Analysis of the TID-I and TID-L Splice Variants’ Expression Profile under In Vitro Differentiation of Human Mesenchymal Bone Marrow Cells into Osteoblasts
Abstract
:1. Introduction
2. Materials and Methods
2.1. Isolation and Culture of B-MSCs
2.2. Proliferation of B-MSCs
2.3. Differentiation of B-MSCs
2.4. Culture of Control Cells
2.5. Analysis of Cell Proliferation
2.6. Flow Cytometry
2.7. Alkaline Phosphatase Activity Assay
2.8. Alizarin Red Staining
2.9. Western Blotting
2.10. Real Time q-PCR
2.11. Silencing of Gene Expression by siRNA
2.12. Statistical Analysis
3. Results
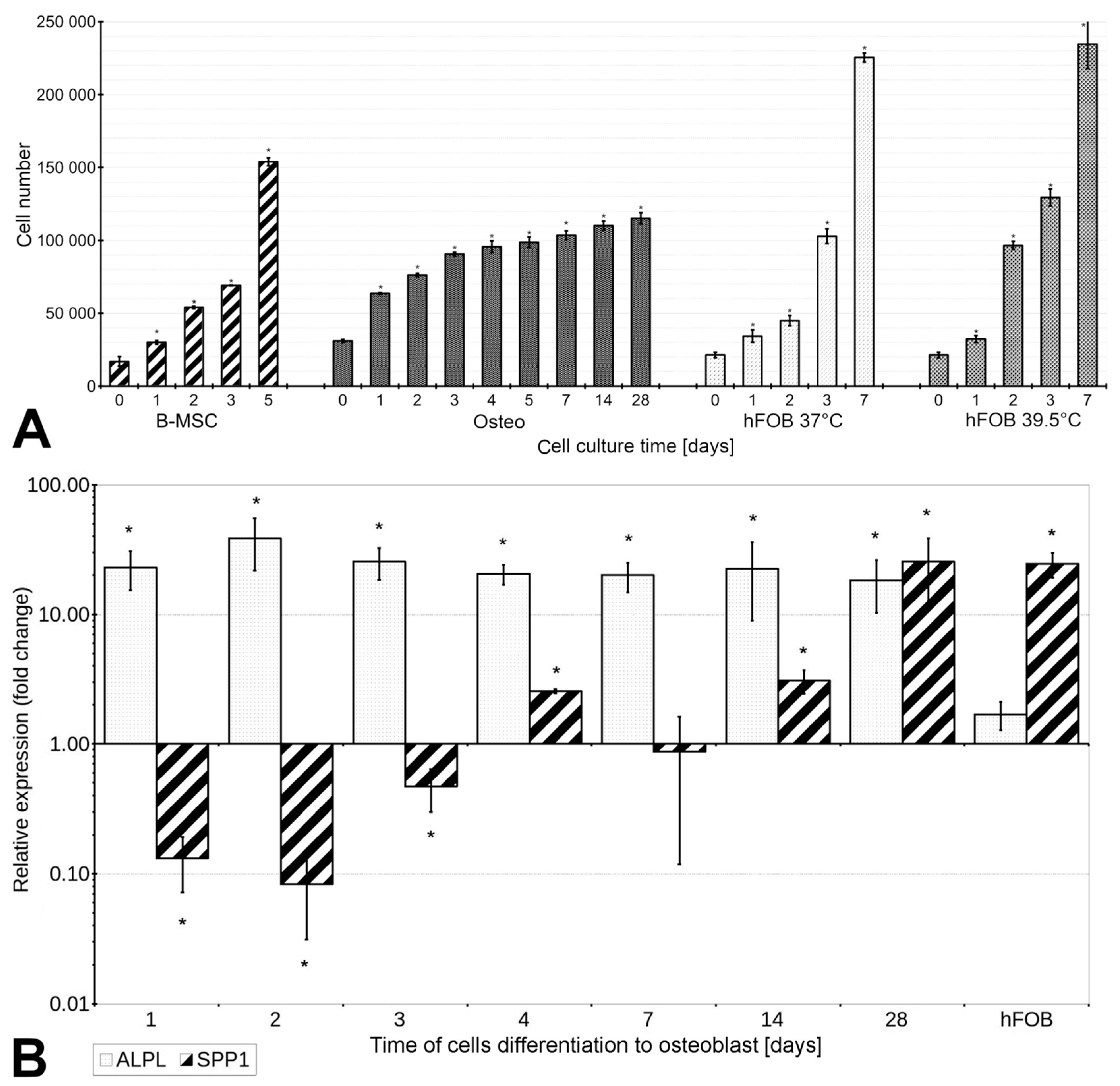
3.1. The Proliferation Rate of B-MSCs Decreases under Conditions of Differentiation
3.2. Specific Surface Antigens’ Expression, Cell Size, and Granularity Change during Differentiation
3.3. B-MSCs’ Differentiation into Osteoblasts Is Associated with Increased Expression of ALPL and SPP1, and the Activity of Alkaline Phosphatase
3.4. B-MSCs’ Differentiation into Osteoblasts Is Accompanied by Appearance of Mineralization Nodules
3.5. TID Proteins Are Detected in B-MSCs and Osteoblasts
3.6. The Expression of Alternative Splice Variants of TID Fluctuates under Differentiation
3.7. Silencing the TID Splice Variants Induces the Expression of SPP1
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- McNamara, L.M. 2.10 Bone as a Material. In Comprehensive Biomaterials II; Ducheyne, P., Ed.; Elsevier: Amsterdam, The Netherlands, 2017; pp. 202–227. [Google Scholar]
- Kulterer, B.; Friedl, G.; Jandrositz, A.; Sanchez-Cabo, F.; Prokesch, A.; Paar, C.; Scheideler, M.; Windhager, R.; Preisegger, K.-H.; Trajanoski, Z. Gene Expression Profiling of Human Mesenchymal Stem Cells Derived from Bone Marrow during Expansion and Osteoblast Differentiation. BMC Genom. 2007, 8, 70. [Google Scholar] [CrossRef] [PubMed]
- Birmingham, E.; Niebur, G.; McHugh, P.; Shaw, G.; Barry, F.; McNamara, L. Osteogenic Differentiation of Mesenchymal Stem Cells Is Regulated by Osteocyte and Osteoblast Cells in a Simplified Bone Niche. Eur. Cell Mater. 2012, 23, 13–27. [Google Scholar] [CrossRef] [PubMed]
- Holm, E.; Gleberzon, J.S.; Liao, Y.; Sørensen, E.S.; Beier, F.; Hunter, G.K.; Goldberg, H.A. Osteopontin Mediates Mineralization and Not Osteogenic Cell Development In Vitro. Biochem. J. 2014, 464, 355–364. [Google Scholar] [CrossRef]
- Tsao, Y.-T.; Huang, Y.-J.; Wu, H.-H.; Liu, Y.-A.; Liu, Y.-S.; Lee, O. Osteocalcin Mediates Biomineralization during Osteogenic Maturation in Human Mesenchymal Stromal Cells. Int. J. Mol. Sci. 2017, 18, 159. [Google Scholar] [CrossRef]
- Witkowska-Zimny, M.; Wróbel, E.; Przybylski, J. Najważniejsze Czynniki Transkrypcyjne Procesu Osteoblastogenezy. Post. Biol. Kom. 2009, 4, 695–705. [Google Scholar]
- Bagdu, N.; Merugu, N. Human Alkaline Phosphatases in Health and Disease: A Mini Review. Int. J. Res. Pharm. Sci. 2013, 4, 371–379. [Google Scholar]
- Boxall, S.A.; Jones, E. Markers for Characterization of Bone Marrow Multipotential Stromal Cells. Stem Cells Int. 2012, 975871, 1–12. [Google Scholar] [CrossRef]
- Tan, K.; Zhu, H.; Zhang, J.; Ouyang, W.; Tang, J.; Zhang, Y.; Qiu, L.; Liu, X.; Ding, Z.; Deng, X. CD73 Expression on Mesenchymal Stem Cells Dictates the Reparative Properties via Its Anti-Inflammatory Activity. Stem Cells Int. 2019, 2019, 8717694. [Google Scholar] [CrossRef]
- Zhou, L.; Hao, Q.; Sugita, S.; Naito, Y.; He, H.; Yeh, C.C.; Lee, J.W. Role of CD44 in increasing the potency of mesenchymal stem cell extracellular vesicles by hyaluronic acid in severe pneumonia. Stem Cell Res. Ther. 2021, 12, 293. [Google Scholar] [CrossRef]
- Spaeth, E.L.; Labaff, A.M.; Toole, B.P.; Klopp, A.; Andreeff, M.; Marini, F.C. Mesenchymal CD44 expression contributes to the acquisition of an activated fibroblast phenotype via TWIST activation in the tumor microenvironment. Cancer Res. 2013, 73, 5347–5359. [Google Scholar] [CrossRef]
- Jin, H.J.; Bae, Y.K.; Kim, M.; Kwon, S.-J.; Jeon, H.B.; Choi, S.J.; Kim, S.W.; Yang, Y.S.; Oh, W.; Chang, J.W. Comparative Analysis of Human Mesenchymal Stem Cells from Bone Marrow, Adipose Tissue, and Umbilical Cord Blood as Sources of Cell Therapy. Int. J. Mol. Sci. 2013, 14, 17986–18001. [Google Scholar] [CrossRef] [PubMed]
- Chung, E.; Rylander, M.N. Response of Preosteoblasts to Thermal Stress Conditioning and Osteoinductive Growth Factors. Cell Stress Chaperones 2011, 17, 203–214. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Shi, Z.-D.; Ji, X.; Morales, J.; Zhang, J.; Kaur, N.; Wang, S. Enhanced Osteogenesis of Human Mesenchymal Stem Cells by Periodic Heat Shock in Self-Assembling Peptide Hydrogel. Tissue Eng. Part A 2013, 19, 716–728. [Google Scholar] [CrossRef] [PubMed]
- Lo, J.F.; Hayashi, M.; Woo-Kim, S.; Tian, B.; Huang, J.F.; Fearns, C.; Takayama, S.; Zapata, J.M.; Yang, Y.; Lee, J.D. Tid1, a Cochaperone of the Heat Shock 70 Protein and the Mammalian Counterpart of the Drosophila Tumor Suppressor L(2)Tid, Is Critical for Early Embryonic Development and Cell Survival. Mol. Cell Biol. 2004, 24, 2226–2236. [Google Scholar] [CrossRef] [PubMed]
- Hageman, J.; Kampinga, H.H. Computational Analysis of the Human HSPH/HSPA/DNAJ Family and Cloning of a Human HSPH/HSPA/DNAJ Expression Library. Cell Stress Chaperones 2008, 14, 1–21. [Google Scholar] [CrossRef] [PubMed]
- Kumada, K.; Fuse, N.; Tamura, T.; Okamori, C.; Kurata, S. HSP70/DNAJA3 Chaperone/Cochaperone Regulates NF-ΚB Activity in Immune Responses. Biochem. Biophys. Res. Commun. 2019, 513, 947–951. [Google Scholar] [CrossRef] [PubMed]
- Qiu, X.-B.; Shao, Y.-M.; Miao, S.; Wang, L. The Diversity of the DnaJ/Hsp40 Family, the Crucial Partners for Hsp70 Chaperones. Cell Mol. Life Sci. 2006, 63, 2560–2570. [Google Scholar] [CrossRef]
- Patra, M.; Weiss, C.; Abu-Libdeh, B.; Ashhab, M.; Abuzer, S.; Elpeleg, O.; Mahajnah, M.; Kessel, A.; Azem, A. A Novel Variant of the Human Mitochondrial DnaJ Protein, Tid1, Associates with a Human Disease Exhibiting Developmental Delay and Polyneuropathy. Eur. J. Hum. Genet. 2019, 27, 1072–1080. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.H.; Choi, D.-K.; Sohn, K.-C.; Kwak, S.S.; Suk, J.; Lim, J.-S.; Shin, I.; Kim, S.-W.; Lee, J.-H.; Joe, C.O. Absence of a Human DnaJ Protein HTid-1S Correlates with Aberrant Actin Cytoskeleton Organization in Lesional Psoriatic Skin. J. Biol. Chem. 2012, 287, 25954–25963. [Google Scholar] [CrossRef]
- Kurzik-Dumke, U.; Czaja, J. Htid-1, the Human Homolog of the Drosophila Melanogaster L(2)Tid Tumor Suppressor, Defines a Novel Physiological Role of APC. Cell Signal 2007, 19, 1973–1985. [Google Scholar] [CrossRef]
- Canamasas, I.; Debes, A.; Natali, P.G.; Kurzik-Dumke, U. Understanding Human Cancer Using Drosophila. J. Biol. Chem. 2003, 278, 30952–30960. [Google Scholar] [CrossRef] [PubMed]
- Lu, B.; Garrido, N.; Spelbrink, J.N.; Suzuki, C.K. Tid1 Isoforms Are Mitochondrial DnaJ-like Chaperones with Unique Carboxyl Termini That Determine Cytosolic Fate. J. Biol. Chem. 2006, 281, 13150–13158. [Google Scholar] [CrossRef] [PubMed]
- Dhennin-Duthille, I.; Nyga, R.; Yahiaoui, S.; Gouilleux-Gruart, V.; Régnier, A.; Lassoued, K.; Gouilleux, F. The Tumor Suppressor HTid1 Inhibits STAT5b Activity via Functional Interaction. J. Biol. Chem. 2010, 286, 5034–5042. [Google Scholar] [CrossRef] [PubMed]
- Sarkar, S.; Pollack, B.P.; Lin, K.-T.; Kotenko, S.V.; Cook, J.R.; Lewis, A.; Pestka, S. HTid-1, a Human DnaJ Protein, Modulates the Interferon Signaling Pathway. J. Biol. Chem. 2001, 276, 49034–49042. [Google Scholar] [CrossRef]
- Maletzko, A.; Key, J.; Wittig, I.; Gispert, S.; Koepf, G.; Canet-Pons, J.; Torres-Odio, S.; West, A.P.; Auburger, G. Increased Presence of Nuclear DNAJA3 and Upregulation of Cytosolic STAT1 and of Nucleic Acid Sensors Trigger Innate Immunity in the ClpP-Null Mouse. Neurogenetics 2021, 22, 297–312. [Google Scholar] [CrossRef] [PubMed]
- Torregroza, I.; Evans, T. Tid1 Is a Smad-Binding Protein That Can Modulate Smad7 Activity in Developing Embryos. Biochem. J. 2005, 393, 311–320. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.-W.; Chao, T.-H.; Xiang, R.; Lo, J.-F.; Campbell, M.J.; Fearns, C.; Lee, J.-D. Tid1, the Human Homologue of a Drosophila Tumor Suppressor, Reduces the Malignant Activity of ErbB-2 in Carcinoma Cells. Cancer Res. 2004, 64, 7732–7739. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.Y.; Jan, C.I.; Lo, J.F.; Yang, S.C.; Chang, Y.L.; Pan, S.H.; Wang, W.L.; Hong, T.M.; Yang, P.C. Tid1-L Inhibits EGFR Signaling in Lung Adenocarcinoma by Enhancing EGFR Ubiquitinylation and Degradation. Cancer Res. 2013, 73, 4009–4019. [Google Scholar] [CrossRef] [PubMed]
- Copeland, E.; Balgobin, S.; Lee, C.M.; Rozakis-Adcock, M. HTID-1 Defines a Novel Regulator of C-Met Receptor Signaling in Renal Cell Carcinomas. Oncogene 2011, 30, 2252–2263. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; MacDonald, J.L.; Hryciw, T.; Li, C.; Meakin, S.O. Human Tumorous Imaginal Disc 1 (TID1) Associates with Trk Receptor Tyrosine Kinases and Regulates Neurite Outgrowth in Nnr5-TrkA Cells. J. Biol. Chem. 2005, 280, 19461–19471. [Google Scholar] [CrossRef]
- Linnoila, J.; Wang, Y.; Yao, Y.; Wang, Z.-Z. A Mammalian Homolog of Drosophila Tumorous Imaginal Discs, Tid1, Mediates Agrin Signaling at the Neuromuscular Junction. Neuron 2008, 60, 625–641. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Balice-Gordon, R. New Dogs in the Dogma: Lrp4 and Tid1 in Neuromuscular Synapse Formation. Neuron 2008, 60, 526–528. [Google Scholar] [CrossRef] [PubMed]
- Niu, G.; Zhang, H.; Liu, D.; Chen, L.; Belani, C.; Wang, H.G.; Cheng, H. Tid1, the Mammalian Homologue of Drosophila Tumor Suppressor Tid56, Mediates Macroautophagy by Interacting with Beclin1-Containing Autophagy Protein Complex. J. Biol. Chem. 2015, 290, 18102–18110. [Google Scholar] [CrossRef] [PubMed]
- Qi, M.; Zhang, L.; Ma, Y.; Shuai, Y.; Li, L.; Luo, K.; Liu, W.; Jin, Y. Autophagy Maintains the Function of Bone Marrow Mesenchymal Stem Cells to Prevent Estrogen Deficiency-Induced Osteoporosis. Theranostics 2017, 7, 4498–4516. [Google Scholar] [CrossRef] [PubMed]
- Traicoff, J.; Hewitt, S.; Chung, J. DNAJA3 (DnaJ (Hsp40) Homolog, Subfamily A, Member 3). Atlas Genet. Cytogenet. Oncol. Haematol. 2012, 16. [Google Scholar] [CrossRef]
- Banerjee, S.; Chaturvedi, R.; Singh, A.; Kushwaha, H.R. Putting Human Tid-1 in Context: An Insight into Its Role in the Cell and in Different Disease States. Cell Commun. Signal. 2022, 20, 109. [Google Scholar] [CrossRef] [PubMed]
- Yin, X.; Rozakis-Adcock, M. Genomic Organization and Expression of the Human Tumorous Imaginal Disc (TID1) Gene. Gene 2001, 278, 201–210. [Google Scholar] [CrossRef] [PubMed]
- Marcol, W.; Slusarczyk, W.; Sieroń, A.L.; Koryciak-Komarska, H.; Lewin-Kowalik, J. Bone Marrow Stem Cells Delivered into the Subarachnoid Space via Cisterna Magna Improve Repair of Injured Rat Spinal Cord White Matter. Int. J. Clin. Exp. Med. 2015, 8, 14680–14692. [Google Scholar] [PubMed]
- Harris, S.A.; Enger, R.J.; Riggs, L.B.; Spelsberg, T.C. Development and Characterization of a Conditionally Immortalized Human Fetal Osteoblastic Cell Line. J. Bone Miner. Res. 2009, 10, 178–186. [Google Scholar] [CrossRef] [PubMed]
- Yen, M.-L.; Chien, C.-C.; Chiu, I.-M.; Huang, H.-I.; Chen, Y.-C.; Hu, H.-I.; Yen, B.L. Multilineage Differentiation and Characterization of the Human Fetal Osteoblastic 1.19 Cell Line: A Possible in vitro Model of Human Mesenchymal Progenitors. Stem Cells 2007, 25, 125–131. [Google Scholar] [CrossRef]
- Marozin, S.; Simon-Nobbe, B.; Irausek, S.; Chung, L.W.K.; Lepperdinger, G. Kinship of Conditionally Immortalized Cells Derived from Fetal Bone to Human Bone-Derived Mesenchymal Stroma Cells. Sci. Rep. 2021, 11, 10933. [Google Scholar] [CrossRef] [PubMed]
- de Souza Malaspina, T.S.; Zambuzzi, W.F.; dos Santos, C.X.; Campanelli, A.P.; Laurindo, F.R.M.; Sogayar, M.C.; Granjeiro, J.M. A Possible Mechanism of Low Molecular Weight Protein Tyrosine Phosphatase (LMW-PTP) Activity Modulation by Glutathione Action during Human Osteoblast Differentiation. Arch. Oral Biol. 2009, 54, 642–650. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Zhou, Z.; Saunders, M.M.; Donahue, H.J. Modulation of Connexin43 Alters Expression of Osteoblastic Differentiation Markers. Am. J. Physiol. Cell Physiol. 2006, 290, C1248–C1255. [Google Scholar] [CrossRef] [PubMed]
- Dominici, M.; Le Blanc, K.; Mueller, I.; Slaper-Cortenbach, I.; Marini, F.C.; Krause, D.S.; Deans, R.J.; Keating, A.; Prockop, D.J.; Horwitz, E.M. Minimal Criteria for Defining Multipotent Mesenchymal Stromal Cells. The International Society for Cellular Therapy Position Statement. Cytotherapy 2006, 8, 315–317. [Google Scholar] [CrossRef] [PubMed]
- Majka, M.; Janeczko, M.; Goździk, J.; Jarocha, D.; Auguściak-Duma, A.; Witecka, J.; Lesiak, M.; Koryciak-Komarska, H.; Sieroń, A.L.; Pietrzyk, J.J. Cell Therapy of a Patient with Type III Osteogenesis Imperfecta Caused by Mutation in COL1A2 Gene and Unstable Collagen Type I. Open J. Genet. 2013, 3, 49–60. [Google Scholar] [CrossRef]
- Untergasser, A.; Nijveen, H.; Rao, X.; Bisseling, T.; Geurts, R.; Leunissen, J.A.M. Primer3Plus, an Enhanced Web Interface to Primer3. Nuc. Ac. Res. 2007, 35, 71–74. [Google Scholar] [CrossRef] [PubMed]
- Kurzik-Dumke, U.; Hörner, M.; Czaja, J.; Nicotra, M.R.; Simiantonaki, N.; Koslowski, M.; Natali, P.G. Progression of Colorectal Cancers Correlates with Overexpression and Loss of Polarization of Expression of the Htid-1 Tumor Suppressor. Int. J. Mol. Med. 2001, 21, 19–31. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Hartmann, I.; Hollweck, T.; Haffner, S.; Krebs, M.; Meiser, B.; Reichart, B.; Eissner, G. Umbilical Cord Tissue-Derived Mesenchymal Stem Cells Grow Best under GMP-Compliant Culture Conditions and Maintain Their Phenotypic and Functional Properties. J. Immunol. Methods 2010, 363, 80–89. [Google Scholar] [CrossRef] [PubMed]
- Rashid, U.; Saba, E.; Yousaf, A.; Tareen, W.A.; Sarfraz, A.; Rhee, M.H.; Sandhu, M.A. Autologous Platelet Lysate Is an Alternative to Fetal Bovine Serum for Canine Adipose-Derived Mesenchymal Stem Cell Culture and Differentiation. Animals 2023, 13, 2655. [Google Scholar] [CrossRef]
- Hanna, H.; Mir, L.M.; Andre, F.M. In Vitro Osteoblastic Differentiation of Mesenchymal Stem Cells Generates Cell Layers with Distinct Properties. Stem Cell Res. Ther. 2018, 9, 203. [Google Scholar] [CrossRef] [PubMed]
- Choi, M.-H.; Noh, W.-C.; Park, J.-W.; Lee, J.-M.; Suh, J.-Y. Gene Expression Pattern during Osteogenic Differentiation of Human Periodontal Ligament Cells in Vitro. J. Periodontal Implant Sci. 2011, 41, 167. [Google Scholar] [CrossRef] [PubMed]
- Mendicino, M.; Bailey, A.M.; Wonnacott, K.; Puri, R.K.; Bauer, S.R. MSC-Based Product Characterization for Clinical Trials: An FDA Perspective. Cell Stem Cell 2014, 14, 141–145. [Google Scholar] [CrossRef] [PubMed]
- Rallapalli, S.; Bishi, D.K.; Verma, R.S.; Cherian, K.M.; Guhathakurta, S. A Multiplex PCR Technique to Characterize Human Bone Marrow Derived Mesenchymal Stem Cells. Biotechnol. Lett. 2009, 31, 1843–1850. [Google Scholar] [CrossRef] [PubMed]
- Delorme, B.; Ringe, J.; Gallay, N.; Vern, Y.L.; Kerboeuf, D.; Jorgensen, C.; Rosset, P.; Sensebé, L.; Layrolle, P.; Häupl, T.; et al. Specific Plasma Membrane Protein Phenotype of Culture-Amplified and Native Human Bone Marrow Mesenchymal Stem Cells. Blood 2008, 111, 2631–2635. [Google Scholar] [CrossRef] [PubMed]
- Jin, H.J.; Park, S.K.; Oh, W.; Yang, Y.S.; Kim, S.W.; Choi, S.J. Down-Regulation of CD105 Is Associated with Multi-Lineage Differentiation in Human Umbilical Cord Blood-Derived Mesenchymal Stem Cells. Biochem. Biophys. Res. Commun. 2009, 381, 676–681. [Google Scholar] [CrossRef]
- Ishibashi, O.; Ikegame, M.; Takizawa, F.; Yoshizawa, T.; Moksed, A.; Izawa, F.; Mera, H.; Matsuda, A.; Kawashima, H. Endoglin Is Involved in BMP-2-Induced Osteogenic Differentiation of Periodontal Ligament Cells through a Pathway Independent of Smad-1/5/8 Phosphorylation. J. Cell Physiol. 2009, 222, 465–473. [Google Scholar] [CrossRef] [PubMed]
- Lévi, B.; Wan, D.C.; Glotzbach, J.P.; Hyun, J.S.; Januszyk, M.; Montoro, D.T.; Sorkin, M.; James, A.W.; Nelson, E.R.; Li, S.; et al. CD105 Protein Depletion Enhances Human Adipose-Derived Stromal Cell Osteogenesis through Reduction of Transforming Growth Factor β1 (TGF-β1) Signaling. J. Biol. Chem. 2011, 286, 39497–39509. [Google Scholar] [CrossRef]
- Lian, J.B.; Stein, G.S. Development of the Osteoblast Phenotype: Molecular Mechanisms Mediating Osteoblast Growth and Differentiation. Iowa Orthop. J. 1995, 15, 118–140. [Google Scholar]
- Ren, L.; Guo, L.; Kou, N.; Lv, J.; Wang, Z.; Yang, K. LncRNA LINC00963 Promotes Osteogenic Differentiation of HBMSCs and Alleviates Osteoporosis Progression by Targeting MiRNA-760/ETS1 Axis. Autoimmunity 2021, 54, 313–325. [Google Scholar] [CrossRef]
- Wang, H.; Fan, M.; An, Y.; He, D. Molecular Mechanism of Long Noncoding RNA SNHG14 in Osteogenic Differentiation of Bone Marrow-Derived Mesenchymal Stem Cells through the NEDD4L/FOXA2/PCP4 Axis. Stem Cells Int. 2023, 2023, 7545635. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Zhang, L.; Liu, H.; Zhang, J.; Tang, P. Exosomes Promote HFOB1.19 Proliferation and Differentiation via LINC00520. J. Orthop. Surg. Res. 2023, 18, 546. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.-T.; Shih, Y.-R.V.; Kuo, T.K.; Lee, O.K.; Wei, Y.-H. Coordinated Changes of Mitochondrial Biogenesis and Antioxidant Enzymes during Osteogenic Differentiation of Human Mesenchymal Stem Cells. Stem Cells 2008, 26, 960–968. [Google Scholar] [CrossRef] [PubMed]
- Ng, A.C.-H.; Baird, S.D.; Screaton, R.A. Essential Role of TID1 in Maintaining Mitochondrial Membrane Potential Homogeneity and Mitochondrial DNA Integrity. Mol. Cell Biol. 2014, 34, 1427–1437. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, M.; Imanaka-Yoshida, K.; Yoshida, T.; Wood, M.; Fearns, C.; Tatake, R.J.; Lee, J.-D. A Crucial Role of Mitochondrial Hsp40 in Preventing Dilated Cardiomyopathy. Nat. Med. 2005, 12, 128–132. [Google Scholar] [CrossRef] [PubMed]
- Ciesielski, G.L.; Plotka, M.; Manicki, M.; Schilke, B.A.; Dutkiewicz, R.; Sahi, C.; Marszalek, J.; Craig, E.A. Nucleoid Localization of Hsp40 Mdj1 Is Important for Its Function in Maintenance of Mitochondrial DNA. Biochim. Biophys. Acta 2013, 1833, 2233–2243. [Google Scholar] [CrossRef] [PubMed]
- Sayson, S.L.; Fan, J.-N.; Ku, C.-L.; Lo, J.-F.; Chou, S.-H. DNAJA3 Regulates B Cell Development and Immune Function. Biomed. J. 2023, 100628. [Google Scholar] [CrossRef] [PubMed]
- Renault, M.-A.; Jalvy, S.; Potier, M.; Belloc, I.; Genot, E.; Dekker, L.V.; Desgranges, C.; Gadeau, A.-P. UTP Induces Osteopontin Expression through a Coordinate Action of NFκB, Activator Protein-1, and Upstream Stimulatory Factor in Arterial Smooth Muscle Cells. J. Biol. Chem. 2005, 280, 2708–2713. [Google Scholar] [CrossRef] [PubMed]
- Cheng, H.; Cenciarelli, C.; Tao, M.; Parks, W.P.; Cheng-Mayer, C. HTLV-1 Tax-Associated HTid-1, a Human DnaJ Protein, Is a Repressor of IκB Kinase β Subunit. J. Biol. Chem. 2002, 277, 20605–20610. [Google Scholar] [CrossRef] [PubMed]
- Si, J.; Wang, C.; Zhang, D.; Wang, B.; Hou, W.; Zhou, Y. Osteopontin in Bone Metabolism and Bone Diseases. Med. Sci. Monit. 2020, 26, e919159. [Google Scholar] [CrossRef]
- Paunovska, K.; Loughrey, D.; Dahlman, J.E. Drug Delivery Systems for RNA Therapeutics. Nat. Rev. Gene 2022, 23, 265–280. [Google Scholar] [CrossRef] [PubMed]



| Cells | B-MSCs | hFBO1.19 | ||
|---|---|---|---|---|
| Experimental Conditions | Proliferation (days) | Differentiation (days) | 37 °C (days) | 39.5 °C (days) |
| Proliferation assay (Alamar Blue) | 1, 2, 3, 5 | 1, 2, 3, 4, 5, 7, 14, 28 | 1, 2, 3, 7 | 1, 2, 3, 7 |
| Flow Cytometry | 5 | 4, 7, 14, 28 | 10 | 10 |
| Alizarin red, alkaline phosphatase activity, Western blotting, RT q-PCR | 5 | 4, 7, 14, 28 | - | 10 |
| siRNA | 1, 2, 3 | 1, 2, 3, 4, 5, 14 | - | - |
| Transcript | Name | Sequence | Primer Length (bps) | Product Length (bps) |
|---|---|---|---|---|
| GAPDH 1 | GAPDH_f | 5′-GAG TCA ACG GAT TTG GTC GTA-3′ | 21 | 245 |
| GAPDH_r | 5′-GCC CCA CTT GAT TTT GGA G-3′ | 19 | 245 | |
| HPRT1 2 | HPRT_s | 5′-TGA CAC TGG CAA AAC AAT GCA-3′ | 21 | 94 |
| HPRT_a | 5′-GGT CCT TTT CAC CAG CAA GCT-3′ | 21 | 94 | |
| SPP1 1 | SSP1_F | 5′-GCA ACC GAA GTT TTC ACT CC-3′ | 20 | 339 |
| SPP1_R | 5′-GCT CTC ATC ATT GGC TTT CC-3′ | 20 | 339 | |
| ALPL 1 | ALPL_F | 5′-CAA GCA CTC CCA CTT CAT CTG-3′ | 21 | 203 |
| ALPL_R | 5′-CCA GCA AGA AGA AGC CTT TG-3′ | 20 | 203 | |
| TID-S 2 | P3f | 5′-CAG CCT CAG GAA GAA ACC ATC-3′ | 21 | 399 |
| P3r | 5′-GGG ATC GTC ACG TTG ATC GTC-3′ | 21 | 399 | |
| TID-I 1&2 | P1f | 5′-CTG GGA TAT CAT GAG GTA AAC-3′ | 21 | 716 |
| P2r | 5′-CCA GTG GAT CTT TTT CCA GAG-3′ | 21 | 716 | |
| TID-L 2 | P1f | 5′-GTT GAC ATT CAA TCA AGC TGC-3′ | 21 | 821 |
| P1r | 5′-CTG GGA TAT CAT GAG GTA AAC-3′ | 21 | 821 | |
| TID-L 1 | TL_F | 5′-TCA CCG TGA ACA TCA TGG AC-3′ | 20 | 733 |
| TL_R | 5′-GAA AGG AAT CCC TCC TCG TC-3′ | 20 | 733 |
| Cell Type | % of Cells with the Antigen in the Population | Value Compared with B-MSCs (%) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| CD105 | CD90 | CD73 | CD90 + CD105 | CD105 + CD73 | CD73 + CD90 | CD44 | HSC Mark. | FSC | SSC | ||
| B-MSCs | 97 | 100 | 100 | 98 | 99 | 100 | 99 | 1 | 100 | 100 | |
| B-MSCs + 2 passages | 93 | 100 | 100 | 94 | 95 | 99 | 99 | 0 | 129 | 137 | |
| Days of differentiation | 4 | 87 * | 98 | 99 | 82 * | 87 * | 97 | 99 | 0 | 82 | 74 |
| 7 | 80 * | 97 * | 99 | 77 * | 82 * | 97 | 99 | 0 | 83 | 63 | |
| 14 | 56 * | 97 | 99 | 54 * | 56 | 95 | 99 | 0 | 65 | 44 | |
| 28 | 19 * | 78 | 86 | 15 * | 25 * | 75 | 87 | 0 | 52 * | 29 * | |
| hFOB1.19 @37 °C | 95 | 91 * | 99 | 88 * | 95 | 91 | 99 | 0 | 113 | 32 * | |
| hFOB1.19 @39.5 °C | 63 * | 28 * | 96 | 1 * | 34 * | 12 * | 91 * | 0 | 79 | 60 | |
| Cell Type | Relative Proliferation | Relative Activity of Alkaline Phosphatase | Relative Number of Calcium Deposits | |
|---|---|---|---|---|
| B-MSCs | 99.7 ± 1.6% | 102.5 ± 2.2% | - | |
| Day of Differentiation | 3rd | 97.7 ± 1.0% | 103.5 ± 4.8% | - |
| 5th | 99.3 ± 0.9% | 100.3 ± 2.6% | - | |
| 14th | - | - | 100.6 ± 4.5% | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Krakowian, D.; Lesiak, M.; Auguściak-Duma, A.; Witecka, J.; Kusz, D.; Sieroń, A.L.; Gawron, K. Analysis of the TID-I and TID-L Splice Variants’ Expression Profile under In Vitro Differentiation of Human Mesenchymal Bone Marrow Cells into Osteoblasts. Cells 2024, 13, 1021. https://doi.org/10.3390/cells13121021
Krakowian D, Lesiak M, Auguściak-Duma A, Witecka J, Kusz D, Sieroń AL, Gawron K. Analysis of the TID-I and TID-L Splice Variants’ Expression Profile under In Vitro Differentiation of Human Mesenchymal Bone Marrow Cells into Osteoblasts. Cells. 2024; 13(12):1021. https://doi.org/10.3390/cells13121021
Chicago/Turabian StyleKrakowian, Daniel, Marta Lesiak, Aleksandra Auguściak-Duma, Joanna Witecka, Damian Kusz, Aleksander L. Sieroń, and Katarzyna Gawron. 2024. "Analysis of the TID-I and TID-L Splice Variants’ Expression Profile under In Vitro Differentiation of Human Mesenchymal Bone Marrow Cells into Osteoblasts" Cells 13, no. 12: 1021. https://doi.org/10.3390/cells13121021
APA StyleKrakowian, D., Lesiak, M., Auguściak-Duma, A., Witecka, J., Kusz, D., Sieroń, A. L., & Gawron, K. (2024). Analysis of the TID-I and TID-L Splice Variants’ Expression Profile under In Vitro Differentiation of Human Mesenchymal Bone Marrow Cells into Osteoblasts. Cells, 13(12), 1021. https://doi.org/10.3390/cells13121021








