Patient-Derived Conditionally Reprogrammed Cells in Prostate Cancer Research
Abstract
:1. Introduction
1.1. Overview of Prostate Cancer (PCa)
1.2. Limitations of Traditional PCa Cell Lines
2. Patient-Derived Primary Cell Cultures in PCa Research
3. Role of CR in Cell Culture
3.1. Overview of CR
3.2. Optimization of CR
3.3. Rapid and Effective Growth of Patient-Derived Normal and Tumor Cells
3.4. Acquisition of Stem Cell Properties
4. Advantages of CR over Traditional Approaches
4.1. Patient-Derived Models for Precision Medicine in PCa
4.2. Revolutionizing Drug Sensitivity Testing in PCa
4.3. Addressing Challenges in Rapid and Continuous Cell Growth
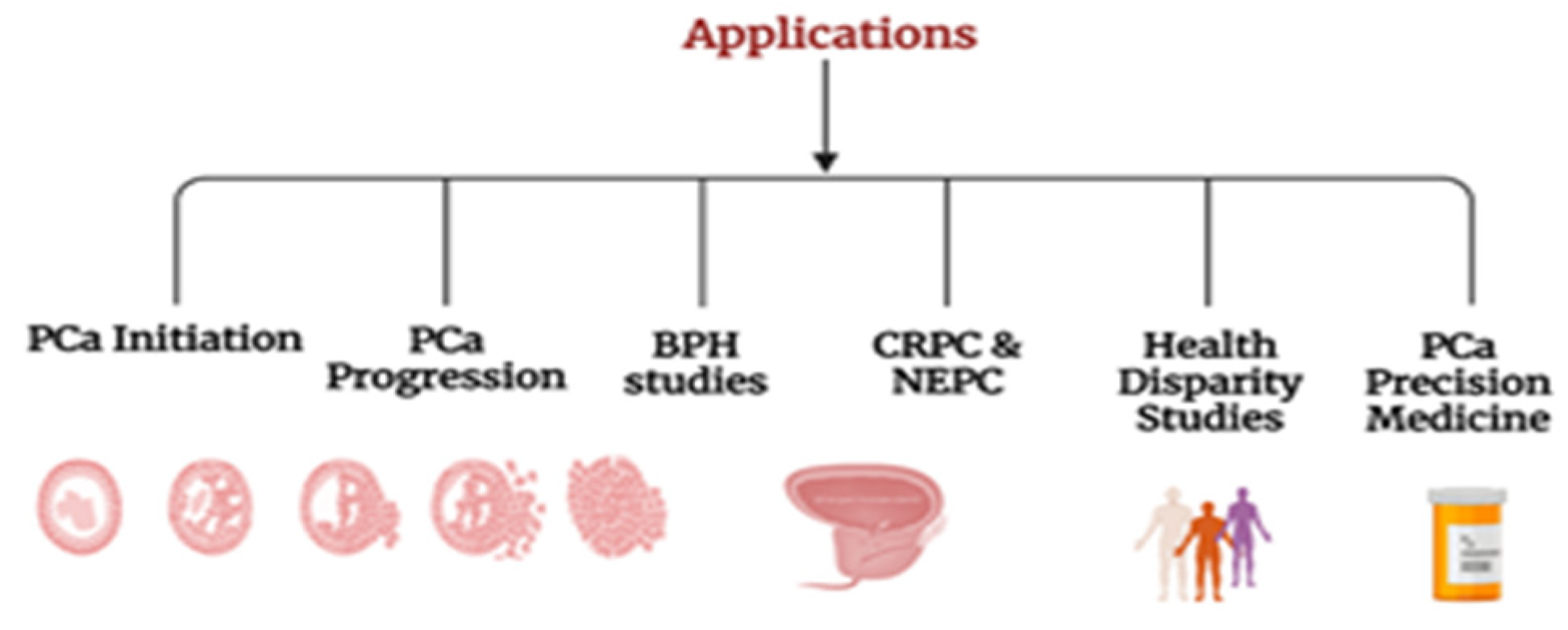
5. Applications of CR Cells in PCa Initiation and Progression
5.1. Insights into PCa Initiation
5.2. Evolution from Localized to Metastatic PCa
5.3. Unraveling the Molecular Pathogenesis of Metastatic PCa and Progression
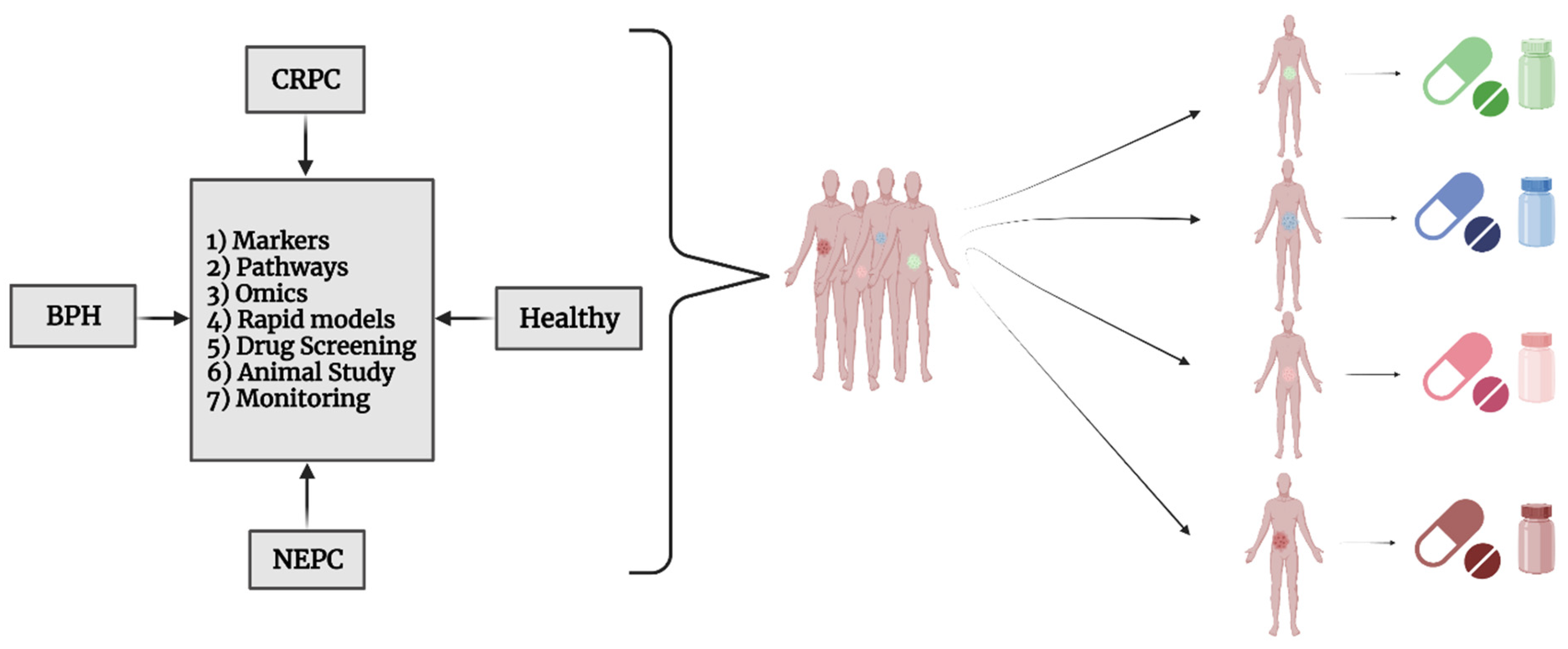
5.4. BPH Studies
5.5. CRPC and NEPC
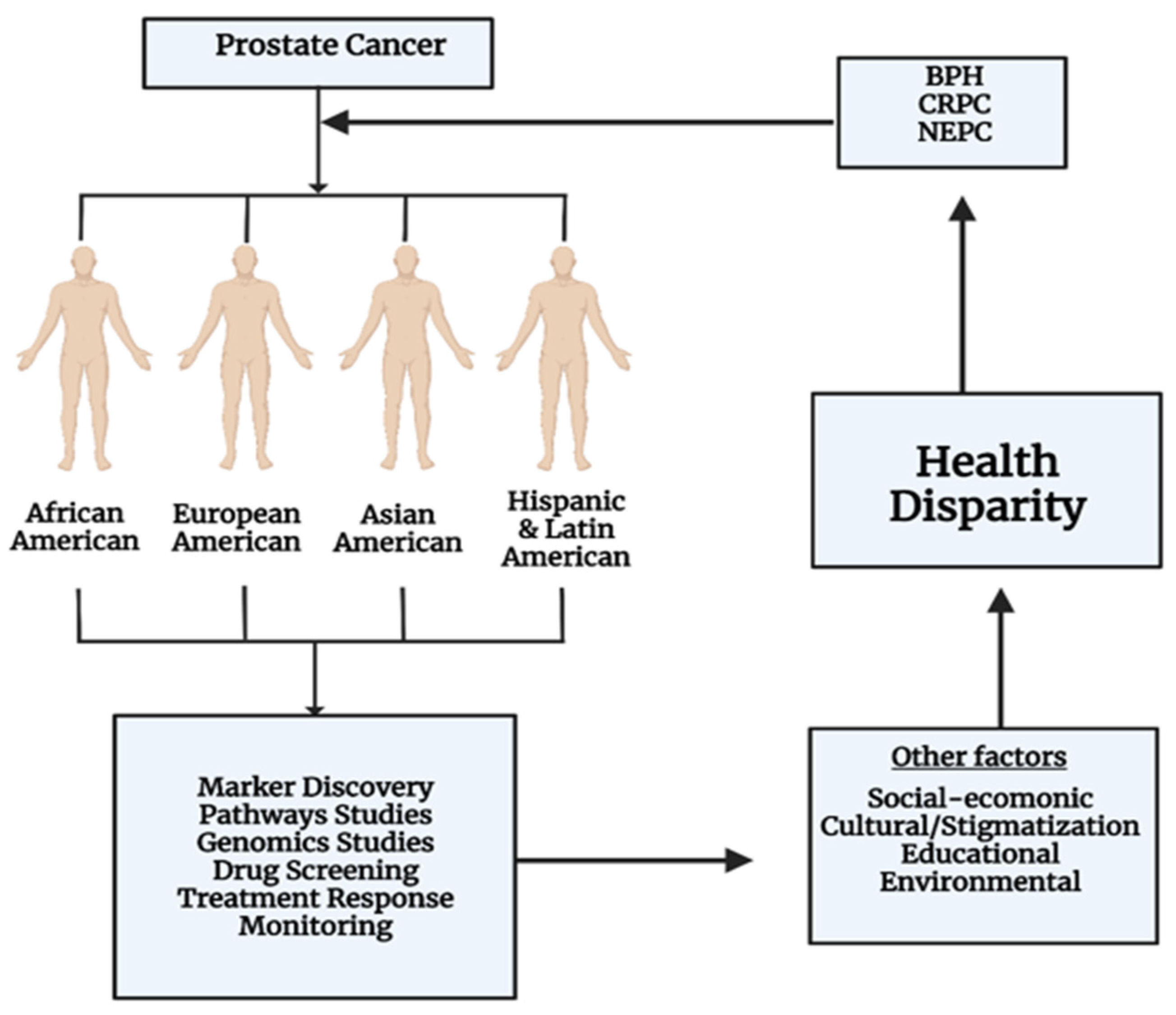
5.6. Health Disparities
5.7. Drug Discovery and Precision Medicine for PCa
5.8. Other Applications of CR
6. Limitations and Future Aspects
7. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Siegel, R.L.; Giaquinto, A.N.; Jemal, A. Cancer statistics, 2024. CA Cancer J. Clin. 2024, 74, 12–49. [Google Scholar] [CrossRef] [PubMed]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Hinata, N.; Fujisawa, M. Racial Differences in Prostate Cancer Characteristics and Cancer-Specific Mortality: An Overview. World J. Men’s Health 2022, 40, 217–227. [Google Scholar] [CrossRef]
- Carceles-Cordon, M.; Kelly, W.K.; Gomella, L.; Knudsen, K.E.; Rodriguez-Bravo, V.; Domingo-Domenech, J. Cellular rewiring in lethal prostate cancer: The architect of drug resistance. Nat. Rev. Urol. 2020, 17, 292–307. [Google Scholar] [CrossRef] [PubMed]
- Quintanal-Villalonga, A.; Chan, J.M.; Yu, H.A.; Pe’er, D.; Sawyers, C.L.; Sen, T.; Rudin, C.M. Lineage plasticity in cancer: A shared pathway of therapeutic resistance. Nat. Rev. Clin. Oncol. 2020, 17, 360–371. [Google Scholar] [CrossRef]
- Zhong, M.; Fu, L. Culture and application of conditionally reprogrammed primary tumor cells. Gastroenterol. Rep. 2020, 8, 224–233. [Google Scholar] [CrossRef]
- Zhao, R.; Li, R.; An, T.; Liu, X. Conditional Cell Reprogramming in Modeling Digestive System Diseases. Front. Cell Dev. Biol. 2021, 9, 669756. [Google Scholar] [CrossRef]
- Wu, X.; Wang, S.; Li, M.; Li, J.; Shen, J.; Zhao, Y.; Pang, J.; Wen, Q.; Chen, M.; Wei, B.; et al. Conditional reprogramming: Next generation cell culture. Acta Pharm. Sin. B 2020, 10, 1360–1381. [Google Scholar] [CrossRef] [PubMed]
- Martinovich, K.M.; Iosifidis, T.; Buckley, A.G.; Looi, K.; Ling, K.-M.; Sutanto, E.N.; Kicic-Starcevich, E.; Garratt, L.W.; Shaw, N.C.; Montgomery, S.; et al. Conditionally reprogrammed primary airway epithelial cells maintain morphology, lineage and disease specific functional characteristics. Sci. Rep. 2017, 7, 17971. [Google Scholar] [CrossRef]
- Moya, L.; Walpole, C.; Rae, F.; Srinivasan, S.; Seim, I.; Lai, J.; Nicol, D.; Williams, E.D.; Clements, J.A.; Batra, J. Characterisation of cell lines derived from prostate cancer patients with localised disease. Prostate Cancer Prostatic Dis. 2023, 26, 614–624. [Google Scholar] [CrossRef]
- Ebhardt, H.A.; Root, A.; Liu, Y.; Gauthier, N.P.; Sander, C.; Aebersold, R. Systems pharmacology using mass spectrometry identifies critical response nodes in prostate cancer. NPJ Syst. Biol. Appl. 2018, 4, 26. [Google Scholar] [CrossRef]
- Xue, J.; Mo, H.; Tian, Y.; Tang, R.; Wu, B. Chapter 15—Tryptophan fluorescence and machine learning to study. In Biophotonics, Tryptophan and Disease; Sordillo, L.A., Sordillo, P.P., Eds.; Academic Press: Cambridge, MA, USA, 2022; pp. 173–183. [Google Scholar] [CrossRef]
- Namekawa, T.; Ikeda, K.; Horie-Inoue, K.; Inoue, S. Application of Prostate Cancer Models for Preclinical Study: Advantages and Limitations of Cell Lines, Patient-Derived Xenografts, and Three-Dimensional Culture of Patient-Derived Cells. Cells 2019, 8, 74. [Google Scholar] [CrossRef]
- Smith, R.; Liu, M.; Liby, T.; Bayani, N.; Bucher, E.; Chiotti, K.; Derrick, D.; Chauchereau, A.; Heiser, L.; Alumkal, J. Enzalutamide response in a panel of prostate cancer cell lines reveals a role for glucocorticoid receptor in enzalutamide resistant disease. Sci. Rep. 2020, 10, 21750. [Google Scholar] [CrossRef]
- Frame, F.M.; Noble, A.R.; O’Toole, P.; Marrison, J.; Godden, T.; O’Brien, A.; Maitland, N.J. Assessing the Advantages, Limitations and Potential of Human Primary Prostate Epithelial Cells as a Pre-clinical Model for Prostate Cancer Research. Adv. Exp. Med. Biol. 2019, 1164, 109–118. [Google Scholar] [CrossRef]
- Tang, D.G. Understanding and targeting prostate cancer cell heterogeneity and plasticity. Semin. Cancer Biol. 2022, 82, 68–93. [Google Scholar] [CrossRef] [PubMed]
- Bishr, M.; Saad, F. Overview of the latest treatments for castration-resistant prostate cancer. Nat. Rev. Urol. 2013, 10, 522–528. [Google Scholar] [CrossRef]
- Idrisova, K.F.; Simon, H.U.; Gomzikova, M.O. Role of Patient-Derived Models of Cancer in Translational Oncology. Cancers 2022, 15, 139. [Google Scholar] [CrossRef] [PubMed]
- Kretzschmar, K.; Clevers, H. Organoids: Modeling Development and the Stem Cell Niche in a Dish. Dev. Cell 2016, 38, 590–600. [Google Scholar] [CrossRef] [PubMed]
- Sato, T.; Vries, R.G.; Snippert, H.J.; van de Wetering, M.; Barker, N.; Stange, D.E.; van Es, J.H.; Abo, A.; Kujala, P.; Peters, P.J.; et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature 2009, 459, 262–265. [Google Scholar] [CrossRef]
- Liu, X.; Krawczyk, E.; Suprynowicz, F.A.; Palechor-Ceron, N.; Yuan, H.; Dakic, A.; Simic, V.; Zheng, Y.L.; Sripadhan, P.; Chen, C.; et al. Conditional reprogramming and long-term expansion of normal and tumor cells from human biospecimens. Nat. Protoc. 2017, 12, 439–451. [Google Scholar] [CrossRef]
- Palechor-Ceron, N.; Krawczyk, E.; Dakic, A.; Simic, V.; Yuan, H.; Blancato, J.; Wang, W.; Hubbard, F.; Zheng, Y.L.; Dan, H.; et al. Conditional Reprogramming for Patient-Derived Cancer Models and Next-Generation Living Biobanks. Cells 2019, 8, 1327. [Google Scholar] [CrossRef]
- Shtivelman, E.; Beer, T.M.; Evans, C.P. Molecular pathways and targets in prostate cancer. Oncotarget 2014, 5, 7217. [Google Scholar] [CrossRef] [PubMed]
- da Silva, H.B.; Amaral, E.P.; Nolasco, E.L.; de Victo, N.C.; Atique, R.; Jank, C.C.; Anschau, V.; Zerbini, L.F.; Correa, R.G. Dissecting major signaling pathways throughout the development of prostate cancer. Prostate Cancer 2013, 2013, 920612. [Google Scholar] [CrossRef]
- Gandhi, J.; Afridi, A.; Vatsia, S.; Joshi, G.; Joshi, G.; Kaplan, S.A.; Smith, N.L.; Khan, S.A. The molecular biology of prostate cancer: Current understanding and clinical implications. Prostate Cancer Prostatic Dis. 2018, 21, 22–36. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Ory, V.; Chapman, S.; Yuan, H.; Albanese, C.; Kallakury, B.; Timofeeva, O.A.; Nealon, C.; Dakic, A.; Simic, V. ROCK inhibitor and feeder cells induce the conditional reprogramming of epithelial cells. Am. J. Pathol. 2012, 180, 599–607. [Google Scholar] [CrossRef] [PubMed]
- Suprynowicz, F.A.; Upadhyay, G.; Krawczyk, E.; Kramer, S.C.; Hebert, J.D.; Liu, X.; Yuan, H.; Cheluvaraju, C.; Clapp, P.W.; Boucher, R.C., Jr.; et al. Conditionally reprogrammed cells represent a stem-like state of adult epithelial cells. Proc. Natl. Acad. Sci. USA 2012, 109, 20035–20040. [Google Scholar] [CrossRef]
- Daneshdoust, D.; Yin, M.; Luo, M.; Sundi, D.; Dang, Y.; Lee, C.; Li, J.; Liu, X. Conditional Reprogramming Modeling of Bladder Cancer for Clinical Translation. Cells 2023, 12, 1714. [Google Scholar] [CrossRef]
- Liu, W.; Ju, L.; Cheng, S.; Wang, G.; Qian, K.; Liu, X.; Xiao, Y.; Wang, X. Conditional reprogramming: Modeling urological cancer and translation to clinics. Clin. Transl. Med. 2020, 10, e95. [Google Scholar] [CrossRef]
- Cao, J.; Chan, W.C.; Chow, M.S. Use of conditional reprogramming cell, patient derived xenograft and organoid for drug screening for individualized prostate cancer therapy: Current and future perspectives. Int. J. Oncol. 2022, 60, 52. [Google Scholar] [CrossRef]
- Alamri, A.M.; Kang, K.; Groeneveld, S.; Wang, W.; Zhong, X.; Kallakury, B.; Hennighausen, L.; Liu, X.; Furth, P.A. Primary cancer cell culture: Mammary-optimized vs. conditional reprogramming. Endocr. Relat. Cancer 2016, 23, 535–554. [Google Scholar] [CrossRef]
- Liu, X.; Wu, Y.; Rong, L. Conditionally Reprogrammed Human Normal Airway Epithelial Cells at ALI: A Physiological Model for Emerging Viruses. Virol. Sin. 2020, 35, 280–289. [Google Scholar] [CrossRef] [PubMed]
- Shen, M.M.; Abate-Shen, C. Molecular genetics of prostate cancer: New prospects for old challenges. Genes. Dev. 2010, 24, 1967–2000. [Google Scholar] [CrossRef] [PubMed]
- Mei, W.; Lin, X.; Kapoor, A.; Gu, Y.; Zhao, K.; Tang, D. The contributions of prostate cancer stem cells in prostate cancer initiation and metastasis. Cancers 2019, 11, 434. [Google Scholar] [CrossRef] [PubMed]
- Sadeghi, M.; Barzegar, A. Precision medicine insight into primary prostate tumor through transcriptomic data and an integrated systems biology approach. Meta Gene 2020, 26, 100787. [Google Scholar] [CrossRef]
- Robinson, D.; Van Allen, E.M.; Wu, Y.M.; Schultz, N.; Lonigro, R.J.; Mosquera, J.M.; Montgomery, B.; Taplin, M.E.; Pritchard, C.C.; Attard, G.; et al. Integrative clinical genomics of advanced prostate cancer. Cell 2015, 161, 1215–1228. [Google Scholar] [CrossRef] [PubMed]
- Fraser, M.; Sabelnykova, V.Y.; Yamaguchi, T.N.; Heisler, L.E.; Livingstone, J.; Huang, V.; Shiah, Y.J.; Yousif, F.; Lin, X.; Masella, A.P.; et al. Genomic hallmarks of localized, non-indolent prostate cancer. Nature 2017, 541, 359–364. [Google Scholar] [CrossRef] [PubMed]
- Mehra, R.; Kumar-Sinha, C.; Shankar, S.; Lonigro, R.J.; Jing, X.; Philips, N.E.; Siddiqui, J.; Han, B.; Cao, X.; Smith, D.C.; et al. Characterization of bone metastases from rapid autopsies of prostate cancer patients. Clin. Cancer Res. 2011, 17, 3924–3932. [Google Scholar] [CrossRef]
- Van Allen, E.M.; Foye, A.; Wagle, N.; Kim, W.; Carter, S.L.; McKenna, A.; Simko, J.P.; Garraway, L.A.; Febbo, P.G. Successful whole-exome sequencing from a prostate cancer bone metastasis biopsy. Prostate Cancer Prostatic Dis. 2014, 17, 23–27. [Google Scholar] [CrossRef]
- Ci, X.; Hao, J.; Dong, X.; Xue, H.; Wu, R.; Choi, S.Y.C.; Haegert, A.M.; Collins, C.C.; Liu, X.; Lin, D.; et al. Conditionally Reprogrammed Cells from Patient-Derived Xenograft to Model Neuroendocrine Prostate Cancer Development. Cells 2020, 9, 1398. [Google Scholar] [CrossRef]
- Schrecengost, R.; Knudsen, K.E. Molecular pathogenesis and progression of prostate cancer. Semin. Oncol. 2013, 40, 244–258. [Google Scholar] [CrossRef]
- Choudhary, S.; Ramasundaram, P.; Dziopa, E.; Mannion, C.; Kissin, Y.; Tricoli, L.; Albanese, C.; Lee, W.; Zilberberg, J. Human ex vivo 3D bone model recapitulates osteocyte response to metastatic prostate cancer. Sci. Rep. 2018, 8, 17975. [Google Scholar] [CrossRef] [PubMed]
- Jung, M.; Kowalczyk, K.; Hankins, R.; Bandi, G.; Kallakury, B.; Carrasquilla, M.A.; Banerjee, P.P.; Grindrod, S.; Dritschilo, A. Novel Paired Normal Prostate and Prostate Cancer Model Cell Systems Derived from African American Patients. Cancer Res. Commun. 2022, 2, 1617–1625. [Google Scholar] [CrossRef] [PubMed]
- Hata, J.; Machida, T.; Matsuoka, K.; Hoshi, S.; Akaihata, H.; Hiraki, H.; Suzuki, T.; Ogawa, S.; Kataoka, M.; Haga, N.; et al. Complement activation by autoantigen recognition in the growth process of benign prostatic hyperplasia. Sci. Rep. 2019, 9, 20357. [Google Scholar] [CrossRef] [PubMed]
- Lorenzo, G.; Hughes, T.J.R.; Dominguez-Frojan, P.; Reali, A.; Gomez, H. Computer simulations suggest that prostate enlargement due to benign prostatic hyperplasia mechanically impedes prostate cancer growth. Proc. Natl. Acad. Sci. USA 2019, 116, 1152–1161. [Google Scholar] [CrossRef] [PubMed]
- Shah, A.; Shah, A.A.; K, N.; Lobo, R. Mechanistic targets for BPH and prostate cancer—A review. Rev. Environ. Health 2021, 36, 261–270. [Google Scholar] [CrossRef] [PubMed]
- Nicholson, T.M.; Ricke, W.A. Androgens and estrogens in benign prostatic hyperplasia: Past, present and future. Differentiation 2011, 82, 184–199. [Google Scholar] [CrossRef] [PubMed]
- Hendriksen, P.J.; Dits, N.F.; Kokame, K.; Veldhoven, A.; van Weerden, W.M.; Bangma, C.H.; Trapman, J.; Jenster, G. Evolution of the androgen receptor pathway during progression of prostate cancer. Cancer Res. 2006, 66, 5012–5020. [Google Scholar] [CrossRef] [PubMed]
- Minutoli, L.; Rinaldi, M.; Marini, H.; Irrera, N.; Crea, G.; Lorenzini, C.; Puzzolo, D.; Valenti, A.; Pisani, A.; Adamo, E.B.; et al. Apoptotic Pathways Linked to Endocrine System as Potential Therapeutic Targets for Benign Prostatic Hyperplasia. Int. J. Mol. Sci. 2016, 17, 1311. [Google Scholar] [CrossRef] [PubMed]
- Soler, R.; Andersson, K.E.; Chancellor, M.B.; Chapple, C.R.; de Groat, W.C.; Drake, M.J.; Gratzke, C.; Lee, R.; Cruz, F. Future direction in pharmacotherapy for non-neurogenic male lower urinary tract symptoms. Eur. Urol. 2013, 64, 610–621. [Google Scholar] [CrossRef]
- Hammarsten, J.; Hogstedt, B. Clinical, haemodynamic, anthropometric, metabolic and insulin profile of men with high-stage and high-grade clinical prostate cancer. Blood Press. 2004, 13, 47–55. [Google Scholar] [CrossRef]
- Udensi, U.K.; Tchounwou, P.B. Oxidative stress in prostate hyperplasia and carcinogenesis. J. Exp. Clin. Cancer Res. 2016, 35, 139. [Google Scholar] [CrossRef] [PubMed]
- Holder, K.G.; Galvan, B.; Knight, A.S.; Ha, F.; Collins, R.; Weaver, P.E.; Brandi, L.; de Riese, W.T. Possible clinical implications of prostate capsule thickness and glandular epithelial cell density in benign prostate hyperplasia. Investig. Clin. Urol. 2021, 62, 423–429. [Google Scholar] [CrossRef] [PubMed]
- McNally, C.J.; Ruddock, M.W.; Moore, T.; McKenna, D.J. Biomarkers That Differentiate Benign Prostatic Hyperplasia from Prostate Cancer: A Literature Review. Cancer Manag. Res. 2020, 12, 5225–5241. [Google Scholar] [CrossRef] [PubMed]
- Jin, B.-R.; Kim, H.-J.; Na, J.-H.; Lee, W.-K.; An, H.-J. Targeting benign prostate hyperplasia treatments: AR/TGF-β/NOX4 inhibition by apocynin suppresses inflammation and proliferation. J. Adv. Res. 2024, 57, 135–147. [Google Scholar] [CrossRef] [PubMed]
- Long, Z.; Deng, L.; Li, C.; He, Q.; He, Y.; Hu, X.; Cai, Y.; Gan, Y. Loss of EHF facilitates the development of treatment-induced neuroendocrine prostate cancer. Cell Death Dis. 2021, 12, 46. [Google Scholar] [CrossRef]
- Liu, S.; Alabi, B.R.; Yin, Q.; Stoyanova, T. Molecular mechanisms underlying the development of neuroendocrine prostate cancer. Semin. Cancer Biol. 2022, 86 Pt 3, 57–68. [Google Scholar] [CrossRef] [PubMed]
- Vlachostergios, P.J.; Puca, L.; Beltran, H. Emerging Variants of Castration-Resistant Prostate Cancer. Curr. Oncol. Rep. 2017, 19, 32. [Google Scholar] [CrossRef] [PubMed]
- Yamada, Y.; Beltran, H. Clinical and Biological Features of Neuroendocrine Prostate Cancer. Curr. Oncol. Rep. 2021, 23, 15. [Google Scholar] [CrossRef] [PubMed]
- Timofeeva, O.A.; Palechor-Ceron, N.; Li, G.; Yuan, H.; Krawczyk, E.; Zhong, X.; Liu, G.; Upadhyay, G.; Dakic, A.; Yu, S.; et al. Conditionally reprogrammed normal and primary tumor prostate epithelial cells: A novel patient-derived cell model for studies of human prostate cancer. Oncotarget 2017, 8, 22741–22758. [Google Scholar] [CrossRef] [PubMed]
- Tricoli, L.; Naeem, A.; Parasido, E.; Mikhaiel, J.P.; Choudhry, M.U.; Berry, D.L.; Abdelgawad, I.A.; Lee, R.J.; Feldman, A.S.; Ihemelandu, C.; et al. Characterization of the effects of defined, multidimensional culture conditions on conditionally reprogrammed primary human prostate cells. Oncotarget 2018, 9, 2193–2207. [Google Scholar] [CrossRef]
- Pedersen, E.A.; Shiozawa, Y.; Pienta, K.J.; Taichman, R.S. The prostate cancer bone marrow niche: More than just ‘fertile soil’. Asian J. Androl. 2012, 14, 423–427. [Google Scholar] [CrossRef] [PubMed]
- Ubaidus, S.; Li, M.; Sultana, S.; de Freitas, P.H.; Oda, K.; Maeda, T.; Takagi, R.; Amizuka, N. FGF23 is mainly synthesized by osteocytes in the regularly distributed osteocytic lacunar canalicular system established after physiological bone remodeling. J. Electron Microsc 2009, 58, 381–392. [Google Scholar] [CrossRef] [PubMed]
- Muller, P.A.; Vousden, K.H. Mutant p53 in cancer: New functions and therapeutic opportunities. Cancer Cell 2014, 25, 304–317. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.Y.; Karsli-Uzunbas, G.; Mathew, R.; Aisner, S.C.; Kamphorst, J.J.; Strohecker, A.M.; Chen, G.; Price, S.; Lu, W.; Teng, X.; et al. Autophagy suppresses progression of K-ras-induced lung tumors to oncocytomas and maintains lipid homeostasis. Genes Dev. 2013, 27, 1447–1461. [Google Scholar] [CrossRef]
- Guo, J.Y.; Xia, B.; White, E. Autophagy-mediated tumor promotion. Cell 2013, 155, 1216–1219. [Google Scholar] [CrossRef] [PubMed]
- Tasdemir, E.; Chiara Maiuri, M.; Morselli, E.; Criollo, A.; D’Amelio, M.; Djavaheri-Mergny, M.; Cecconi, F.; Tavernarakis, N.; Kroemer, G. A dual role of p53 in the control of autophagy. Autophagy 2008, 4, 810–814. [Google Scholar] [CrossRef] [PubMed]
- Nikoletopoulou, V.; Markaki, M.; Palikaras, K.; Tavernarakis, N. Crosstalk between apoptosis, necrosis and autophagy. Biochim. Biophys. Acta 2013, 1833, 3448–3459. [Google Scholar] [CrossRef] [PubMed]
- Ringer, L.; Sirajuddin, P.; Tricoli, L.; Waye, S.; Choudhry, M.U.; Parasido, E.; Sivakumar, A.; Heckler, M.; Naeem, A.; Abdelgawad, I.; et al. The induction of the p53 tumor suppressor protein bridges the apoptotic and autophagic signaling pathways to regulate cell death in prostate cancer cells. Oncotarget 2014, 5, 10678–10691. [Google Scholar] [CrossRef]
- Liu, C.; Lou, W.; Yang, J.C.; Liu, L.; Armstrong, C.M.; Lombard, A.P.; Zhao, R.; Noel, O.D.V.; Tepper, C.G.; Chen, H.W.; et al. Proteostasis by STUB1/HSP70 complex controls sensitivity to androgen receptor targeted therapy in advanced prostate cancer. Nat. Commun. 2018, 9, 4700. [Google Scholar] [CrossRef]
- Yang, J.C.; Xu, P.; Ning, S.; Wasielewski, L.J.; Adomat, H.; Hwang, S.H.; Morisseau, C.; Gleave, M.; Corey, E.; Gao, A.C.; et al. Novel inhibition of AKR1C3 and androgen receptor axis by PTUPB synergizes enzalutamide treatment in advanced prostate cancer. Oncogene 2023, 42, 693–707. [Google Scholar] [CrossRef]
- Ather, M.H.; Siddiqui, T. The genetics of neuroendocrine prostate cancers: A review of current and emerging candidates. Appl. Clin. Genet. 2012, 5, 105–110. [Google Scholar] [CrossRef] [PubMed]
- Parimi, V.; Goyal, R.; Poropatich, K.; Yang, X.J. Neuroendocrine differentiation of prostate cancer: A review. Am. J. Clin. Exp. Urol. 2014, 2, 273–285. [Google Scholar] [PubMed]
- Gupta, K.; Gupta, S. Neuroendocrine differentiation in prostate cancer: Key epigenetic players. Transl. Cancer Res. 2017, 6 (Suppl. S1), S104–S108. [Google Scholar] [CrossRef] [PubMed]
- Komiya, A.; Yasuda, K.; Watanabe, A.; Fujiuchi, Y.; Tsuzuki, T.; Fuse, H. The prognostic significance of loss of the androgen receptor and neuroendocrine differentiation in prostate biopsy specimens among castration-resistant prostate cancer patients. Mol. Clin. Oncol. 2013, 1, 257–262. [Google Scholar] [CrossRef] [PubMed]
- Ather, M.H.; Abbas, F.; Faruqui, N.; Israr, M.; Pervez, S. Correlation of three immunohistochemically detected markers of neuroendocrine differentiation with clinical predictors of disease progression in prostate cancer. BMC Urol. 2008, 8, 21. [Google Scholar] [CrossRef] [PubMed]
- Bostwick, D.G.; Qian, J.; Pacelli, A.; Zincke, H.; Blute, M.; Bergstralh, E.J.; Slezak, J.M.; Cheng, L. Neuroendocrine expression in node positive prostate cancer: Correlation with systemic progression and patient survival. J. Urol. 2002, 168, 1204–1211. [Google Scholar] [CrossRef] [PubMed]
- Lin, D.; Wyatt, A.W.; Xue, H.; Wang, Y.; Dong, X.; Haegert, A.; Wu, R.; Brahmbhatt, S.; Mo, F.; Jong, L.; et al. High fidelity patient-derived xenografts for accelerating prostate cancer discovery and drug development. Cancer Res. 2014, 74, 1272–1283. [Google Scholar] [CrossRef] [PubMed]
- Chornokur, G.; Dalton, K.; Borysova, M.E.; Kumar, N.B. Disparities at presentation, diagnosis, treatment, and survival in African American men, affected by prostate cancer. Prostate 2011, 71, 985–997. [Google Scholar] [CrossRef] [PubMed]
- Lillard, J.W., Jr.; Moses, K.A.; Mahal, B.A.; George, D.J. Racial disparities in Black men with prostate cancer: A literature review. Cancer 2022, 128, 3787–3795. [Google Scholar] [CrossRef]
- Rais-Bahrami, S.; Zhu, Y. Disparities in prostate cancer diagnosis and management: Recognizing that disparities exist at all junctures along the prostate cancer journey. Prostate Cancer Prostatic Dis. 2023, 26, 441–442. [Google Scholar] [CrossRef]
- Mahal, B.A.; Gerke, T.; Awasthi, S.; Soule, H.R.; Simons, J.W.; Miyahira, A.; Halabi, S.; George, D.; Platz, E.A.; Mucci, L.; et al. Prostate cancer racial disparities: A systematic review by the prostate cancer foundation panel. Eur. Urol. Oncol. 2022, 5, 18–29. [Google Scholar] [CrossRef] [PubMed]
- Lynch, S.M.; Sorice, K.; Tagai, E.K.; Handorf, E.A. Use of empiric methods to inform prostate cancer health disparities: Comparison of neighborhood-wide association study “hits” in black and white men. Cancer 2020, 126, 1949–1957. [Google Scholar] [CrossRef] [PubMed]
- Wall, N.R.; Fuller, R.N.; Morcos, A.; De Leon, M. Pancreatic Cancer Health Disparity: Pharmacologic Anthropology. Cancers 2023, 15, 5070. [Google Scholar] [CrossRef] [PubMed]
- Nicolas, N.; Upadhyay, G.; Velena, A.; Kallakury, B.; Rhim, J.S.; Dritschilo, A.; Jung, M. African-American Prostate Normal and Cancer Cells for Health Disparities Research. Adv. Exp. Med. Biol. 2019, 1164, 101–108. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.M.; Cieslik, M.; Lonigro, R.J.; Vats, P.; Reimers, M.A.; Cao, X.; Ning, Y.; Wang, L.; Kunju, L.P.; de Sarkar, N.; et al. Inactivation of CDK12 Delineates a Distinct Immunogenic Class of Advanced Prostate Cancer. Cell 2018, 173, 1770–1782. [Google Scholar] [CrossRef] [PubMed]
- Qian, K.; Wang, G.; Ju, L.; Liu, J.; Luo, Y.; Wang, Y.; Peng, T.; Chen, F.; Zhang, Y.; Xiao, Y.; et al. A novel germline EGFR variant p.R831H causes predisposition to familial CDK12-mutant prostate cancer with tandem duplicator phenotype. Oncogene 2020, 39, 6871–6878. [Google Scholar] [CrossRef] [PubMed]
- Antonarakis, E.S.; Isaacsson Velho, P.; Fu, W.; Wang, H.; Agarwal, N.; Sacristan Santos, V.; Maughan, B.L.; Pili, R.; Adra, N.; Sternberg, C.N.; et al. CDK12-Altered Prostate Cancer: Clinical Features and Therapeutic Outcomes to Standard Systemic Therapies, Poly (ADP-Ribose) Polymerase Inhibitors, and PD-1 Inhibitors. JCO Precis. Oncol. 2020, 4, 370–381. [Google Scholar] [CrossRef] [PubMed]
- Meacham, C.E.; Morrison, S.J. Tumour heterogeneity and cancer cell plasticity. Nature 2013, 501, 328–337. [Google Scholar] [CrossRef] [PubMed]
- Kruglyak, L.; Nickerson, D.A. Variation is the spice of life. Nat. Genet. 2001, 27, 234–236. [Google Scholar] [CrossRef]
- Abida, W.; Cheng, M.L.; Armenia, J.; Middha, S.; Autio, K.A.; Vargas, H.A.; Rathkopf, D.; Morris, M.J.; Danila, D.C.; Slovin, S.F.; et al. Analysis of the Prevalence of Microsatellite Instability in Prostate Cancer and Response to Immune Checkpoint Blockade. JAMA Oncol. 2019, 5, 471–478. [Google Scholar] [CrossRef]
- Casak, S.J.; Marcus, L.; Fashoyin-Aje, L.; Mushti, S.L.; Cheng, J.; Shen, Y.L.; Pierce, W.F.; Her, L.; Goldberg, K.B.; Theoret, M.R.; et al. FDA Approval Summary: Pembrolizumab for the First-line Treatment of Patients with MSI-H/dMMR Advanced Unresectable or Metastatic Colorectal Carcinoma. Clin. Cancer Res. 2021, 27, 4680–4684. [Google Scholar] [CrossRef] [PubMed]
- Marcus, L.; Fashoyin-Aje, L.A.; Donoghue, M.; Yuan, M.; Rodriguez, L.; Gallagher, P.S.; Philip, R.; Ghosh, S.; Theoret, M.R.; Beaver, J.A.; et al. FDA Approval Summary: Pembrolizumab for the Treatment of Tumor Mutational Burden-High Solid Tumors. Clin. Cancer Res. 2021, 27, 4685–4689. [Google Scholar] [CrossRef] [PubMed]
- Alkhilaiwi, F. Conditionally Reprogrammed Cells and Robotic High-Throughput Screening for Precision Cancer Therapy. Front. Oncol. 2021, 11, 761986. [Google Scholar] [CrossRef] [PubMed]
- Hawkey, N.M.; Broderick, A.; George, D.J.; Sartor, O.; Armstrong, A.J. The Value of Phenotypic Precision Medicine in Prostate Cancer. Oncologist 2022, 28, 93–104. [Google Scholar] [CrossRef] [PubMed]
- Zeevi, D.; Korem, T.; Zmora, N.; Israeli, D.; Rothschild, D.; Weinberger, A.; Ben-Yacov, O.; Lador, D.; Avnit-Sagi, T.; Lotan-Pompan, M.; et al. Personalized Nutrition by Prediction of Glycemic Responses. Cell 2015, 163, 1079–1094. [Google Scholar] [CrossRef] [PubMed]
- Yuan, H.; Myers, S.; Wang, J.; Zhou, D.; Woo, J.A.; Kallakury, B.; Ju, A.; Bazylewicz, M.; Carter, Y.M.; Albanese, C.; et al. Use of reprogrammed cells to identify therapy for respiratory papillomatosis. N. Engl. J. Med. 2012, 367, 1220–1227. [Google Scholar] [CrossRef] [PubMed]
- Saeed, K.; Rahkama, V.; Eldfors, S.; Bychkov, D.; Mpindi, J.P.; Yadav, B.; Paavolainen, L.; Aittokallio, T.; Heckman, C.; Wennerberg, K.; et al. Comprehensive Drug Testing of Patient-derived Conditionally Reprogrammed Cells from Castration-resistant Prostate Cancer. Eur. Urol. 2017, 71, 319–327. [Google Scholar] [CrossRef] [PubMed]
- Bonollo, F.; Thalmann, G.N.; Kruithof-de Julio, M.; Karkampouna, S. The role of cancer-associated fibroblasts in prostate cancer tumorigenesis. Cancers 2020, 12, 1887. [Google Scholar] [CrossRef] [PubMed]
- Moffat, J.G.; Rudolph, J.; Bailey, D. Phenotypic screening in cancer drug discovery—Past, present and future. Nat. Rev. Drug Discov. 2014, 13, 588–602. [Google Scholar] [CrossRef]
- Lee, J.A.; Uhlik, M.T.; Moxham, C.M.; Tomandl, D.; Sall, D.J. Modern phenotypic drug discovery is a viable, neoclassic pharma strategy. J. Med. Chem. 2012, 55, 4527–4538. [Google Scholar] [CrossRef]
- Swinney, D.C. Phenotypic vs. target-based drug discovery for first-in-class medicines. Clin. Pharmacol. Ther. 2013, 93, 299–301. [Google Scholar] [CrossRef] [PubMed]
- Fang, Y. Label-free drug discovery. Front. Pharmacol. 2014, 5, 52. [Google Scholar] [CrossRef] [PubMed]
- Eggert, U.S. The why and how of phenotypic small-molecule screens. Nat. Chem. Biol. 2013, 9, 206–209. [Google Scholar] [CrossRef] [PubMed]
- Hillger, J.M.; Lieuw, W.L.; Heitman, L.H.; AP, I.J. Label-free technology and patient cells: From early drug development to precision medicine. Drug Discov. Today 2017, 22, 1808–1815. [Google Scholar] [CrossRef] [PubMed]
- Abassi, Y.A.; Xi, B.; Zhang, W.; Ye, P.; Kirstein, S.L.; Gaylord, M.R.; Feinstein, S.C.; Wang, X.; Xu, X. Kinetic cell-based morphological screening: Prediction of mechanism of compound action and off-target effects. Chem. Biol. 2009, 16, 712–723. [Google Scholar] [CrossRef]
- Dowling, C.M.; Herranz Ors, C.; Kiely, P.A. Using real-time impedance-based assays to monitor the effects of fibroblast-derived media on the adhesion, proliferation, migration and invasion of colon cancer cells. Biosci. Rep. 2014, 34, 415–427. [Google Scholar] [CrossRef] [PubMed]
- Fajardo, C.A.; Guedan, S.; Rojas, L.A.; Moreno, R.; Arias-Badia, M.; de Sostoa, J.; June, C.H.; Alemany, R. Oncolytic Adenoviral Delivery of an EGFR-Targeting T-cell Engager Improves Antitumor Efficacy. Cancer Res. 2017, 77, 2052–2063. [Google Scholar] [CrossRef]
- Scott, C.W.; Peters, M.F. Label-free whole-cell assays: Expanding the scope of GPCR screening. Drug Discov. Today 2010, 15, 704–716. [Google Scholar] [CrossRef]
- Gibson, C.C.; Zhu, W.; Davis, C.T.; Bowman-Kirigin, J.A.; Chan, A.C.; Ling, J.; Walker, A.E.; Goitre, L.; Delle Monache, S.; Retta, S.F.; et al. Strategy for identifying repurposed drugs for the treatment of cerebral cavernous malformation. Circulation 2015, 131, 289–299. [Google Scholar] [CrossRef]
- Kho, D.; MacDonald, C.; Johnson, R.; Unsworth, C.P.; O’Carroll, S.J.; du Mez, E.; Angel, C.E.; Graham, E.S. Application of xCELLigence RTCA Biosensor Technology for Revealing the Profile and Window of Drug Responsiveness in Real Time. Biosensors 2015, 5, 199–222. [Google Scholar] [CrossRef]
- Tahtouh, M.; Despland, P.; Shimmon, R.; Kalman, J.R.; Reedy, B.J. The application of infrared chemical imaging to the detection and enhancement of latent fingerprints: Method optimization and further findings. J. Forensic Sci. 2007, 52, 1089–1096. [Google Scholar] [CrossRef] [PubMed]
- Atala, A. Regenerative medicine strategies. J. Pediatr. Surg. 2012, 47, 17–28. [Google Scholar] [CrossRef] [PubMed]
- Riazi, A.M.; Kwon, S.Y.; Stanford, W.L. Stem cell sources for regenerative medicine. Methods Mol. Biol. 2009, 482, 55–90. [Google Scholar] [CrossRef] [PubMed]
- Hamilton, N.J.I.; Hynds, R.E.; Gowers, K.H.C.; Tait, A.; Butler, C.R.; Hopper, C.; Burns, A.J.; Birchall, M.A.; Lowdell, M.; Janes, S.M. Using a Three-Dimensional Collagen Matrix to Deliver Respiratory Progenitor Cells to Decellularized Trachea In Vivo. Tissue Eng. Part. C Methods 2019, 25, 93–102. [Google Scholar] [CrossRef]
- Wang, Z.; Li, Y.; Zhao, W.; Jiang, S.; Huang, Y.; Hou, J.; Zhang, X.; Zhai, Z.; Yang, C.; Wang, J. Integrative multi-omics and drug–response characterization of patient-derived prostate cancer primary cells. Signal Transduct. Target. Ther. 2023, 8, 175. [Google Scholar] [CrossRef]
| Sample Origin | Conventional Cell Lines | Primary Cells | PDX Model | 3D Organoid | CR Cells |
|---|---|---|---|---|---|
| FNA | − | − | − | −/+ | +++ |
| Core biopsy | − | + | − | + | +++ |
| Surgical specimens | + | ++ | ++ | +++ | +++ |
| Cryopreserved tissue | −/+ | +/++ | −/+ | +++ | +++ |
| Cancerous tissue | +++ | ++ | ++ | +++ | +++ |
| Noncancerous tissue | − | −/+ | − | + | +++ |
| Urine derived cells | − | − | − | − | + |
| Timing | Several days | 1 to 4 weeks | 1 to 5 months | 1 to 4 weeks | 1 to 10 days |
| Success rate | + | ++ | ++ | ++ | +++ |
| Rapid expansion | +++ | ++ | + | ++ | +++ |
| Genetic Stability | + | ++ | ++ | ++ | ++ |
| Cost | + | ++ | +++ | ++ | + |
| Life span | +++ | + | + | ++ | +++ |
| Difficulty of differentiation | +++ | + | +++ | + | + |
| Biobanking | − | + | ++ | +++ | +++ |
| Tissue-specific | + | +++ | +++ | +++ | +++ |
| Genetic manipulation | +++ | −/+ | − | ++ | ++ |
| Tumor–stromal interaction | − | − | ++ | + | − |
| Representation of primary tissue | + | ++ | ++ | ++ | ++ |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Elbialy, A.; Kappala, D.; Desai, D.; Wang, P.; Fadiel, A.; Wang, S.-J.; Makary, M.S.; Lenobel, S.; Sood, A.; Gong, M.; et al. Patient-Derived Conditionally Reprogrammed Cells in Prostate Cancer Research. Cells 2024, 13, 1005. https://doi.org/10.3390/cells13121005
Elbialy A, Kappala D, Desai D, Wang P, Fadiel A, Wang S-J, Makary MS, Lenobel S, Sood A, Gong M, et al. Patient-Derived Conditionally Reprogrammed Cells in Prostate Cancer Research. Cells. 2024; 13(12):1005. https://doi.org/10.3390/cells13121005
Chicago/Turabian StyleElbialy, Abdalla, Deepthi Kappala, Dhruv Desai, Peng Wang, Ahmed Fadiel, Shang-Jui Wang, Mina S. Makary, Scott Lenobel, Akshay Sood, Michael Gong, and et al. 2024. "Patient-Derived Conditionally Reprogrammed Cells in Prostate Cancer Research" Cells 13, no. 12: 1005. https://doi.org/10.3390/cells13121005
APA StyleElbialy, A., Kappala, D., Desai, D., Wang, P., Fadiel, A., Wang, S. -J., Makary, M. S., Lenobel, S., Sood, A., Gong, M., Dason, S., Shabsigh, A., Clinton, S., Parwani, A. V., Putluri, N., Shvets, G., Li, J., & Liu, X. (2024). Patient-Derived Conditionally Reprogrammed Cells in Prostate Cancer Research. Cells, 13(12), 1005. https://doi.org/10.3390/cells13121005







