Injection Drug Use Alters Plasma Regulation of the B Cell Response
Abstract
:1. Introduction
2. Materials and Methods
2.1. Clinical Samples
2.2. DIA-Mass Spectrometry
2.3. ELISA
2.4. In Vitro Lymphocyte Proliferation Assays
2.5. RNA Sequencing
2.6. Statistical Analysis
3. Results
3.1. Cohort Description
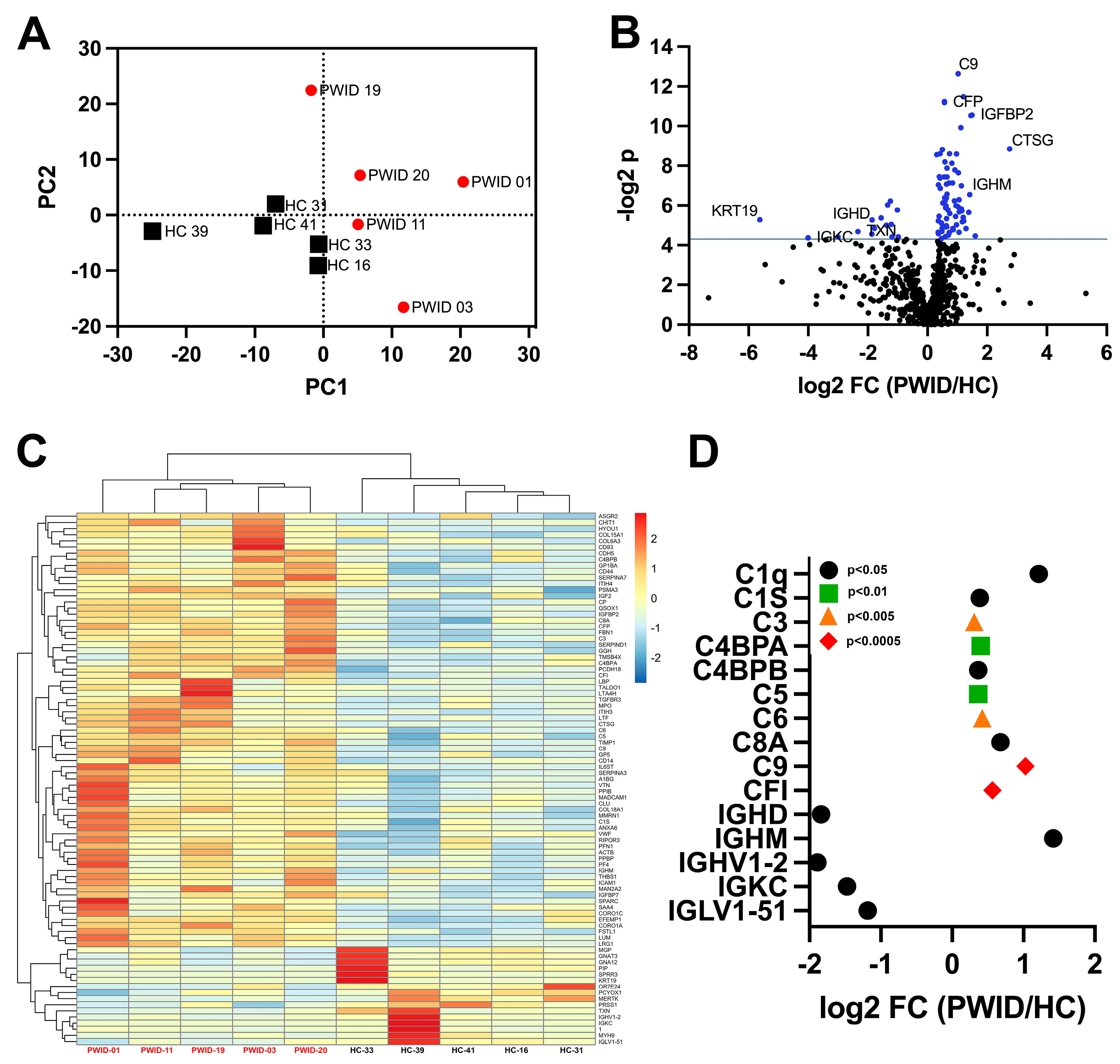
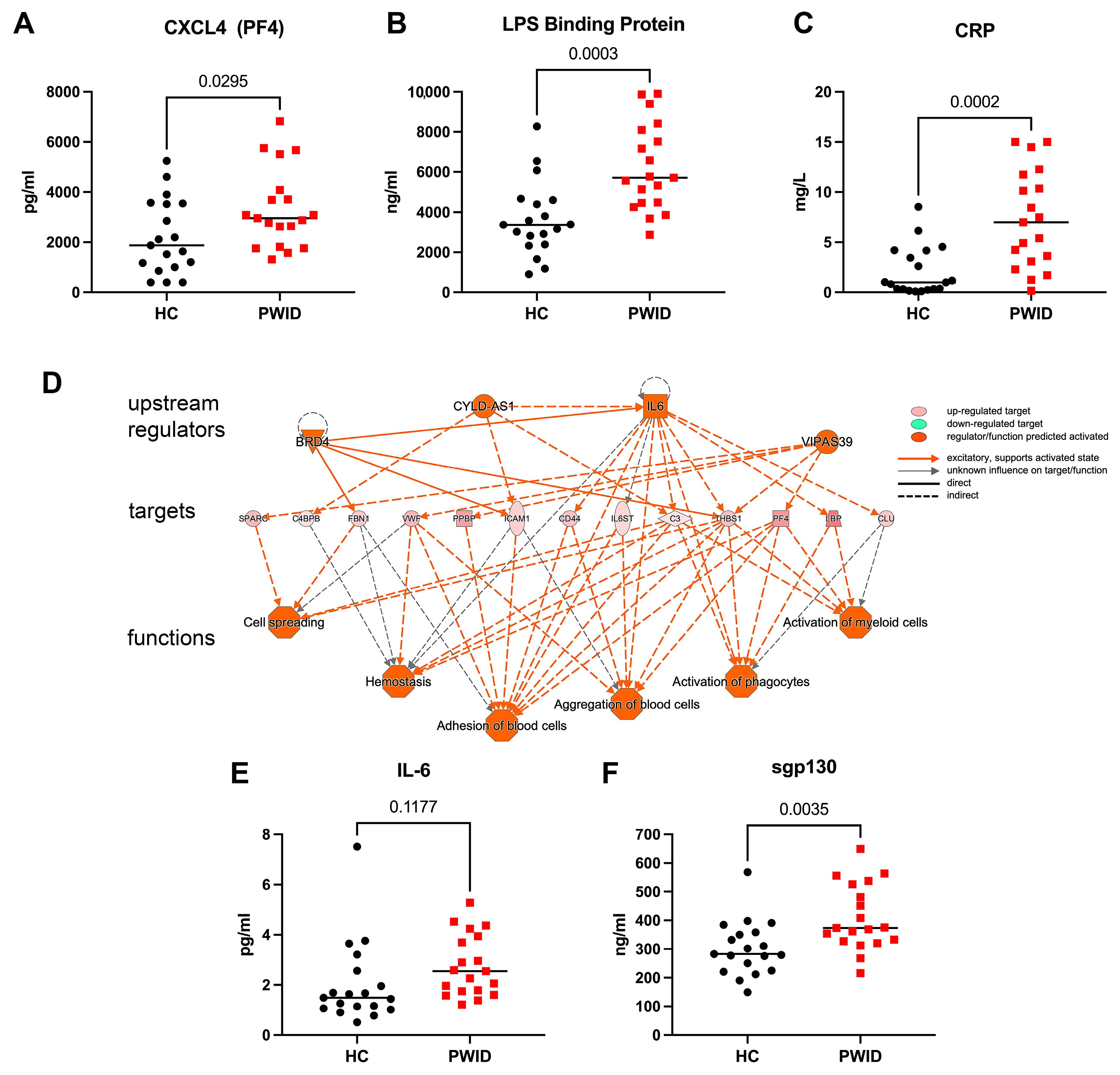
3.2. Distinct Plasma Proteomics Profile of PWID
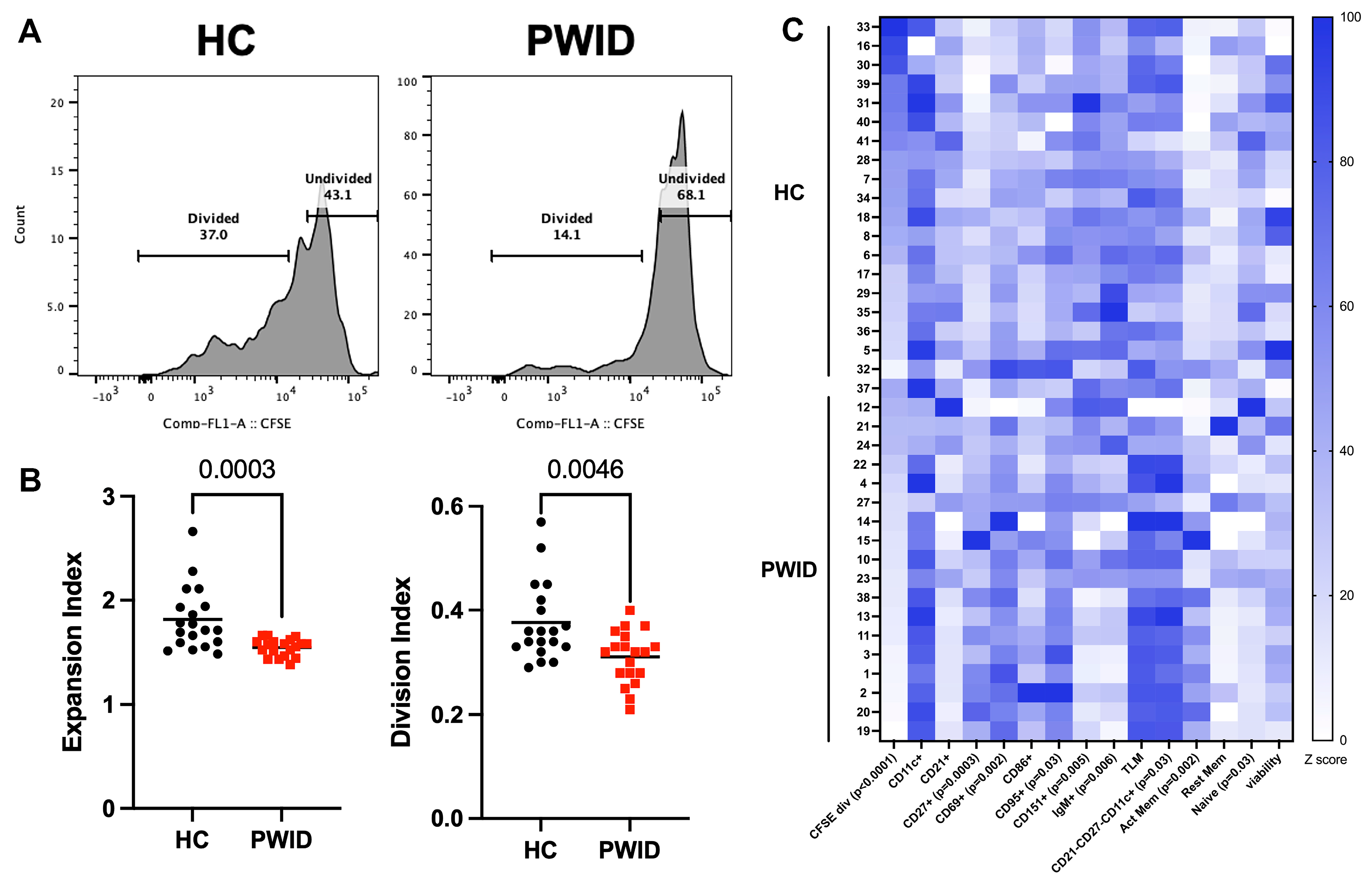
3.3. Inhibition of B Cell Proliferation by PWID Plasma
3.4. PWID Plasma Alters B Cell Transcriptional Response to Activation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jenkins, R.A. The fourth wave of the US opioid epidemic and its implications for the rural US: A federal perspective. Prev. Med. 2021, 152 Pt 2, 106541. [Google Scholar] [CrossRef] [PubMed]
- Rangachari, P.; Govindarajan, A.; Mehta, R.; Seehusen, D.; Rethemeyer, R.K. The relationship between Social Determinants of Health (SDoH) and death from cardiovascular disease or opioid use in counties across the United States (2009–2018). BMC Public Health 2022, 22, 236. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Vallecillo, G.; Pedro-Botet, J.; Fernandez, S.; Roman, I.; Elosua, R.; Camps, A.; Torrens, M.; Marrugat, J. High cardiovascular risk in older patients with opioid use disorder: Differences with the general population. Drug Alcohol. Rev. 2022, 41, 1078–1084. [Google Scholar] [CrossRef] [PubMed]
- Sharma, A.; Govindan, P.; Toukatly, M.; Healy, J.; Henry, C.; Senter, S.; Najafian, B.; Kestenbaum, B. Heroin Use Is Associated with AA-Type Kidney Amyloidosis in the Pacific Northwest. Clin. J. Am. Soc. Nephrol. 2018, 13, 1030–1036. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Machowska, A.; Carrero, J.J.; Lindholm, B.; Stenvinkel, P. Therapeutics targeting persistent inflammation in chronic kidney disease. Transl. Res. 2016, 167, 204–213. [Google Scholar] [CrossRef] [PubMed]
- Weiss, A.J.O.P.; Karaca, Z.; Heslin, K.C.; Henke, R.M.; McDermott, K.W.; Pickens, G.; Barrett, M.L. County-Level Determinants of High Opioid-Related Hospitalization Rates. 2022. Available online: https://hcup-us.ahrq.gov/reports.jsp (accessed on 26 March 2024).
- Govitrapong, P.; Suttitum, T.; Kotchabhakdi, N.; Uneklabh, T. Alterations of immune functions in heroin addicts and heroin withdrawal subjects. J. Pharmacol. Exp. Ther. 1998, 286, 883–889. [Google Scholar] [PubMed]
- Garcia, J.B.; Cardoso, M.G.; Dos-Santos, M.C. Opioids and the immune system: Clinical relevance. Rev. Bras. Anestesiol. 2012, 62, 709–718. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.W.; Liu, F.L.; Mu, D.; Deng, D.Y.; Zheng, Y.T. Increased expression and dysregulated association of restriction factors and type I interferon in HIV, HCV mono- and co-infected patients. J. Med. Virol. 2016, 88, 987–995. [Google Scholar] [CrossRef] [PubMed]
- Sacerdote, P.; Franchi, S.; Gerra, G.; Leccese, V.; Panerai, A.E.; Somaini, L. Buprenorphine and methadone maintenance treatment of heroin addicts preserves immune function. Brain Behav. Immun. 2008, 22, 606–613. [Google Scholar] [CrossRef] [PubMed]
- McCarthy, L.; Wetzel, M.; Sliker, J.K.; Eisenstein, T.K.; Rogers, T.J. Opioids, opioid receptors, and the immune response. Drug Alcohol. Depend. 2001, 62, 111–123. [Google Scholar] [CrossRef] [PubMed]
- Al-Hashimi, M.; Scott, S.W.; Thompson, J.P.; Lambert, D.G. Opioids and immune modulation: More questions than answers. Br. J. Anaesth. 2013, 111, 80–88. [Google Scholar] [CrossRef] [PubMed]
- Happel, C.; Kutzler, M.; Rogers, T.J. Opioid-induced chemokine expression requires NF-kappaB activity: The role of PKCzeta. J. Leukoc. Biol. 2011, 89, 301–309. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Saurer, T.B.; Carrigan, K.A.; Ijames, S.G.; Lysle, D.T. Morphine-induced alterations of immune status are blocked by the dopamine D2-like receptor agonist 7-OH-DPAT. J. Neuroimmunol. 2004, 148, 54–62. [Google Scholar] [CrossRef] [PubMed]
- Roy, S.; Ninkovic, J.; Banerjee, S.; Charboneau, R.G.; Das, S.; Dutta, R.; Kirchner, V.A.; Koodie, L.; Ma, J.; Meng, J.; et al. Opioid drug abuse and modulation of immune function: Consequences in the susceptibility to opportunistic infections. J. Neuroimmune Pharmacol. 2011, 6, 442–465. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Ninkovic, J.; Roy, S. Role of the mu-opioid receptor in opioid modulation of immune function. Amino Acids 2013, 45, 9–24. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Brown, J.N.; Ortiz, G.M.; Angel, T.E.; Jacobs, J.M.; Gritsenko, M.; Chan, E.Y.; Purdy, D.E.; Murnane, R.D.; Larsen, K.; Palermo, R.E.; et al. Morphine produces immunosuppressive effects in nonhuman primates at the proteomic and cellular levels. Mol. Cell Proteom. 2012, 11, 605–618. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Meng, J.; Yu, H.; Ma, J.; Wang, J.; Banerjee, S.; Charboneau, R.; Barke, R.A.; Roy, S. Morphine induces bacterial translocation in mice by compromising intestinal barrier function in a TLR-dependent manner. PLoS ONE 2013, 8, e54040. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Rogers, D.F.; Barnes, P.J. Opioid inhibition of neurally mediated mucus secretion in human bronchi. Lancet 1989, 1, 930–932. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, S.; Sindberg, G.; Wang, F.; Meng, J.; Sharma, U.; Zhang, L.; Dauer, P.; Chen, C.; Dalluge, J.; Johnson, T.; et al. Opioid-induced gut microbial disruption and bile dysregulation leads to gut barrier compromise and sustained systemic inflammation. Mucosal Immunol. 2016, 9, 1418–1428. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Wang, F.; Roy, S. Gut Homeostasis, Microbial Dysbiosis, and Opioids. Toxicol. Pathol. 2017, 45, 150–156. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Zhang, P.; Yang, M.; Chen, C.; Liu, L.; Wei, X.; Zeng, S. Toll-Like Receptor 4 (TLR4)/Opioid Receptor Pathway Crosstalk and Impact on Opioid Analgesia, Immune Function, and Gastrointestinal Motility. Front. Immunol. 2020, 11, 1455. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Eisenstein, T.K. The Role of Opioid Receptors in Immune System Function. Front. Immunol. 2019, 10, 2904. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Piepenbrink, M.S.; Samuel, M.; Zheng, B.; Carter, B.; Fucile, C.; Bunce, C.; Kiebala, M.; Khan, A.A.; Thakar, J.; Maggirwar, S.B.; et al. Humoral Dysregulation Associated with Increased Systemic Inflammation among Injection Heroin Users. PLoS ONE 2016, 11, e0158641. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Hileman, C.O.; Bowman, E.R.; Gabriel, J.; Kettelhut, A.; Labbato, D.; Smith, C.; Avery, A.; Parran, T.; Funderburg, N.; McComsey, G.A. Impact of Heroin and HIV on Gut Integrity and Immune Activation. J. Acquir. Immune Defic. Syndr. 2022, 89, 519–526. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Mehandru, S.; Deren, S.; Kang, S.Y.; Banfield, A.; Garg, A.; Garmon, D.; LaMar, M.; Evering, T.H.; Markowitz, M. Behavioural, Mucosal and Systemic Immune Parameters in HIV-infected and Uninfected Injection Drug Users. J. Addict. Res. Ther. 2015, 6, 1–8. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Taub, D.D.; Eisenstein, T.K.; Geller, E.B.; Adler, M.W.; Rogers, T.J. Immunomodulatory activity of mu- and kappa-selective opioid agonists. Proc. Natl. Acad. Sci. USA 1991, 88, 360–364. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Jamali, A.; Roostaee, M.H.; Soleimanjahi, H.; Ghaderi Pakdel, F.; Bamdad, T. DNA vaccine-encoded glycoprotein B of HSV-1 fails to protect chronic morphine-treated mice against HSV-1 challenge. Comp. Immunol. Microbiol. Infect. Dis. 2007, 30, 71–80. [Google Scholar] [CrossRef] [PubMed]
- Meacham, M.C.; Rudolph, A.E.; Strathdee, S.A.; Rusch, M.L.; Brouwer, K.C.; Patterson, T.L.; Vera, A.; Rangel, G.; Roesch, S.C. Polydrug Use and HIV Risk Among People Who Inject Heroin in Tijuana, Mexico: A Latent Class Analysis. Subst. Use Misuse 2015, 50, 1351–1359. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Kuramoto, S.J.; Bohnert, A.S.; Latkin, C.A. Understanding subtypes of inner-city drug users with a latent class approach. Drug Alcohol. Depend. 2011, 118, 237–243. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Roederer, M. Interpretation of cellular proliferation data: Avoid the panglossian. Cytom. A 2011, 79, 95–101. [Google Scholar] [CrossRef] [PubMed]
- Krueger, F.; Trim Galore. Github2024. Available online: https://github.com/FelixKrueger/TrimGalore (accessed on 25 April 2024).
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq—A Python framework to work with high-throughput sequencing data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Liberzon, A. A description of the Molecular Signatures Database (MSigDB) Web site. Methods Mol. Biol. 2014, 1150, 153–160. [Google Scholar] [CrossRef] [PubMed]
- Gabr, M.M.; Saeed, I.; Miles, J.A.; Ross, B.P.; Shaw, P.N.; Hollmann, M.W.; Parat, M.O. Interaction of Opioids with TLR4-Mechanisms and Ramifications. Cancers 2021, 13, 5274. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Cruz-Lebron, A.; Johnson, R.; Mazahery, C.; Troyer, Z.; Joussef-Pina, S.; Quinones-Mateu, M.E.; Strauch, C.M.; Hazen, S.L.; Levine, A.D. Chronic opioid use modulates human enteric microbiota and intestinal barrier integrity. Gut Microbes. 2021, 13, 1946368. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Azzoni, L.; Giron, L.B.; Vadrevu, S.; Zhao, L.; Lalley-Chareczko, L.; Hiserodt, E.; Fair, M.; Lynn, K.; Trooskin, S.; Mounzer, K.; et al. Methadone use is associated with increased levels of sCD14, immune activation, and inflammation during suppressed HIV infection. J. Leukoc. Biol. 2022, 112, 733–744. [Google Scholar] [CrossRef] [PubMed]
- Dam, K.A.; Barnes, C.O.; Gristick, H.B.; Schoofs, T.; Gnanapragasam, P.N.P.; Nussenzweig, M.C.; Bjorkman, P.J. HIV-1 CD4-binding site germline antibody-Env structures inform vaccine design. Nat. Commun. 2022, 13, 6123. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Prechel, M.M.; Walenga, J.M. Emphasis on the Role of PF4 in the Incidence, Pathophysiology and Treatment of Heparin Induced Thrombocytopenia. Thromb. J. 2013, 11, 7. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Lee, G.M.; Arepally, G.M. Heparin-induced thrombocytopenia. Hematol. Am. Soc. Hematol. Educ. Program 2013, 2013, 668–674. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Kopp, F.; Kupsch, S.; Schromm, A.B. Lipopolysaccharide-binding protein is bound and internalized by host cells and colocalizes with LPS in the cytoplasm: Implications for a role of LBP in intracellular LPS-signaling. Biochim. Biophys. Acta 2016, 1863, 660–672. [Google Scholar] [CrossRef] [PubMed]
- Park, B.S.; Lee, J.O. Recognition of lipopolysaccharide pattern by TLR4 complexes. Exp. Mol. Med. 2013, 45, e66. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Cao, W.; Rosen, D.B.; Ito, T.; Bover, L.; Bao, M.; Watanabe, G.; Zhengbin, Y.; Li, Z.; Lewis, L.L.; Yong-Jun, L. Plasmacytoid dendritic cell-specific receptor ILT7-Fc epsilonRI gamma inhibits Toll-like receptor-induced interferon production. J. Exp. Med. 2006, 203, 1399–1405. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Liu, B.; Lin, Y.; Yan, J.; Yao, J.; Liu, D.; Ma, W.; Wang, J.; Liu, W.; Wang, C.; Zhang, L.; et al. Affinity-coupled CCL22 promotes positive selection in germinal centres. Nature 2021, 592, 133–137. [Google Scholar] [CrossRef] [PubMed]
- Trezise, S.; Karnowski, A.; Fedele, P.L.; Mithraprabhu, S.; Liao, Y.; D’Costa, K.; Kueh, A.J.; Hardy, M.P.; Owczarek, C.M.; Herold, M.J.; et al. Mining the Plasma Cell Transcriptome for Novel Cell Surface Proteins. Int. J. Mol. Sci. 2018, 19, 2161. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Cheng, R.Y.; de Rutte, J.; Ito, C.E.K.; Ott, A.R.; Bosler, L.; Kuo, W.Y.; Cheng, R.Y.; de Rutte, J.; Ito, C.E.K.; Ott, A.R.; et al. SEC-seq: Association of molecular signatures with antibody secretion in thousands of single human plasma cells. Nat. Commun. 2023, 14, 3567. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Karnell, J.L.; Karnell, F.G., 3rd; Stephens, G.L.; Rajan, B.; Morehouse, C.; Li, Y.; Swerdlow, B.; Wilson, M.; Goldbach-Mansky, R.; Groves, C.; et al. Mycophenolic acid differentially impacts B cell function depending on the stage of differentiation. J. Immunol. 2011, 187, 3603–3612. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Calise, S.J.; Abboud, G.; Kasahara, H.; Morel, L.; Chan, E.K.L. Immune Response-Dependent Assembly of IMP Dehydrogenase Filaments. Front. Immunol. 2018, 9, 2789. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Zuo, Z.; Kania, A.K.; Patterson, D.G.; Hicks, S.L.; Maurer, J.; Gupta, M.; Boss, J.M.; Scharer, C.D. CRISPR/Cas9 editing reveals IRF8 regulated gene signatures restraining plasmablast differentiation. Heliyon 2023, 9, e17527. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Chan, Y.Y.; Yang, S.N.; Lin, J.C.; Chang, J.L.; Lin, J.G.; Lo, W.Y. Inflammatory response in heroin addicts undergoing methadone maintenance treatment. Psychiatry Res. 2015, 226, 230–234. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.L.; Lee, S.Y.; Tao, P.L.; Chang, Y.H.; Chen, S.H.; Chu, C.H.; Chen, P.S.; Lee, I.H.; Yeh, T.L.; Yang, Y.K.; et al. Dextromethorphan attenuated inflammation and combined opioid use in humans undergoing methadone maintenance treatment. J. Neuroimmune Pharmacol. 2012, 7, 1025–1033. [Google Scholar] [CrossRef] [PubMed]
- Warshow, U.M.; Riva, A.; Hegazy, D.; Thurairajah, P.H.; Kaminski, E.R.; Chokshi, S.; Cramp, M.E. Cytokine profiles in high risk injection drug users suggests innate as opposed to adaptive immunity in apparent resistance to hepatitis C virus infection. J. Viral Hepat. 2012, 19, 501–508. [Google Scholar] [CrossRef] [PubMed]
- Leng, S.X.; Dandorf, S.; Li, H.; Carlson, J.; Hui, J.; Mehta, S.H.; Piggott, D.; Islam, S.; Manwani, B.; Kirk, G.D. Associations of Circulating Soluble Tumor Necrosis Factor-alpha Receptors 1 and 2 with Interleukin-6 Levels in an Aging Cohort of Injection Drug Users with or at High Risk for HIV Infection. AIDS Res. Hum. Retroviruses 2015, 31, 1257–1264. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Neri, M.; Panata, L.; Bacci, M.; Fiore, C.; Riezzo, I.; Turillazzi, E.; Fineschi, V. Cytokines, chaperones and neuroinflammatory responses in heroin-related death: What can we learn from different patterns of cellular expression? Int. J. Mol. Sci. 2013, 14, 19831–19845. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Buettner, M.; Toennes, S.W.; Buettner, S.; Bickel, M.; Allwinn, R.; Geiger, H.; Bratzke, H.; Amann, K.; Jung, O. Nephropathy in illicit drug abusers: A postmortem analysis. Am. J. Kidney Dis. 2014, 63, 945–953. [Google Scholar] [CrossRef] [PubMed]
- Leung-Hagesteijn, C.; Erdmann, N.; Cheung, G.; Keats, J.J.; Stewart, A.K.; Reece, D.E.; Chung, K.C.; Tiedemann, R.E. Xbp1s-negative tumor B cells and pre-plasmablasts mediate therapeutic proteasome inhibitor resistance in multiple myeloma. Cancer Cell. 2013, 24, 289–304. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Cheng, G.L.; Zeng, H.; Leung, M.K.; Zhang, H.J.; Lau, B.W.; Liu, Y.P.; Liu, G.X.; Sham, P.C.; Chan, C.C.; So, K.F.; et al. Heroin abuse accelerates biological aging: A novel insight from telomerase and brain imaging interaction. Transl. Psychiatry 2013, 3, e260. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Anthony, I.C.; Norrby, K.E.; Dingwall, T.; Carnie, F.W.; Millar, T.; Arango, J.C.; Robertson, R.; Bell, J.E. Predisposition to accelerated Alzheimer-related changes in the brains of human immunodeficiency virus negative opiate abusers. Brain 2010, 133 Pt 12, 3685–3698. [Google Scholar] [CrossRef] [PubMed]
- Reece, A.S.; Hulse, G.K. Duration of opiate exposure as a determinant of arterial stiffness and vascular age in male opiate dependence: A longitudinal study. J. Clin. Pharm. Ther. 2014, 39, 158–167. [Google Scholar] [CrossRef] [PubMed]
- Bode, J.G.; Albrecht, U.; Haussinger, D.; Heinrich, P.C.; Schaper, F. Hepatic acute phase proteins--regulation by IL-6- and IL-1-type cytokines involving STAT3 and its crosstalk with NF-kappaB-dependent signaling. Eur. J. Cell Biol. 2012, 91, 496–505. [Google Scholar] [CrossRef] [PubMed]
- Kasperska-Zajac, A.; Grzanka, A.; Damasiewicz-Bodzek, A. IL-6 Transsignaling in Patients with Chronic Spontaneous Urticaria. PLoS ONE 2015, 10, e0145751. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Ravid, K.; Kuter, D.J.; Rosenberg, R.D. rmIL-6 stimulates the transcriptional activity of the rat PF4 gene. Exp. Hematol. 1995, 23, 397–401. [Google Scholar] [PubMed]
- Kievlan, D.R.; Gukasyan, M.; Gesch, J.; Rodriguez, R.M. Clinical profile of injection drug users presenting to the ED. Am. J. Emerg. Med. 2015, 33, 674–676. [Google Scholar] [CrossRef] [PubMed]
- Burbano, X.; Miguez, M.J.; Lecusay, R.; Rodriguez, A.; Ruiz, P.; Morales, G.; Castillo, G.; Baum, M.; Shor-Posner, G. Thrombocytopenia in HIV-infected drug users in the HAART era. Platelets 2001, 12, 456–461. [Google Scholar] [CrossRef] [PubMed]
- Koury, M.J. Thrombocytopenic purpura in HIV-seronegative users of intravenous cocaine. Am. J. Hematol. 1990, 35, 134–135. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sarkar, S.; Hill, D.D.; Rosenberg, A.F.; Eaton, E.F.; Kutsch, O.; Kobie, J.J. Injection Drug Use Alters Plasma Regulation of the B Cell Response. Cells 2024, 13, 1011. https://doi.org/10.3390/cells13121011
Sarkar S, Hill DD, Rosenberg AF, Eaton EF, Kutsch O, Kobie JJ. Injection Drug Use Alters Plasma Regulation of the B Cell Response. Cells. 2024; 13(12):1011. https://doi.org/10.3390/cells13121011
Chicago/Turabian StyleSarkar, Sanghita, Dave D. Hill, Alexander F. Rosenberg, Ellen F. Eaton, Olaf Kutsch, and James J. Kobie. 2024. "Injection Drug Use Alters Plasma Regulation of the B Cell Response" Cells 13, no. 12: 1011. https://doi.org/10.3390/cells13121011




