ human progenitor cell expansion. Oncogene 2019, 38, 261–272. [Google Scholar] [CrossRef] [PubMed]
- Yan, M.; Kanbe, E.; Peterson, L.F.; Boyapati, A.; Miao, Y.; Wang, Y.; Chen, I.M.; Chen, Z.; Rowley, J.D.; Willman, C.L.; et al. A previously unidentified alternatively spliced isoform of t(8;21) transcript promotes leukemogenesis. Nat. Med. 2006, 12, 945–949. [Google Scholar] [CrossRef] [PubMed]
- Link, K.A.; Lin, S.; Shrestha, M.; Bowman, M.; Wunderlich, M.; Bloomfield, C.D.; Huang, G.; Mulloy, J.C. Supraphysiologic levels of the AML1-ETO isoform AE9a are essential for transformation. Proc. Natl. Acad. Sci. USA 2016, 113, 9075–9080. [Google Scholar] [CrossRef] [PubMed]
- DeKelver, R.C.; Yan, M.; Ahn, E.Y.; Shia, W.J.; Speck, N.A.; Zhang, D.E. Attenuation of AML1-ETO cellular dysregulation correlates with increased leukemogenic potential. Blood 2013, 121, 3714–3717. [Google Scholar] [CrossRef] [PubMed]
- Castilla, L.H.; Wijmenga, C.; Wang, Q.; Stacy, T.; Speck, N.A.; Eckhaus, M.; Marin-Padilla, M.; Collins, F.S.; Wynshaw-Boris, A.; Liu, P.P. Failure of embryonic hematopoiesis and lethal hemorrhages in mouse embryos heterozygous for a knocked-in leukemia gene CBFB-MYH11. Cell 1996, 87, 687–696. [Google Scholar] [CrossRef] [PubMed]
- Gough, S.M.; Slape, C.I.; Aplan, P.D. NUP98 gene fusions and hematopoietic malignancies: Common themes and new biologic insights. Blood 2011, 118, 6247–6257. [Google Scholar] [CrossRef]
- Ghannam, G.; Takeda, A.; Camarata, T.; Moore, M.A.; Viale, A.; Yaseen, N.R. The oncogene Nup98-HOXA9 induces gene transcription in myeloid cells. J. Biol. Chem. 2004, 279, 866–875. [Google Scholar] [CrossRef]
- Takeda, A.; Goolsby, C.; Yaseen, N.R. NUP98-HOXA9 induces long-term proliferation and blocks differentiation of primary human CD34+ hematopoietic cells. Cancer Res. 2006, 66, 6628–6637. [Google Scholar] [CrossRef]
- Abraham, A.; Kim, Y.S.; Zhao, H.; Humphries, K.; Persons, D.A. Increased Engraftment of Human Short Term Repopulating Hematopoietic Cells in NOD/SCID/IL2rgammanull Mice by Lentiviral Expression of NUP98-HOXA10HD. PLoS ONE 2016, 11, e0147059. [Google Scholar] [CrossRef]
- Zhang, X.; Sun, J.; Yu, W.; Jin, J. Current views on the genetic landscape and management of variant acute promyelocytic leukemia. Biomark. Res. 2021, 9, 33. [Google Scholar] [CrossRef] [PubMed]
- Duong, V.; Rochette-Egly, C. The molecular physiology of nuclear retinoic acid receptors. From health to disease. Biochim. Biophys. Acta Mol. Basis Dis. 2011, 1812, 1023–1031. [Google Scholar] [CrossRef] [PubMed]
- Lavudi, K.; Nuguri, S.M.; Olverson, Z.; Dhanabalan, A.K.; Patnaik, S.; Kokkanti, R.R. Targeting the retinoic acid signaling pathway as a modern precision therapy against cancers. Front. Cell Dev. Biol. 2023, 11, 1254612. [Google Scholar] [CrossRef] [PubMed]
- Altucci, L.; Leibowitz, M.D.; Ogilvie, K.M.; de Lera, A.R.; Gronemeyer, H. RAR and RXR modulation in cancer and metabolic disease. Nat. Rev. Drug Discov. 2007, 6, 793–810. [Google Scholar] [CrossRef] [PubMed]
- Guan, D.; Kao, H.Y. The function, regulation and therapeutic implications of the tumor suppressor protein, PML. Cell Biosci. 2015, 5, 60. [Google Scholar] [CrossRef]
- Noguera, N.I.; Catalano, G.; Banella, C.; Divona, M.; Faraoni, I.; Ottone, T.; Arcese, W.; Voso, M.T. Acute Promyelocytic Leukemia: Update on the Mechanisms of Leukemogenesis, Resistance and on Innovative Treatment Strategies. Cancers 2019, 11, 1591. [Google Scholar] [CrossRef]
- Bain, B.J.; Bene, M.C. Morphological and Immunophenotypic Clues to the WHO Categories of Acute Myeloid Leukaemia. Acta Haematol. 2019, 141, 232–244. [Google Scholar] [CrossRef]
- Turhan, A.G.; Lemoine, F.M.; Debert, C.; Bonnet, M.L.; Baillou, C.; Picard, F.; Macintyre, E.A.; Varet, B. Highly purified primitive hematopoietic stem cells are PML-RARA negative and generate nonclonal progenitors in acute promyelocytic leukemia. Blood 1995, 85, 2154–2161. [Google Scholar] [CrossRef]
- Grignani, F.; Valtieri, M.; Gabbianelli, M.; Gelmetti, V.; Botta, R.; Luchetti, L.; Masella, B.; Morsilli, O.; Pelosi, E.; Samoggia, P.; et al. PML/RAR alpha fusion protein expression in normal human hematopoietic progenitors dictates myeloid commitment and the promyelocytic phenotype. Blood 2000, 96, 1531–1537. [Google Scholar] [CrossRef]
- Abdul-Nabi, A.M.; Yassin, E.R.; Varghese, N.; Deshmukh, H.; Yaseen, N.R. In vitro transformation of primary human CD34+ cells by AML fusion oncogenes: Early gene expression profiling reveals possible drug target in AML. PLoS ONE 2010, 5, e12464. [Google Scholar] [CrossRef]
- Du, C.; Redner, R.L.; Cooke, M.P.; Lavau, C. Overexpression of wild-type retinoic acid receptor alpha (RARalpha) recapitulates retinoic acid-sensitive transformation of primary myeloid progenitors by acute promyelocytic leukemia RARalpha-fusion genes. Blood 1999, 94, 793–802. [Google Scholar] [CrossRef] [PubMed]
- Ito, M.; Kobayashi, K.; Nakahata, T. NOD/Shi-scid IL2rgamma(null) (NOG) mice more appropriate for humanized mouse models. Curr. Top Microbiol. Immunol. 2008, 324, 53–76. [Google Scholar] [CrossRef] [PubMed]
- Matsushita, H.; Yahata, T.; Sheng, Y.; Nakamura, Y.; Muguruma, Y.; Matsuzawa, H.; Tanaka, M.; Hayashi, H.; Sato, T.; Damdinsuren, A.; et al. Establishment of a humanized APL model via the transplantation of PML-RARA-transduced human common myeloid progenitors into immunodeficient mice. PLoS ONE 2014, 9, e111082. [Google Scholar] [CrossRef]
- Lessard, J.; Sauvageau, G. Bmi-1 determines the proliferative capacity of normal and leukaemic stem cells. Nature 2003, 423, 255–260. [Google Scholar] [CrossRef] [PubMed]
- Hidalgo, I.; Wahlestedt, M.; Yuan, O.; Zhang, Q.; Bryder, D.; Pronk, C.J. Bmi1 induction protects hematopoietic stem cells against pronounced long-term hematopoietic stress. Exp. Hematol. 2022, 109, 35–44. [Google Scholar] [CrossRef] [PubMed]
- Sahasrabuddhe, A.A. BMI1: A Biomarker of Hematologic Malignancies. Biomark. Cancer 2016, 8, 65–75. [Google Scholar] [CrossRef]
- Rizo, A.; Dontje, B.; Vellenga, E.; de Haan, G.; Schuringa, J.J. Long-term maintenance of human hematopoietic stem/progenitor cells by expression of BMI1. Blood 2008, 111, 2621–2630. [Google Scholar] [CrossRef]
- Edling, C.E.; Hallberg, B. c-Kit—A hematopoietic cell essential receptor tyrosine kinase. Int. J. Biochem. Cell Biol. 2007, 39, 1995–1998. [Google Scholar] [CrossRef]
- Malaise, M.; Steinbach, D.; Corbacioglu, S. Clinical implications of c-Kit mutations in acute myelogenous leukemia. Curr. Hematol. Malig. Rep. 2009, 4, 77–82. [Google Scholar] [CrossRef]
- Wichmann, C.; Quagliano-Lo Coco, I.; Yildiz, O.; Chen-Wichmann, L.; Weber, H.; Syzonenko, T.; Doring, C.; Brendel, C.; Ponnusamy, K.; Kinner, A.; et al. Activating c-KIT mutations confer oncogenic cooperativity and rescue RUNX1/ETO-induced DNA damage and apoptosis in human primary CD34+ hematopoietic progenitors. Leukemia 2015, 29, 279–289. [Google Scholar] [CrossRef]
- Chin, P.S.; Assi, S.A.; Ptasinska, A.; Imperato, M.R.; Cockerill, P.N.; Bonifer, C. RUNX1/ETO and mutant KIT both contribute to programming the transcriptional and chromatin landscape in t(8;21) acute myeloid leukemia. Exp. Hematol. 2020, 92, 62–74. [Google Scholar] [CrossRef] [PubMed]
- Higuchi, M.; O’Brien, D.; Kumaravelu, P.; Lenny, N.; Yeoh, E.J.; Downing, J.R. Expression of a conditional AML1-ETO oncogene bypasses embryonic lethality and establishes a murine model of human t(8;21) acute myeloid leukemia. Cancer Cell 2002, 1, 63–74. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Y.; Zhou, L.; Miyamoto, T.; Iwasaki, H.; Harakawa, N.; Hetherington, C.J.; Burel, S.A.; Lagasse, E.; Weissman, I.L.; Akashi, K.; et al. AML1-ETO expression is directly involved in the development of acute myeloid leukemia in the presence of additional mutations. Proc. Natl. Acad. Sci. USA 2001, 98, 10398–10403. [Google Scholar] [CrossRef] [PubMed]
- Rhoades, K.L.; Hetherington, C.J.; Harakawa, N.; Yergeau, D.A.; Zhou, L.; Liu, L.Q.; Little, M.T.; Tenen, D.G.; Zhang, D.E. Analysis of the role of AML1-ETO in leukemogenesis, using an inducible transgenic mouse model. Blood 2000, 96, 2108–2115. [Google Scholar] [CrossRef] [PubMed]
- Nick, H.J.; Kim, H.G.; Chang, C.W.; Harris, K.W.; Reddy, V.; Klug, C.A. Distinct classes of c-Kit-activating mutations differ in their ability to promote RUNX1-ETO-associated acute myeloid leukemia. Blood 2012, 119, 1522–1531. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.Y.; Zhao, L.J.; Wu, C.F.; Liu, P.; Shi, L.; Liang, Y.; Xiong, S.M.; Mi, J.Q.; Chen, Z.; Ren, R.; et al. C-KIT mutation cooperates with full-length AML1-ETO to induce acute myeloid leukemia in mice. Proc. Natl. Acad. Sci. USA 2011, 108, 2450–2455. [Google Scholar] [CrossRef]
- Goyama, S.; Schibler, J.; Gasilina, A.; Shrestha, M.; Lin, S.; Link, K.A.; Chen, J.; Whitman, S.P.; Bloomfield, C.D.; Nicolet, D.; et al. UBASH3B/Sts-1-CBL axis regulates myeloid proliferation in human preleukemia induced by AML1-ETO. Leukemia 2016, 30, 728–739. [Google Scholar] [CrossRef]
- Basecke, J.; Schwieger, M.; Griesinger, F.; Schiedlmeier, B.; Wulf, G.; Trumper, L.; Stocking, C. AML1/ETO promotes the maintenance of early hematopoietic progenitors in NOD/SCID mice but does not abrogate their lineage specific differentiation. Leuk. Lymphoma 2005, 46, 265–272. [Google Scholar] [CrossRef]
- Chou, F.S.; Wunderlich, M.; Griesinger, A.; Mulloy, J.C. N-Ras(G12D) induces features of stepwise transformation in preleukemic human umbilical cord blood cultures expressing the AML1-ETO fusion gene. Blood 2011, 117, 2237–2240. [Google Scholar] [CrossRef][Green Version]
- Pinto, C.; Estrada, M.F.; Brito, C. In Vitro and Ex Vivo Models—The Tumor Microenvironment in a Flask. Adv. Exp. Med. Biol. 2020, 1219, 431–443. [Google Scholar] [CrossRef]
- Jagannathan, L.; Cuddapah, S.; Costa, M. Oxidative stress under ambient and physiological oxygen tension in tissue culture. Curr. Pharmacol. Rep. 2016, 2, 64–72. [Google Scholar] [CrossRef] [PubMed]
- Teague, R.M.; Kline, J. Immune evasion in acute myeloid leukemia: Current concepts and future directions. J. Immunother. Cancer 2013, 1, 1. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Lamble, A.J.; Lind, E.F. Targeting the Immune Microenvironment in Acute Myeloid Leukemia: A Focus on T Cell Immunity. Front. Oncol. 2018, 8, 213. [Google Scholar] [CrossRef] [PubMed]
- Austin, R.; Smyth, M.J.; Lane, S.W. Harnessing the immune system in acute myeloid leukaemia. Crit. Rev. Oncol. Hematol. 2016, 103, 62–77. [Google Scholar] [CrossRef]
- Fischer, M. Mice Are Not Humans: The Case of p53. Trends Cancer 2021, 7, 12–14. [Google Scholar] [CrossRef]
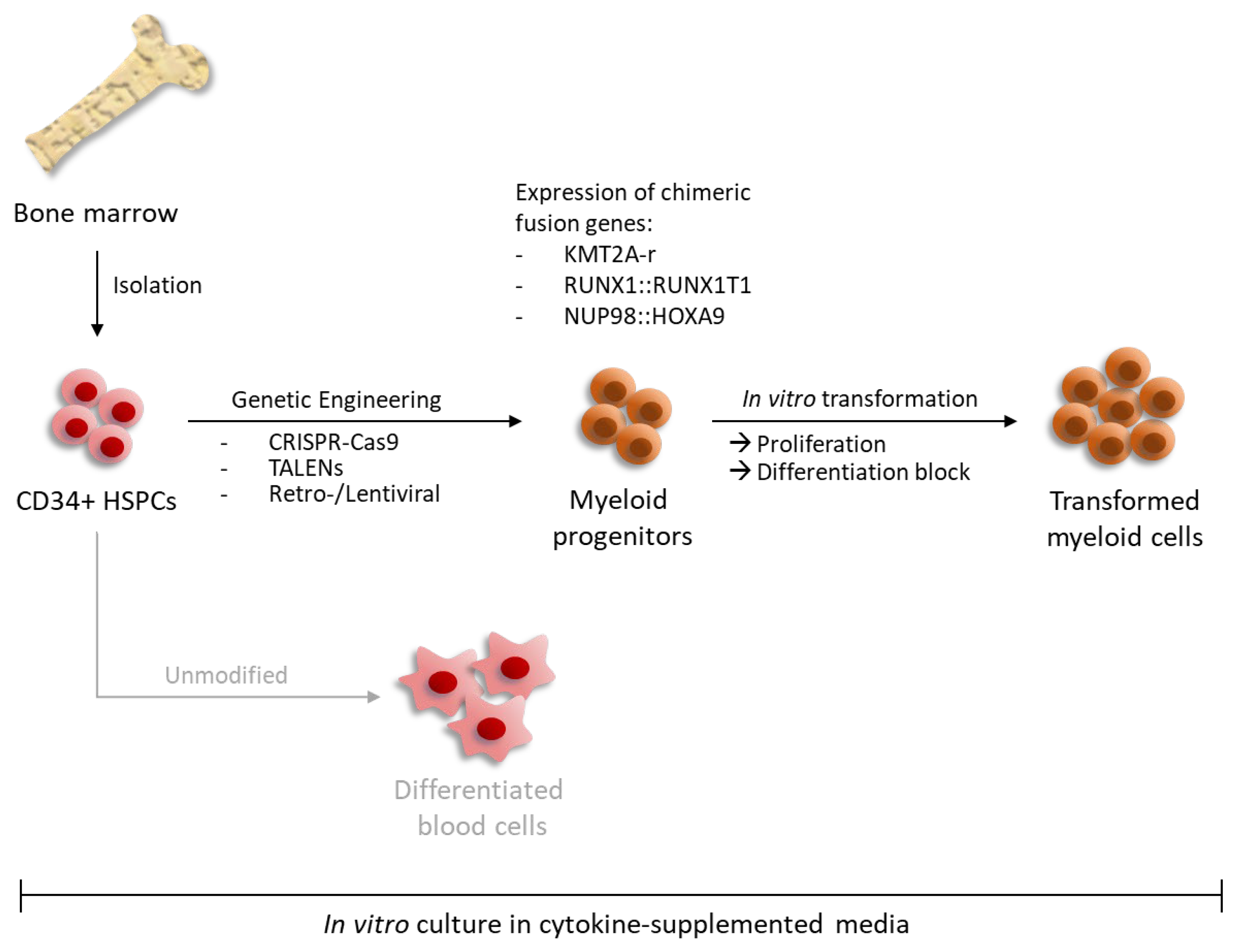

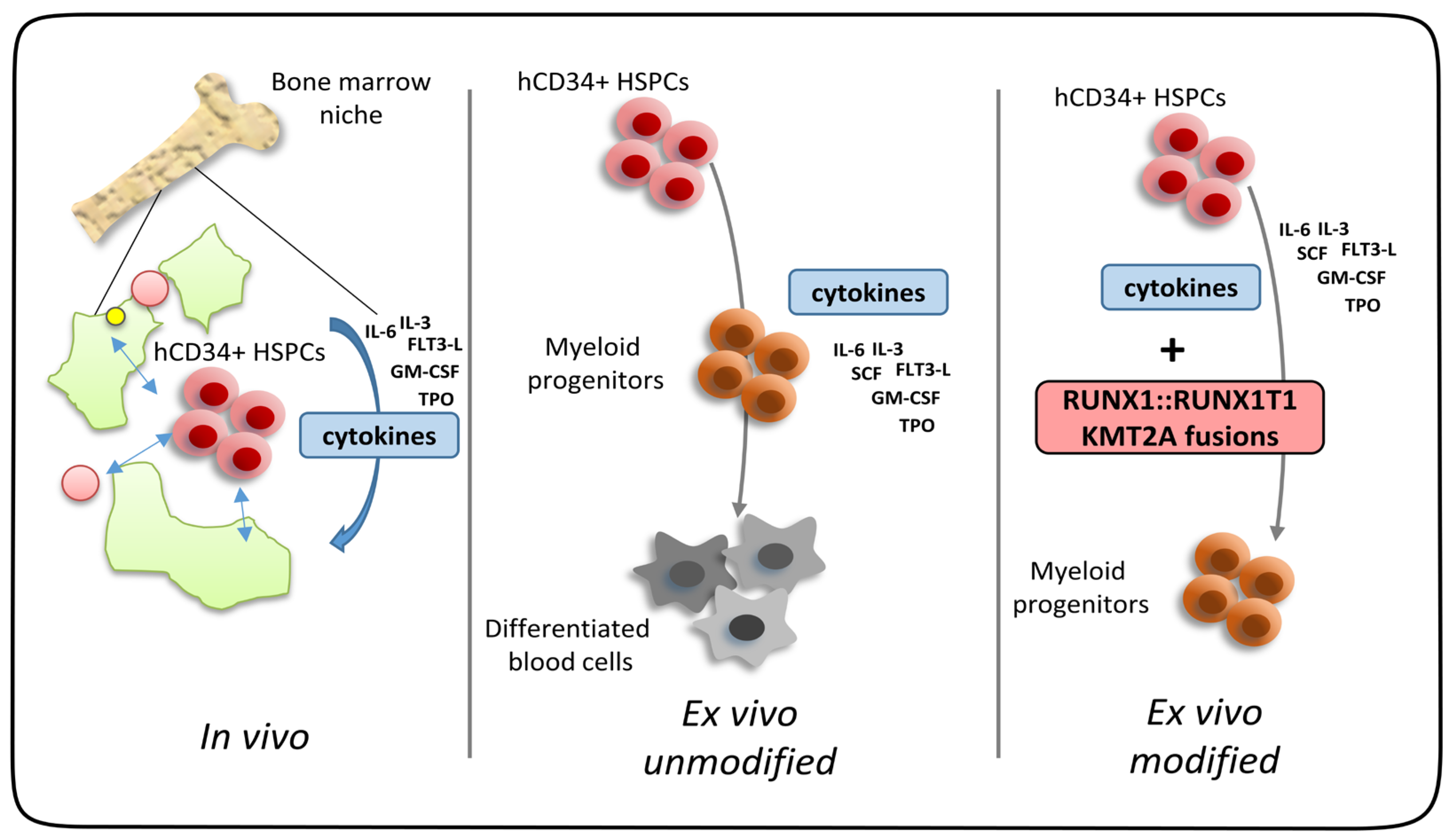
| Genetic Abnormality | Resulting Fusion Gene/Rearrangement |
|---|---|
| t(8;21) | RUNX1::RUNX1T1 |
| t(16;16) or inv(16) | CBFB::MYH11 |
| t(6;9) | DEK::NUP214 |
| t(3;3) or inv(3) | RPN1::EVI1 |
| t(1;22) | RBM15::MRTFA |
| t(9;22) | BCR::ABL1 |
| t(15;17) | PML::RARA |
| t(11q23) | KMT2A (MLL1, MLL)-r |
| t(3q26) | MECOM-r |
| t(11p15) | NUP98-r |
| biallelic mutations of the CEBPA gene | - |
| with other defined genetic alterations | - |
| Phenotype | Gene | Culture Conditions | References |
|---|---|---|---|
| Myeloid, CD34+ subpopulation and mature cells | RUNX1::RUNX1T1 | SCF, IL-3, IL-6, GM-CSF, FLT3-L, erythropoietin | [12,13] |
| CBFβ::MYH11 | SCF, IL-3, IL-6, FLT3-L, thrombopoietin | [14] | |
| KMT2A::AF4 | IL-3, SCF, FLT3-L | [15] | |
| BCR::ABL + BMI1 | IL-3, IL-6, G-CSF, SCF, FLT3-L MS5 stromal feeder cells | [16] | |
| Myeloid progenitors, no remaining CD34+ subpopulation | KMT2A::AF9 | SCF, IL-3, IL-6, FLT3-L and TPO | [17] |
| NUP98::HOXA9 | SCF, FLT3-L, thrombopoietin | [18] | |
| Immature granulocyte-macrophage progenitor-like cells | RUNX1 mutations e.g., RUNX1-S291fs300X | IL-3, IL-6, G-CSF, SCF, FLT3-L, erythropoietin | [19] |
| Both myeloid and lymphoid progenitors | KMT2A::ENL | IL-3, SCF, FLT3-L, IL-7 | [20] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kreissig, S.; Windisch, R.; Wichmann, C. Deciphering Acute Myeloid Leukemia Associated Transcription Factors in Human Primary CD34+ Hematopoietic Stem/Progenitor Cells. Cells 2024, 13, 78. https://doi.org/10.3390/cells13010078
Kreissig S, Windisch R, Wichmann C. Deciphering Acute Myeloid Leukemia Associated Transcription Factors in Human Primary CD34+ Hematopoietic Stem/Progenitor Cells. Cells. 2024; 13(1):78. https://doi.org/10.3390/cells13010078
Chicago/Turabian StyleKreissig, Sophie, Roland Windisch, and Christian Wichmann. 2024. "Deciphering Acute Myeloid Leukemia Associated Transcription Factors in Human Primary CD34+ Hematopoietic Stem/Progenitor Cells" Cells 13, no. 1: 78. https://doi.org/10.3390/cells13010078
APA StyleKreissig, S., Windisch, R., & Wichmann, C. (2024). Deciphering Acute Myeloid Leukemia Associated Transcription Factors in Human Primary CD34+ Hematopoietic Stem/Progenitor Cells. Cells, 13(1), 78. https://doi.org/10.3390/cells13010078





