Epigenetic Regulation of Neuroinflammation in Alzheimer’s Disease
Abstract
:1. Introduction
2. Retrieval Strategy
3. Neuroinflammatory Response in AD
4. Epigenetic Regulation in Inflammation
4.1. DNA Modifications in Neuroinflammation
4.1.1. DNA Methylation in Neuroinflammation
4.1.2. DNA Demethylation in Neuroinflammation
4.2. Histone Modification in Neuroinflammation
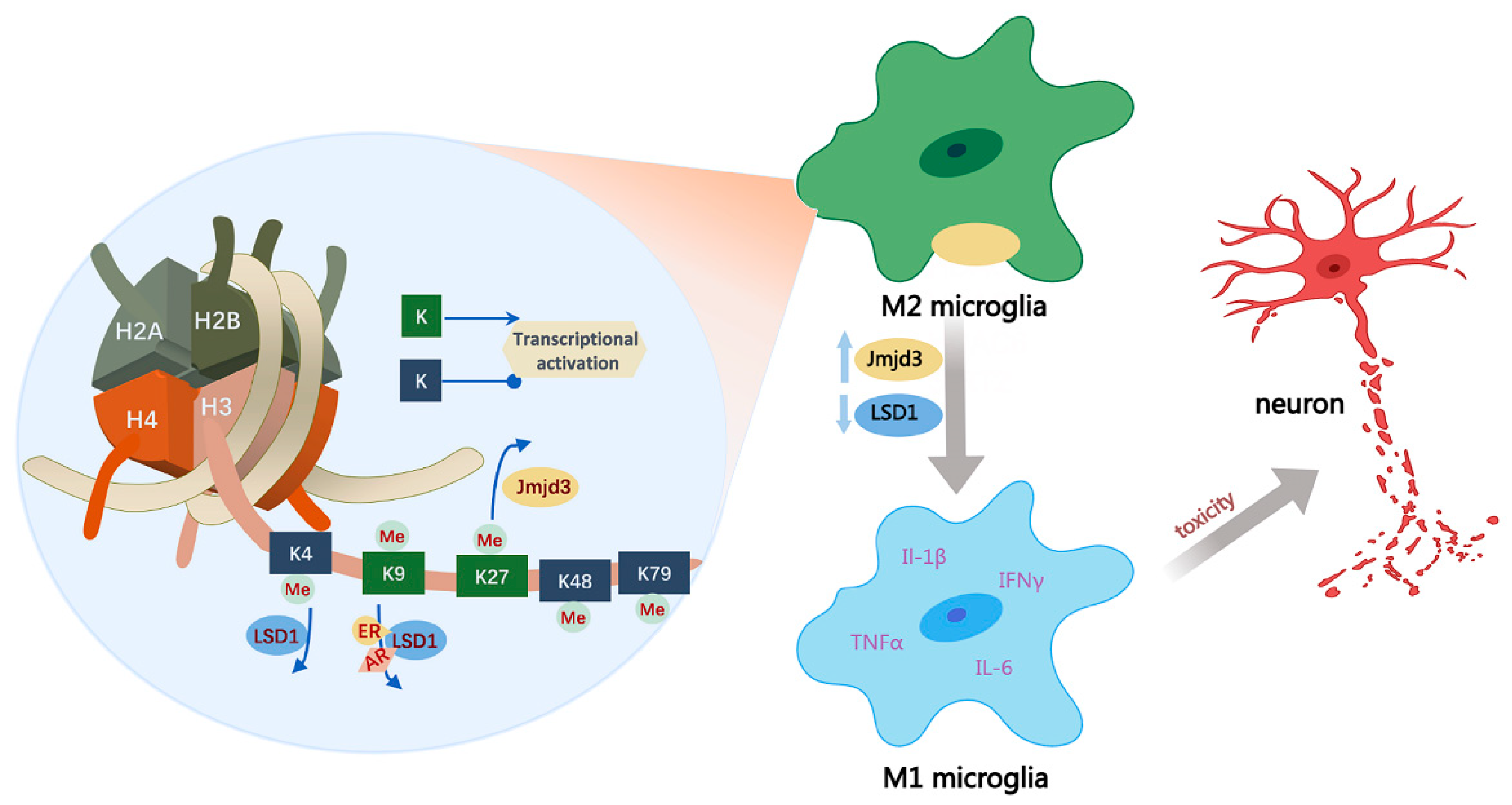
4.2.1. Histone Methylation in Neuroinflammation
4.2.2. Histone Acetylation in Neuroinflammation
4.2.3. Histone Ubiquitination in Neuroinflammation
4.3. Histone Deacetylase and Histone Demethylase Inhibitors in Clinical Trials
5. Conclusions and Prospects
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Long, J.M.; Holtzman, D.M. Alzheimer Disease: An Update on Pathobiology and Treatment Strategies. Cell 2019, 179, 312–339. [Google Scholar] [CrossRef] [PubMed]
- Ju, Y.; Tam, K.Y. Pathological mechanisms and therapeutic strategies for Alzheimer’s disease. Neural Regen. Res. 2022, 17, 543–549. [Google Scholar] [CrossRef] [PubMed]
- Leng, F.; Edison, P. Neuroinflammation and microglial activation in Alzheimer disease: Where do we go from here? Nat. Rev. Neurol. 2021, 17, 157–172. [Google Scholar] [CrossRef] [PubMed]
- Cornejo, F.; von Bernhardi, R. Role of scavenger receptors in glia-mediated neuroinflammatory response associated with Alzheimer’s disease. Mediat. Inflamm. 2013, 2013, 895651. [Google Scholar] [CrossRef]
- Kinney, J.W.; Bemiller, S.M.; Murtishaw, A.S.; Leisgang, A.M.; Salazar, A.M.; Lamb, B.T. Inflammation as a central mechanism in Alzheimer’s disease. Alzheimer’s Dement. 2018, 4, 575–590. [Google Scholar] [CrossRef]
- Grubman, A.; Chew, G.; Ouyang, J.F.; Sun, G.; Choo, X.Y.; McLean, C.; Simmons, R.K.; Buckberry, S.; Vargas-Landin, D.B.; Poppe, D.; et al. A single-cell atlas of entorhinal cortex from individuals with Alzheimer’s disease reveals cell-type-specific gene expression regulation. Nat. Neurosci. 2019, 22, 2087–2097. [Google Scholar] [CrossRef]
- Spangenberg, E.E.; Green, K.N. Inflammation in Alzheimer’s disease: Lessons learned from microglia-depletion models. Brain Behav. Immun. 2017, 61, 1–11. [Google Scholar] [CrossRef]
- Newcombe, E.A.; Camats-Perna, J.; Silva, M.L.; Valmas, N.; Huat, T.J.; Medeiros, R. Inflammation: The link between comorbidities, genetics, and Alzheimer’s disease. J. Neuroinflamm. 2018, 15, 276. [Google Scholar] [CrossRef]
- Wang, H.; Shen, Y.; Chuang, H.; Chiu, C.; Ye, Y.; Zhao, L. Neuroinflammation in Alzheimer’s Disease: Microglia, Molecular Participants and Therapeutic Choices. Curr. Alzheimer Res. 2019, 16, 659–674. [Google Scholar] [CrossRef]
- Panza, F.; Lozupone, M.; Logroscino, G.; Imbimbo, B.P. A critical appraisal of amyloid-beta-targeting therapies for Alzheimer disease. Nat. Rev. Neurol. 2019, 15, 73–88. [Google Scholar] [CrossRef]
- Chen, L.; Deng, H.; Cui, H.; Fang, J.; Zuo, Z.; Deng, J.; Li, Y.; Wang, X.; Zhao, L. Inflammatory responses and inflammation-associated diseases in organs. Oncotarget 2018, 9, 7204–7218. [Google Scholar] [CrossRef] [PubMed]
- Ferrero-Miliani, L.; Nielsen, O.H.; Andersen, P.S.; Girardin, S.E. Chronic inflammation: Importance of NOD2 and NALP3 in interleukin-1beta generation. Clin. Exp. Immunol. 2007, 147, 227–235. [Google Scholar] [CrossRef] [PubMed]
- Paouri, E.; Georgopoulos, S. Systemic and CNS Inflammation Crosstalk: Implications for Alzheimer’s Disease. Curr. Alzheimer Res. 2019, 16, 559–574. [Google Scholar] [CrossRef] [PubMed]
- Doroszkiewicz, J.; Mroczko, P.; Kulczynska-Przybik, A. Inflammation in the CNS: Understanding Various Aspects of the Pathogenesis of Alzheimer’s Disease. Curr. Alzheimer Res. 2022, 19, 16–31. [Google Scholar] [CrossRef] [PubMed]
- Meraz-Rios, M.A.; Toral-Rios, D.; Franco-Bocanegra, D.; Villeda-Hernandez, J.; Campos-Pena, V. Inflammatory process in Alzheimer’s Disease. Front. Integr. Neurosci. 2013, 7, 59. [Google Scholar] [CrossRef] [PubMed]
- Sawikr, Y.; Yarla, N.S.; Peluso, I.; Kamal, M.A.; Aliev, G.; Bishayee, A. Neuroinflammation in Alzheimer’s Disease: The Preventive and Therapeutic Potential of Polyphenolic Nutraceuticals. Adv. Protein Chem. Struct. Biol. 2017, 108, 33–57. [Google Scholar] [CrossRef]
- Coppede, F. Epigenetic regulation in Alzheimer’s disease: Is it a potential therapeutic target? Expert Opin. Ther. Targets 2021, 25, 283–298. [Google Scholar] [CrossRef]
- Van Roy, Z.; Kielian, T. Exploring epigenetic reprogramming during central nervous system infection. Immunol. Rev. 2022, 311, 112–129. [Google Scholar] [CrossRef]
- Ferrari, R.; Hernandez, D.G.; Nalls, M.A.; Rohrer, J.D.; Ramasamy, A.; Kwok, J.B.; Dobson-Stone, C.; Brooks, W.S.; Schofield, P.R.; Halliday, G.M.; et al. Frontotemporal dementia and its subtypes: A genome-wide association study. Lancet. Neurol. 2014, 13, 686–699. [Google Scholar] [CrossRef]
- Greenberg, M.V.C.; Bourc’his, D. The diverse roles of DNA methylation in mammalian development and disease. Nat. Rev. Mol. Cell Biol. 2019, 20, 590–607. [Google Scholar] [CrossRef]
- Yao, B.; Christian, K.M.; He, C.; Jin, P.; Ming, G.L.; Song, H. Epigenetic mechanisms in neurogenesis. Nat. Rev. Neurosci. 2016, 17, 537–549. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Jin, P. Roles of small regulatory RNAs in determining neuronal identity. Nat. Rev. Neurosci. 2010, 11, 329–338. [Google Scholar] [CrossRef] [PubMed]
- Azizi, G.; Navabi, S.S.; Al-Shukaili, A.; Seyedzadeh, M.H.; Yazdani, R.; Mirshafiey, A. The Role of Inflammatory Mediators in the Pathogenesis of Alzheimer’s Disease. Sultan Qaboos Univ. Med. J. 2015, 15, e305–e316. [Google Scholar] [CrossRef] [PubMed]
- Oldenburg, K.S.; O’Shea, T.M.; Fry, R.C. Genetic and epigenetic factors and early life inflammation as predictors of neurodevelopmental outcomes. Semin. Fetal Neonatal Med. 2020, 25, 101115. [Google Scholar] [CrossRef] [PubMed]
- Fenoglio, C.; Scarpini, E.; Serpente, M.; Galimberti, D. Role of Genetics and Epigenetics in the Pathogenesis of Alzheimer’s Disease and Frontotemporal Dementia. J. Alzheimer’s Dis. JAD 2018, 62, 913–932. [Google Scholar] [CrossRef] [PubMed]
- Armstrong, M.J.; Jin, Y.; Allen, E.G.; Jin, P. Diverse and dynamic DNA modifications in brain and diseases. Hum. Mol. Genet. 2019, 28, R241–R253. [Google Scholar] [CrossRef] [PubMed]
- Ji, K.; Akgul, G.; Wollmuth, L.P.; Tsirka, S.E. Microglia actively regulate the number of functional synapses. PLoS ONE 2013, 8, e56293. [Google Scholar] [CrossRef]
- El Khoury, J.; Toft, M.; Hickman, S.E.; Means, T.K.; Terada, K.; Geula, C.; Luster, A.D. Ccr2 deficiency impairs microglial accumulation and accelerates progression of Alzheimer-like disease. Nat. Med. 2007, 13, 432–438. [Google Scholar] [CrossRef]
- Heneka, M.T.; Kummer, M.P.; Stutz, A.; Delekate, A.; Schwartz, S.; Vieira-Saecker, A.; Griep, A.; Axt, D.; Remus, A.; Tzeng, T.C.; et al. NLRP3 is activated in Alzheimer’s disease and contributes to pathology in APP/PS1 mice. Nature 2013, 493, 674–678. [Google Scholar] [CrossRef]
- Combs, C.K.; Karlo, J.C.; Kao, S.C.; Landreth, G.E. beta-Amyloid stimulation of microglia and monocytes results in TNFalpha-dependent expression of inducible nitric oxide synthase and neuronal apoptosis. J. Neurosci. Off. J. Soc. Neurosci. 2001, 21, 1179–1188. [Google Scholar] [CrossRef]
- Stewart, C.R.; Stuart, L.M.; Wilkinson, K.; van Gils, J.M.; Deng, J.; Halle, A.; Rayner, K.J.; Boyer, L.; Zhong, R.; Frazier, W.A.; et al. CD36 ligands promote sterile inflammation through assembly of a Toll-like receptor 4 and 6 heterodimer. Nat. Immunol. 2010, 11, 155–161. [Google Scholar] [CrossRef] [PubMed]
- Hansen, D.V.; Hanson, J.E.; Sheng, M. Microglia in Alzheimer’s disease. J. Cell Biol. 2018, 217, 459–472. [Google Scholar] [CrossRef] [PubMed]
- Fakhoury, M. Microglia and Astrocytes in Alzheimer’s Disease: Implications for Therapy. Curr. Neuropharmacol. 2018, 16, 508–518. [Google Scholar] [CrossRef] [PubMed]
- Pascoal, T.A.; Benedet, A.L.; Ashton, N.J.; Kang, M.S.; Therriault, J.; Chamoun, M.; Savard, M.; Lussier, F.Z.; Tissot, C.; Karikari, T.K.; et al. Microglial activation and tau propagate jointly across Braak stages. Nat. Med. 2021, 27, 1592–1599. [Google Scholar] [CrossRef]
- Heneka, M.T.; Carson, M.J.; El Khoury, J.; Landreth, G.E.; Brosseron, F.; Feinstein, D.L.; Jacobs, A.H.; Wyss-Coray, T.; Vitorica, J.; Ransohoff, R.M.; et al. Neuroinflammation in Alzheimer’s disease. Lancet Neurol. 2015, 14, 388–405. [Google Scholar] [CrossRef] [PubMed]
- Tan, X.; Sun, L.; Chen, J.; Chen, Z.J. Detection of Microbial Infections through Innate Immune Sensing of Nucleic Acids. Annu. Rev. Microbiol. 2018, 72, 447–478. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.; Zhao, W. NLRP3 Inflammasome-A Key Player in Antiviral Responses. Front. Immunol. 2020, 11, 211. [Google Scholar] [CrossRef]
- Konsman, J.P. Cytokines in the Brain and Neuroinflammation: We Didn’t Starve the Fire! Pharmaceuticals 2022, 15, 140. [Google Scholar] [CrossRef]
- Jeon, S.G.; Yoo, A.; Chun, D.W.; Hong, S.B.; Chung, H.; Kim, J.I.; Moon, M. The Critical Role of Nurr1 as a Mediator and Therapeutic Target in Alzheimer’s Disease-Related Pathogenesis. Aging Dis. 2020, 11, 705–724. [Google Scholar] [CrossRef]
- Oh, M.; Kim, S.Y.; Gil, J.E.; Byun, J.S.; Cha, D.W.; Ku, B.; Lee, W.; Kim, W.K.; Oh, K.J.; Lee, E.W.; et al. Nurr1 performs its anti-inflammatory function by regulating RasGRP1 expression in neuro-inflammation. Sci. Rep. 2020, 10, 10755. [Google Scholar] [CrossRef]
- Law, S.W.; Conneely, O.M.; DeMayo, F.J.; O’Malley, B.W. Identification of a new brain-specific transcription factor, NURR1. Mol. Endocrinol. 1992, 6, 2129–2135. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Seok, M.J.; Kim, Y.E.; Choi, Y.; Song, J.J.; Sulistio, Y.A.; Kim, S.H.; Chang, M.Y.; Oh, S.J.; Nam, M.H.; et al. Adeno-associated virus (AAV) 9-mediated gene delivery of Nurr1 and Foxa2 ameliorates symptoms and pathologies of Alzheimer disease model mice by suppressing neuro-inflammation and glial pathology. Mol. Psychiatry 2022. [Google Scholar] [CrossRef] [PubMed]
- Weaver, D.F. Beta-Amyloid is an Immunopeptide and Alzheimer’s is an Autoimmune Disease. Curr. Alzheimer Res. 2021, 18, 849–857. [Google Scholar] [CrossRef] [PubMed]
- Gosztyla, M.L.; Brothers, H.M.; Robinson, S.R. Alzheimer’s Amyloid-beta is an Antimicrobial Peptide: A Review of the Evidence. J. Alzheimer’s Dis. JAD 2018, 62, 1495–1506. [Google Scholar] [CrossRef] [PubMed]
- Hur, J.Y.; Frost, G.R.; Wu, X.; Crump, C.; Pan, S.J.; Wong, E.; Barros, M.; Li, T.; Nie, P.; Zhai, Y.; et al. The innate immunity protein IFITM3 modulates gamma-secretase in Alzheimer’s disease. Nature 2020, 586, 735–740. [Google Scholar] [CrossRef] [PubMed]
- Halder, R.; Hennion, M.; Vidal, R.O.; Shomroni, O.; Rahman, R.U.; Rajput, A.; Centeno, T.P.; van Bebber, F.; Capece, V.; Garcia Vizcaino, J.C.; et al. DNA methylation changes in plasticity genes accompany the formation and maintenance of memory. Nat. Neurosci. 2016, 19, 102–110. [Google Scholar] [CrossRef] [PubMed]
- Horsburgh, S.; Robson-Ansley, P.; Adams, R.; Smith, C. Exercise and inflammation-related epigenetic modifications: Focus on DNA methylation. Exerc. Immunol. Rev. 2015, 21, 26–41. [Google Scholar]
- Tulloch, J.; Leong, L.; Thomson, Z.; Chen, S.; Lee, E.G.; Keene, C.D.; Millard, S.P.; Yu, C.E. Glia-specific APOE epigenetic changes in the Alzheimer’s disease brain. Brain Res. 2018, 1698, 179–186. [Google Scholar] [CrossRef]
- Velmeshev, D.; Magistri, M.; Mazza, E.M.C.; Lally, P.; Khoury, N.; D’Elia, E.R.; Bicciato, S.; Faghihi, M.A. Cell-Type-Specific Analysis of Molecular Pathology in Autism Identifies Common Genes and Pathways Affected Across Neocortical Regions. Mol. Neurobiol. 2020, 57, 2279–2289. [Google Scholar] [CrossRef]
- Hernandez, D.G.; Nalls, M.A.; Gibbs, J.R.; Arepalli, S.; van der Brug, M.; Chong, S.; Moore, M.; Longo, D.L.; Cookson, M.R.; Traynor, B.J.; et al. Distinct DNA methylation changes highly correlated with chronological age in the human brain. Hum. Mol. Genet. 2011, 20, 1164–1172. [Google Scholar] [CrossRef]
- Qin, Q.; Teng, Z.; Liu, C.; Li, Q.; Yin, Y.; Tang, Y. TREM2, microglia, and Alzheimer’s disease. Mech. Ageing Dev. 2021, 195, 111438. [Google Scholar] [CrossRef] [PubMed]
- Ulland, T.K.; Song, W.M.; Huang, S.C.; Ulrich, J.D.; Sergushichev, A.; Beatty, W.L.; Loboda, A.A.; Zhou, Y.; Cairns, N.J.; Kambal, A.; et al. TREM2 Maintains Microglial Metabolic Fitness in Alzheimer’s Disease. Cell 2017, 170, 649–663.e613. [Google Scholar] [CrossRef] [PubMed]
- Jiang, T.; Zhang, Y.D.; Chen, Q.; Gao, Q.; Zhu, X.C.; Zhou, J.S.; Shi, J.Q.; Lu, H.; Tan, L.; Yu, J.T. TREM2 modifies microglial phenotype and provides neuroprotection in P301S tau transgenic mice. Neuropharmacology 2016, 105, 196–206. [Google Scholar] [CrossRef] [PubMed]
- Jiang, T.; Tan, L.; Zhu, X.C.; Zhang, Q.Q.; Cao, L.; Tan, M.S.; Gu, L.Z.; Wang, H.F.; Ding, Z.Z.; Zhang, Y.D.; et al. Upregulation of TREM2 ameliorates neuropathology and rescues spatial cognitive impairment in a transgenic mouse model of Alzheimer’s disease. Neuropsychopharmacology 2014, 39, 2949–2962. [Google Scholar] [CrossRef] [PubMed]
- Jiang, T.; Zhang, Y.D.; Gao, Q.; Ou, Z.; Gong, P.Y.; Shi, J.Q.; Wu, L.; Zhou, J.S. TREM2 Ameliorates Neuronal Tau Pathology Through Suppression of Microglial Inflammatory Response. Inflammation 2018, 41, 811–823. [Google Scholar] [CrossRef]
- Wang, Y.; Cella, M.; Mallinson, K.; Ulrich, J.D.; Young, K.L.; Robinette, M.L.; Gilfillan, S.; Krishnan, G.M.; Sudhakar, S.; Zinselmeyer, B.H.; et al. TREM2 lipid sensing sustains the microglial response in an Alzheimer’s disease model. Cell 2015, 160, 1061–1071. [Google Scholar] [CrossRef]
- Ozaki, Y.; Yoshino, Y.; Yamazaki, K.; Sao, T.; Mori, Y.; Ochi, S.; Yoshida, T.; Mori, T.; Iga, J.I.; Ueno, S.I. DNA methylation changes at TREM2 intron 1 and TREM2 mRNA expression in patients with Alzheimer’s disease. J. Psychiatr. Res. 2017, 92, 74–80. [Google Scholar] [CrossRef]
- Jones, P.A. Functions of DNA methylation: Islands, start sites, gene bodies and beyond. Nat. Rev. Genet. 2012, 13, 484–492. [Google Scholar] [CrossRef]
- Cui, D.; Xu, X. DNA Methyltransferases, DNA Methylation, and Age-Associated Cognitive Function. Int. J. Mol. Sci. 2018, 19, 1315. [Google Scholar] [CrossRef]
- Oliveira, A.M.; Hemstedt, T.J.; Bading, H. Rescue of aging-associated decline in Dnmt3a2 expression restores cognitive abilities. Nat. Neurosci. 2012, 15, 1111–1113. [Google Scholar] [CrossRef]
- Di Francesco, A.; Arosio, B.; Falconi, A.; Micioni Di Bonaventura, M.V.; Karimi, M.; Mari, D.; Casati, M.; Maccarrone, M.; D’Addario, C. Global changes in DNA methylation in Alzheimer’s disease peripheral blood mononuclear cells. Brain Behav. Immun. 2015, 45, 139–144. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.C.; Alinaghi, S.; Tafakhori, A.; Sikora, E.; Azcona, L.J.; Karkheiran, S.; Goate, A.; Paisan-Ruiz, C.; Darvish, H. Genetic screening in two Iranian families with early-onset Alzheimer’s disease identified a novel PSEN1 mutation. Neurobiol. Aging 2018, 62, 244.e215–244.e217. [Google Scholar] [CrossRef] [PubMed]
- Brohede, J.; Rinde, M.; Winblad, B.; Graff, C. A DNA methylation study of the amyloid precursor protein gene in several brain regions from patients with familial Alzheimer disease. J. Neurogenet. 2010, 24, 179–181. [Google Scholar] [CrossRef]
- Foraker, J.; Millard, S.P.; Leong, L.; Thomson, Z.; Chen, S.; Keene, C.D.; Bekris, L.M.; Yu, C.E. The APOE Gene is Differentially Methylated in Alzheimer’s Disease. J. Alzheimers Dis. 2015, 48, 745–755. [Google Scholar] [CrossRef] [PubMed]
- Huls, A.; Robins, C.; Conneely, K.N.; Edgar, R.; De Jager, P.L.; Bennett, D.A.; Wingo, A.P.; Epstein, M.P.; Wingo, T.S. Brain DNA Methylation Patterns in CLDN5 Associated with Cognitive Decline. Biol. Psychiatry 2022, 91, 389–398. [Google Scholar] [CrossRef] [PubMed]
- Stallings, N.R.; O’Neal, M.A.; Hu, J.; Kavalali, E.T.; Bezprozvanny, I.; Malter, J.S. Pin1 mediates Abeta(42)-induced dendritic spine loss. Sci. Signal. 2018, 11, eaap8734. [Google Scholar] [CrossRef] [PubMed]
- Malter, J.S. Pin1 and Alzheimer’s disease. Transl. Res. J. Lab. Clin. Med. 2022, 254, 24–33. [Google Scholar] [CrossRef]
- Liou, Y.C.; Sun, A.; Ryo, A.; Zhou, X.Z.; Yu, Z.X.; Huang, H.K.; Uchida, T.; Bronson, R.; Bing, G.; Li, X.; et al. Role of the prolyl isomerase Pin1 in protecting against age-dependent neurodegeneration. Nature 2003, 424, 556–561. [Google Scholar] [CrossRef]
- Ma, S.L.; Tang, N.L.S.; Lam, L.C.W. Promoter Methylation and Gene Expression of Pin1 Associated with the Risk of Alzheimer’s Disease in Southern Chinese. Curr. Alzheimer Res. 2020, 17, 1232–1237. [Google Scholar] [CrossRef]
- Vakifahmetoglu-Norberg, H.; Ouchida, A.T.; Norberg, E. The role of mitochondria in metabolism and cell death. Biochem. Biophys. Res. Commun. 2017, 482, 426–431. [Google Scholar] [CrossRef]
- Hroudova, J.; Singh, N.; Fisar, Z. Mitochondrial dysfunctions in neurodegenerative diseases: Relevance to Alzheimer’s disease. BioMed Res. Int. 2014, 2014, 175062. [Google Scholar] [CrossRef] [PubMed]
- Shock, L.S.; Thakkar, P.V.; Peterson, E.J.; Moran, R.G.; Taylor, S.M. DNA methyltransferase 1, cytosine methylation, and cytosine hydroxymethylation in mammalian mitochondria. Proc. Natl. Acad. Sci. USA 2011, 108, 3630–3635. [Google Scholar] [CrossRef] [PubMed]
- Bellizzi, D.; D’Aquila, P.; Scafone, T.; Giordano, M.; Riso, V.; Riccio, A.; Passarino, G. The control region of mitochondrial DNA shows an unusual CpG and non-CpG methylation pattern. DNA Res. Int. J. Rapid Publ. Rep. Genes Genomes 2013, 20, 537–547. [Google Scholar] [CrossRef] [PubMed]
- Weidling, I.; Swerdlow, R.H. Mitochondrial Dysfunction and Stress Responses in Alzheimer’s Disease. Biology 2019, 8, 39. [Google Scholar] [CrossRef] [PubMed]
- Weidling, I.W.; Swerdlow, R.H. Mitochondria in Alzheimer’s disease and their potential role in Alzheimer’s proteostasis. Exp. Neurol. 2020, 330, 113321. [Google Scholar] [CrossRef] [PubMed]
- Tahiliani, M.; Koh, K.P.; Shen, Y.; Pastor, W.A.; Bandukwala, H.; Brudno, Y.; Agarwal, S.; Iyer, L.M.; Liu, D.R.; Aravind, L.; et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science 2009, 324, 930–935. [Google Scholar] [CrossRef] [PubMed]
- Ito, S.; Shen, L.; Dai, Q.; Wu, S.C.; Collins, L.B.; Swenberg, J.A.; He, C.; Zhang, Y. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science 2011, 333, 1300–1303. [Google Scholar] [CrossRef]
- Coulter, J.B.; O’Driscoll, C.M.; Bressler, J.P. Hydroquinone increases 5-hydroxymethylcytosine formation through ten eleven translocation 1 (TET1) 5-methylcytosine dioxygenase. J. Biol. Chem. 2013, 288, 28792–28800. [Google Scholar] [CrossRef]
- Zhang, R.R.; Cui, Q.Y.; Murai, K.; Lim, Y.C.; Smith, Z.D.; Jin, S.; Ye, P.; Rosa, L.; Lee, Y.K.; Wu, H.P.; et al. Tet1 regulates adult hippocampal neurogenesis and cognition. Cell Stem Cell 2013, 13, 237–245. [Google Scholar] [CrossRef]
- Li, L.; Qiu, Y.; Miao, M.; Liu, Z.; Li, W.; Zhu, Y.; Wang, Q. Reduction of Tet2 exacerbates early stage Alzheimer’s pathology and cognitive impairments in 2xTg-AD mice. Hum. Mol. Genet. 2020, 29, 1833–1852. [Google Scholar] [CrossRef]
- Zhang, Q.; Zhao, K.; Shen, Q.; Han, Y.; Gu, Y.; Li, X.; Zhao, D.; Liu, Y.; Wang, C.; Zhang, X.; et al. Tet2 is required to resolve inflammation by recruiting Hdac2 to specifically repress IL-6. Nature 2015, 525, 389–393. [Google Scholar] [CrossRef] [PubMed]
- Szulwach, K.E.; Li, X.; Li, Y.; Song, C.X.; Wu, H.; Dai, Q.; Irier, H.; Upadhyay, A.K.; Gearing, M.; Levey, A.I.; et al. 5-hmC-mediated epigenetic dynamics during postnatal neurodevelopment and aging. Nat. Neurosci. 2011, 14, 1607–1616. [Google Scholar] [CrossRef] [PubMed]
- Pastor, W.A.; Pape, U.J.; Huang, Y.; Henderson, H.R.; Lister, R.; Ko, M.; McLoughlin, E.M.; Brudno, Y.; Mahapatra, S.; Kapranov, P.; et al. Genome-wide mapping of 5-hydroxymethylcytosine in embryonic stem cells. Nature 2011, 473, 394–397. [Google Scholar] [CrossRef] [PubMed]
- Nikolac Perkovic, M.; Videtic Paska, A.; Konjevod, M.; Kouter, K.; Svob Strac, D.; Nedic Erjavec, G.; Pivac, N. Epigenetics of Alzheimer’s Disease. Biomolecules 2021, 11, 195. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Yuan, C.W.; Xu, S.; Zu, T.; Woappi, Y.; Lee, C.A.A.; Abarzua, P.; Wells, M.; Ramsey, M.R.; Frank, N.Y.; et al. Loss of the Epigenetic Mark 5-hmC in Psoriasis: Implications for Epidermal Stem Cell Dysregulation. J. Investig. Dermatol. 2020, 140, 1266–1275.e1263. [Google Scholar] [CrossRef]
- Zhao, J.; Zhu, Y.; Yang, J.; Li, L.; Wu, H.; De Jager, P.L.; Jin, P.; Bennett, D.A. A genome-wide profiling of brain DNA hydroxymethylation in Alzheimer’s disease. Alzheimer’s Dement. J. Alzheimer’s Assoc. 2017, 13, 674–688. [Google Scholar] [CrossRef]
- Ellison, E.M.; Bradley-Whitman, M.A.; Lovell, M.A. Single-Base Resolution Mapping of 5-Hydroxymethylcytosine Modifications in Hippocampus of Alzheimer’s Disease Subjects. J. Mol. Neurosci. MN 2017, 63, 185–197. [Google Scholar] [CrossRef]
- Shu, L.; Sun, W.; Li, L.; Xu, Z.; Lin, L.; Xie, P.; Shen, H.; Huang, L.; Xu, Q.; Jin, P.; et al. Genome-wide alteration of 5-hydroxymenthylcytosine in a mouse model of Alzheimer’s disease. BMC Genom. 2016, 17, 381. [Google Scholar] [CrossRef]
- Bernstein, A.I.; Lin, Y.; Street, R.C.; Lin, L.; Dai, Q.; Yu, L.; Bao, H.; Gearing, M.; Lah, J.J.; Nelson, P.T.; et al. 5-Hydroxymethylation-associated epigenetic modifiers of Alzheimer’s disease modulate Tau-induced neurotoxicity. Hum. Mol. Genet. 2016, 25, 2437–2450. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, Z.; Li, L.; Xu, K.; Ma, Z.; Chow, H.M.; Herrup, K.; Li, J. Selective loss of 5hmC promotes neurodegeneration in the mouse model of Alzheimer’s disease. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2020, 34, 16364–16382. [Google Scholar] [CrossRef]
- Cosgrove, M.S.; Boeke, J.D.; Wolberger, C. Regulated nucleosome mobility and the histone code. Nat. Struct. Mol. Biol. 2004, 11, 1037–1043. [Google Scholar] [CrossRef] [PubMed]
- Jakovcevski, M.; Akbarian, S. Epigenetic mechanisms in neurological disease. Nat. Med. 2012, 18, 1194–1204. [Google Scholar] [CrossRef] [PubMed]
- Zusso, M.; Barbierato, M.; Facci, L.; Skaper, S.D.; Giusti, P. Neuroepigenetics and Alzheimer’s Disease: An Update. J. Alzheimer’s Dis. JAD 2018, 64, 671–688. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Xie, Z.S.; Song, J.Y.; Zeng, H.H.; Dai, L.P.; E, H.C.; Ye, Z.P.; Gao, S.; Xu, J.Y.; Zhang, Z.Q. Four new sesquiterpene lactones from Atractylodes macrocephala and their CREB agonistic activities. Fitoterapia 2020, 147, 104730. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Zhang, J.; Xu, Y. Epigenetic Basis of Lead-Induced Neurological Disorders. Int. J. Environ. Res. Public Health 2020, 17, 4878. [Google Scholar] [CrossRef] [PubMed]
- Surace, A.E.A.; Hedrich, C.M. The Role of Epigenetics in Autoimmune/Inflammatory Disease. Front. Immunol. 2019, 10, 1525. [Google Scholar] [CrossRef] [PubMed]
- Herre, M.; Korb, E. The chromatin landscape of neuronal plasticity. Curr. Opin. Neurobiol. 2019, 59, 79–86. [Google Scholar] [CrossRef]
- Fischer, A.; Sananbenesi, F.; Wang, X.; Dobbin, M.; Tsai, L.H. Recovery of learning and memory is associated with chromatin remodelling. Nature 2007, 447, 178–182. [Google Scholar] [CrossRef]
- Habibi, E.; Masoudi-Nejad, A.; Abdolmaleky, H.M.; Haggarty, S.J. Emerging roles of epigenetic mechanisms in Parkinson’s disease. Funct. Integr. Genom. 2011, 11, 523–537. [Google Scholar] [CrossRef]
- Arrowsmith, C.H.; Bountra, C.; Fish, P.V.; Lee, K.; Schapira, M. Epigenetic protein families: A new frontier for drug discovery. Nat. Rev. Drug Discov. 2012, 11, 384–400. [Google Scholar] [CrossRef]
- Saijo, K.; Winner, B.; Carson, C.T.; Collier, J.G.; Boyer, L.; Rosenfeld, M.G.; Gage, F.H.; Glass, C.K. A Nurr1/CoREST pathway in microglia and astrocytes protects dopaminergic neurons from inflammation-induced death. Cell 2009, 137, 47–59. [Google Scholar] [CrossRef] [PubMed]
- Le, W.; Rowe, D.; Xie, W.; Ortiz, I.; He, Y.; Appel, S.H. Microglial activation and dopaminergic cell injury: An in vitro model relevant to Parkinson’s disease. J. Neurosci. 2001, 21, 8447–8455. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Li, T.; Li, J.; Yang, J.; Liu, H.; Zhang, X.J.; Le, W. Jmjd3 is essential for the epigenetic modulation of microglia phenotypes in the immune pathogenesis of Parkinson’s disease. Cell Death Differ. 2014, 21, 369–380. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Lan, F.; Matson, C.; Mulligan, P.; Whetstine, J.R.; Cole, P.A.; Casero, R.A.; Shi, Y. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell 2004, 119, 941–953. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Xu, D.; Yuan, L.; Sun, Y.; Xu, Z. Epigenetic regulation of Atrophin1 by lysine-specific demethylase 1 is required for cortical progenitor maintenance. Nat. Commun. 2014, 5, 5815. [Google Scholar] [CrossRef] [PubMed]
- Zibetti, C.; Adamo, A.; Binda, C.; Forneris, F.; Toffolo, E.; Verpelli, C.; Ginelli, E.; Mattevi, A.; Sala, C.; Battaglioli, E. Alternative splicing of the histone demethylase LSD1/KDM1 contributes to the modulation of neurite morphogenesis in the mammalian nervous system. J. Neurosci. 2010, 30, 2521–2532. [Google Scholar] [CrossRef]
- Christopher, M.A.; Myrick, D.A.; Barwick, B.G.; Engstrom, A.K.; Porter-Stransky, K.A.; Boss, J.M.; Weinshenker, D.; Levey, A.I.; Katz, D.J. LSD1 protects against hippocampal and cortical neurodegeneration. Nat. Commun. 2017, 8, 805. [Google Scholar] [CrossRef]
- Kim, D.; Nam, H.J.; Lee, W.; Yim, H.Y.; Ahn, J.Y.; Park, S.W.; Shin, H.R.; Yu, R.; Won, K.J.; Bae, J.S.; et al. PKCalpha-LSD1-NF-kappaB-Signaling Cascade Is Crucial for Epigenetic Control of the Inflammatory Response. Mol. Cell 2018, 69, 398–411.e396. [Google Scholar] [CrossRef]
- Jingjing, W.; Zhikai, W.; Xingyi, Z.; Peixuan, L.; Yiwu, F.; Xia, W.; Youpeng, S.; Ershun, Z.; Zhengtao, Y. Lysine-specific demethylase 1 (LSD1) serves as an potential epigenetic determinant to regulate inflammatory responses in mastitis. Int. Immunopharmacol. 2021, 91, 107324. [Google Scholar] [CrossRef]
- Tan, M.; Luo, H.; Lee, S.; Jin, F.; Yang, J.S.; Montellier, E.; Buchou, T.; Cheng, Z.; Rousseaux, S.; Rajagopal, N.; et al. Identification of 67 histone marks and histone lysine crotonylation as a new type of histone modification. Cell 2011, 146, 1016–1028. [Google Scholar] [CrossRef]
- Tie, F.; Banerjee, R.; Saiakhova, A.R.; Howard, B.; Monteith, K.E.; Scacheri, P.C.; Cosgrove, M.S.; Harte, P.J. Trithorax monomethylates histone H3K4 and interacts directly with CBP to promote H3K27 acetylation and antagonize Polycomb silencing. Development 2014, 141, 1129–1139. [Google Scholar] [CrossRef] [PubMed]
- Marzi, S.J.; Leung, S.K.; Ribarska, T.; Hannon, E.; Smith, A.R.; Pishva, E.; Poschmann, J.; Moore, K.; Troakes, C.; Al-Sarraj, S.; et al. A histone acetylome-wide association study of Alzheimer’s disease identifies disease-associated H3K27ac differences in the entorhinal cortex. Nat. Neurosci. 2018, 21, 1618–1627. [Google Scholar] [CrossRef] [PubMed]
- Peleg, S.; Sananbenesi, F.; Zovoilis, A.; Burkhardt, S.; Bahari-Javan, S.; Agis-Balboa, R.C.; Cota, P.; Wittnam, J.L.; Gogol-Doering, A.; Opitz, L.; et al. Altered histone acetylation is associated with age-dependent memory impairment in mice. Science 2010, 328, 753–756. [Google Scholar] [CrossRef] [PubMed]
- Miller, G. Epigenetics. A role for epigenetics in cognition. Science 2010, 329, 27. [Google Scholar] [CrossRef] [PubMed]
- Nativio, R.; Lan, Y.; Donahue, G.; Sidoli, S.; Berson, A.; Srinivasan, A.R.; Shcherbakova, O.; Amlie-Wolf, A.; Nie, J.; Cui, X.; et al. An integrated multi-omics approach identifies epigenetic alterations associated with Alzheimer’s disease. Nat. Genet. 2020, 52, 1024–1035. [Google Scholar] [CrossRef] [PubMed]
- Fang, P.; Chen, C.; Zheng, F.; Jia, J.; Chen, T.; Zhu, J.; Chang, J.; Zhang, Z. NLRP3 inflammasome inhibition by histone acetylation ameliorates sevoflurane-induced cognitive impairment in aged mice by activating the autophagy pathway. Brain Res. Bull. 2021, 172, 79–88. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.Y.; Zhong, Y.; Zhao, Z.; Miao, Y. Epigenetic suppression of hippocampal BDNF mediates the memory deficiency induced by amyloid fibrils. Pharmacol. Biochem. Behav. 2014, 126, 83–89. [Google Scholar] [CrossRef] [PubMed]
- Govindarajan, N.; Rao, P.; Burkhardt, S.; Sananbenesi, F.; Schluter, O.M.; Bradke, F.; Lu, J.; Fischer, A. Reducing HDAC6 ameliorates cognitive deficits in a mouse model for Alzheimer’s disease. EMBO Mol. Med. 2013, 5, 52–63. [Google Scholar] [CrossRef]
- Cook, C.; Carlomagno, Y.; Gendron, T.F.; Dunmore, J.; Scheffel, K.; Stetler, C.; Davis, M.; Dickson, D.; Jarpe, M.; DeTure, M.; et al. Acetylation of the KXGS motifs in tau is a critical determinant in modulation of tau aggregation and clearance. Hum. Mol. Genet. 2014, 23, 104–116. [Google Scholar] [CrossRef]
- Leyk, J.; Goldbaum, O.; Noack, M.; Richter-Landsberg, C. Inhibition of HDAC6 modifies tau inclusion body formation and impairs autophagic clearance. J. Mol. Neurosci. MN 2015, 55, 1031–1046. [Google Scholar] [CrossRef]
- Gomes, B.A.Q.; Silva, J.P.B.; Romeiro, C.F.R.; Dos Santos, S.M.; Rodrigues, C.A.; Goncalves, P.R.; Sakai, J.T.; Mendes, P.F.S.; Varela, E.L.P.; Monteiro, M.C. Neuroprotective Mechanisms of Resveratrol in Alzheimer’s Disease: Role of SIRT1. Oxidative Med. Cell. Longev. 2018, 2018, 8152373. [Google Scholar] [CrossRef] [PubMed]
- Chuang, Y.; Van, I.; Zhao, Y.; Xu, Y. Icariin ameliorate Alzheimer’s disease by influencing SIRT1 and inhibiting Abeta cascade pathogenesis. J. Chem. Neuroanat. 2021, 117, 102014. [Google Scholar] [CrossRef]
- Pais, T.F.; Szego, E.M.; Marques, O.; Miller-Fleming, L.; Antas, P.; Guerreiro, P.; de Oliveira, R.M.; Kasapoglu, B.; Outeiro, T.F. The NAD-dependent deacetylase sirtuin 2 is a suppressor of microglial activation and brain inflammation. EMBO J. 2013, 32, 2603–2616. [Google Scholar] [CrossRef] [PubMed]
- Popovic, D.; Vucic, D.; Dikic, I. Ubiquitination in disease pathogenesis and treatment. Nat. Med. 2014, 20, 1242–1253. [Google Scholar] [CrossRef] [PubMed]
- Sommer, T.; Wolf, D.H. The ubiquitin-proteasome-system. Biochim. Biophys. Acta 2014, 1843, 1. [Google Scholar] [CrossRef]
- Zinngrebe, J.; Montinaro, A.; Peltzer, N.; Walczak, H. Ubiquitin in the immune system. EMBO Rep. 2014, 15, 28–45. [Google Scholar] [CrossRef]
- Lam, Y.A.; Xu, W.; DeMartino, G.N.; Cohen, R.E. Editing of ubiquitin conjugates by an isopeptidase in the 26S proteasome. Nature 1997, 385, 737–740. [Google Scholar] [CrossRef]
- Amerik, A.Y.; Hochstrasser, M. Mechanism and function of deubiquitinating enzymes. Biochim. Biophys. Acta 2004, 1695, 189–207. [Google Scholar] [CrossRef]
- Pohl, C.; Dikic, I. Cellular quality control by the ubiquitin-proteasome system and autophagy. Science 2019, 366, 818–822. [Google Scholar] [CrossRef]
- Chen, R.; Pang, X.; Li, L.; Zeng, Z.; Chen, M.; Zhang, S. Ubiquitin-specific proteases in inflammatory bowel disease-related signalling pathway regulation. Cell Death Dis. 2022, 13, 139. [Google Scholar] [CrossRef]
- Ejarque-Ortiz, A.; Medina, M.G.; Tusell, J.M.; Perez-Gonzalez, A.P.; Serratosa, J.; Saura, J. Upregulation of CCAAT/enhancer binding protein beta in activated astrocytes and microglia. Glia 2007, 55, 178–188. [Google Scholar] [CrossRef] [PubMed]
- Straccia, M.; Gresa-Arribas, N.; Dentesano, G.; Ejarque-Ortiz, A.; Tusell, J.M.; Serratosa, J.; Sola, C.; Saura, J. Pro-inflammatory gene expression and neurotoxic effects of activated microglia are attenuated by absence of CCAAT/enhancer binding protein beta. J. Neuroinflammation 2011, 8, 156. [Google Scholar] [CrossRef] [PubMed]
- Niehrs, C.; Calkhoven, C.F. Emerging Role of C/EBPbeta and Epigenetic DNA Methylation in Ageing. Trends Genet. TIG 2020, 36, 71–80. [Google Scholar] [CrossRef] [PubMed]
- Ndoja, A.; Reja, R.; Lee, S.H.; Webster, J.D.; Ngu, H.; Rose, C.M.; Kirkpatrick, D.S.; Modrusan, Z.; Chen, Y.J.; Dugger, D.L.; et al. Ubiquitin Ligase COP1 Suppresses Neuroinflammation by Degrading c/EBPbeta in Microglia. Cell 2020, 182, 1156–1169.e1112. [Google Scholar] [CrossRef] [PubMed]
- Shang, L.; Peng, T.; Chen, X.; Yan, Z.; Wang, J.; Gao, X.; Chang, C. miR-590-5p Overexpression Alleviates Beta-Amyloid-Induced Neuron Damage via Targeting Pellino-1. Anal. Cell. Pathol. 2022, 2022, 7657995. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Yu, T.; Pietronigro, E.C.; Yuan, J.; Arioli, J.; Pei, Y.; Luo, X.; Ye, J.; Constantin, G.; Mao, C.; et al. Peli1 impairs microglial Abeta phagocytosis through promoting C/EBPbeta degradation. PLoS Biol. 2020, 18, e3000837. [Google Scholar] [CrossRef]
- Han, C.; Yan, P.; He, T.; Cheng, J.; Zheng, W.; Zheng, L.T.; Zhen, X. PHLDA1 promotes microglia-mediated neuroinflammation via regulating K63-linked ubiquitination of TRAF6. Brain Behav. Immun. 2020, 88, 640–653. [Google Scholar] [CrossRef]
- Cao, L.L.; Guan, P.P.; Zhang, S.Q.; Yang, Y.; Huang, X.S.; Wang, P. Downregulating expression of OPTN elevates neuroinflammation via AIM2 inflammasome- and RIPK1-activating mechanisms in APP/PS1 transgenic mice. J. Neuroinflammation 2021, 18, 281. [Google Scholar] [CrossRef]
- Yao, J.; Yang, J.; Yang, Z.; Wang, X.P.; Yang, T.; Ji, B.; Zhang, Z.Y. FBXW11 contributes to stem-cell-like features and liver metastasis through regulating HIC1-mediated SIRT1 transcription in colorectal cancer. Cell Death Dis. 2021, 12, 930. [Google Scholar] [CrossRef]
- Chen, C.; Zhou, H.; Zhang, X.; Liu, Z.; Ma, X. Association of FBXW11 levels with tumor development and prognosis in chondrosarcoma. Cancer Biomark. Sect. A Dis. Markers 2022, 35, 429–437. [Google Scholar] [CrossRef]
- Sun, J.; Qin, X.; Zhang, X.; Wang, Q.; Zhang, W.; Wang, M. FBXW11 deletion alleviates Alzheimer’s disease by reducing neuroinflammation and amyloid-beta plaque formation via repression of ASK1 signaling. Biochem. Biophys. Res. Commun. 2021, 548, 104–111. [Google Scholar] [CrossRef] [PubMed]
- Kramer, O.H.; Zhu, P.; Ostendorff, H.P.; Golebiewski, M.; Tiefenbach, J.; Peters, M.A.; Brill, B.; Groner, B.; Bach, I.; Heinzel, T.; et al. The histone deacetylase inhibitor valproic acid selectively induces proteasomal degradation of HDAC2. EMBO J. 2003, 22, 3411–3420. [Google Scholar] [CrossRef] [PubMed]
- Qing, H.; He, G.; Ly, P.T.; Fox, C.J.; Staufenbiel, M.; Cai, F.; Zhang, Z.; Wei, S.; Sun, X.; Chen, C.H.; et al. Valproic acid inhibits Abeta production, neuritic plaque formation, and behavioral deficits in Alzheimer’s disease mouse models. J. Exp. Med. 2008, 205, 2781–2789. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.Z.; Li, X.J.; Zhang, H.Y. Valproic acid as a promising agent to combat Alzheimer’s disease. Brain Res. Bull. 2010, 81, 3–6. [Google Scholar] [CrossRef] [PubMed]
- Kuendgen, A.; Schmid, M.; Schlenk, R.; Knipp, S.; Hildebrandt, B.; Steidl, C.; Germing, U.; Haas, R.; Dohner, H.; Gattermann, N. The histone deacetylase (HDAC) inhibitor valproic acid as monotherapy or in combination with all-trans retinoic acid in patients with acute myeloid leukemia. Cancer 2006, 106, 112–119. [Google Scholar] [CrossRef]
- Xu, K.; Dai, X.L.; Huang, H.C.; Jiang, Z.F. Targeting HDACs: A promising therapy for Alzheimer’s disease. Oxidative Med. Cell. Longev. 2011, 2011, 143269. [Google Scholar] [CrossRef]
- Thone, J.; Ellrichmann, G.; Faustmann, P.M.; Gold, R.; Haghikia, A. Anti-inflammatory effects of levetiracetam in experimental autoimmune encephalomyelitis. Int. Immunopharmacol. 2012, 14, 9–12. [Google Scholar] [CrossRef]
- Belcastro, V.; Pierguidi, L.; Tambasco, N. Levetiracetam in brain ischemia: Clinical implications in neuroprotection and prevention of post-stroke epilepsy. Brain Dev. 2011, 33, 289–293. [Google Scholar] [CrossRef]
- Rabal, O.; Sanchez-Arias, J.A.; Cuadrado-Tejedor, M.; de Miguel, I.; Perez-Gonzalez, M.; Garcia-Barroso, C.; Ugarte, A.; Estella-Hermoso de Mendoza, A.; Saez, E.; Espelosin, M.; et al. Multitarget Approach for the Treatment of Alzheimer’s Disease: Inhibition of Phosphodiesterase 9 (PDE9) and Histone Deacetylases (HDACs) Covering Diverse Selectivity Profiles. ACS Chem. Neurosci. 2019, 10, 4076–4101. [Google Scholar] [CrossRef]
- Bufill, E.; Ribosa-Nogue, R.; Blesa, R. The Therapeutic Potential of Epigenetic Modifications in Alzheimer’s Disease. In Alzheimer’s Disease: Drug Discovery; Huang, X., Ed.; Exon Publications: Brisbane, Australia, 2020. [Google Scholar] [CrossRef]
- Maes, T.; Mascaro, C.; Rotllant, D.; Lufino, M.M.P.; Estiarte, A.; Guibourt, N.; Cavalcanti, F.; Grinan-Ferre, C.; Pallas, M.; Nadal, R.; et al. Modulation of KDM1A with vafidemstat rescues memory deficit and behavioral alterations. PLoS ONE 2020, 15, e0233468. [Google Scholar] [CrossRef]
- Knight, Z.A.; Shokat, K.M. Chemical genetics: Where genetics and pharmacology meet. Cell 2007, 128, 425–430. [Google Scholar] [CrossRef] [PubMed]





| Drug | Epigenetic Target | Phase | Trial Identifier | Outcomes in (Pre)Clinical Studies |
|---|---|---|---|---|
| Levetiracetam | HDACs | 2 | NCT04004702 | Reduce proinflammatory factors, decrease cell death and oxidative stress, mitigate cognitive impairment. |
| 2 | NCT03875638 | |||
| 2 | NCT02002819 | |||
| 2 | NCT03461861 | |||
| 4 | NCT05969054 | |||
| 2 | NCT03489044 | |||
| 3 | NCT05986721 | |||
| Topiramate | HDACs | 1 | NCT02884050 | Inhibit microglia activation, activate Akt and AMPK, facilitate Aβ transport and clearance. |
| Valproic acid | HDACs | 3 | NCT00071721 | Reduce inflammation, inhibit Aβ deposition, improve cognitive impairments in APP23 and APPswe/PS1ΔE9 AD mice. |
| 1 | NCT01729598 | |||
| 2 | NCT00088387 | |||
| Vorinostat | HDACs | 1 | NCT03056495 | Regulate synaptic plasticity and improve cognitive function, restore gamma oscillation deficits in PSAPP mice, reduce inflammation. |
| RDN-929 | HDAC-CoREST | 1 | NCT03668314 | Reactivate neuronal gene expression, strengthen synaptic function, promote synapse formation. |
| 1 | NCT03963973 | |||
| Vafidemstat | LSD1, MAO-B | 2 | NCT03867253 | Improve learning and mitigate memory deficit in SAMP8 model and decrease inflammation in the hippocampus. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ma, Y.; Wang, W.; Liu, S.; Qiao, X.; Xing, Y.; Zhou, Q.; Zhang, Z. Epigenetic Regulation of Neuroinflammation in Alzheimer’s Disease. Cells 2024, 13, 79. https://doi.org/10.3390/cells13010079
Ma Y, Wang W, Liu S, Qiao X, Xing Y, Zhou Q, Zhang Z. Epigenetic Regulation of Neuroinflammation in Alzheimer’s Disease. Cells. 2024; 13(1):79. https://doi.org/10.3390/cells13010079
Chicago/Turabian StyleMa, Yajing, Wang Wang, Sufang Liu, Xiaomeng Qiao, Ying Xing, Qingfeng Zhou, and Zhijian Zhang. 2024. "Epigenetic Regulation of Neuroinflammation in Alzheimer’s Disease" Cells 13, no. 1: 79. https://doi.org/10.3390/cells13010079
APA StyleMa, Y., Wang, W., Liu, S., Qiao, X., Xing, Y., Zhou, Q., & Zhang, Z. (2024). Epigenetic Regulation of Neuroinflammation in Alzheimer’s Disease. Cells, 13(1), 79. https://doi.org/10.3390/cells13010079






