Preliminary Single-Cell RNA-Sequencing Analysis Uncovers Adipocyte Heterogeneity in Lipedema
Abstract
:1. Introduction
2. Materials and Methods
2.1. Patient Recruitment and Sample Collection
2.2. Adipose Tissue Homogenization, Enzymatic Cell Dissociation, and Single-Cell RNA Sequencing
2.3. Complementary DNA Synthesis and Amplification
2.4. Data Processing and Statistical Analysis
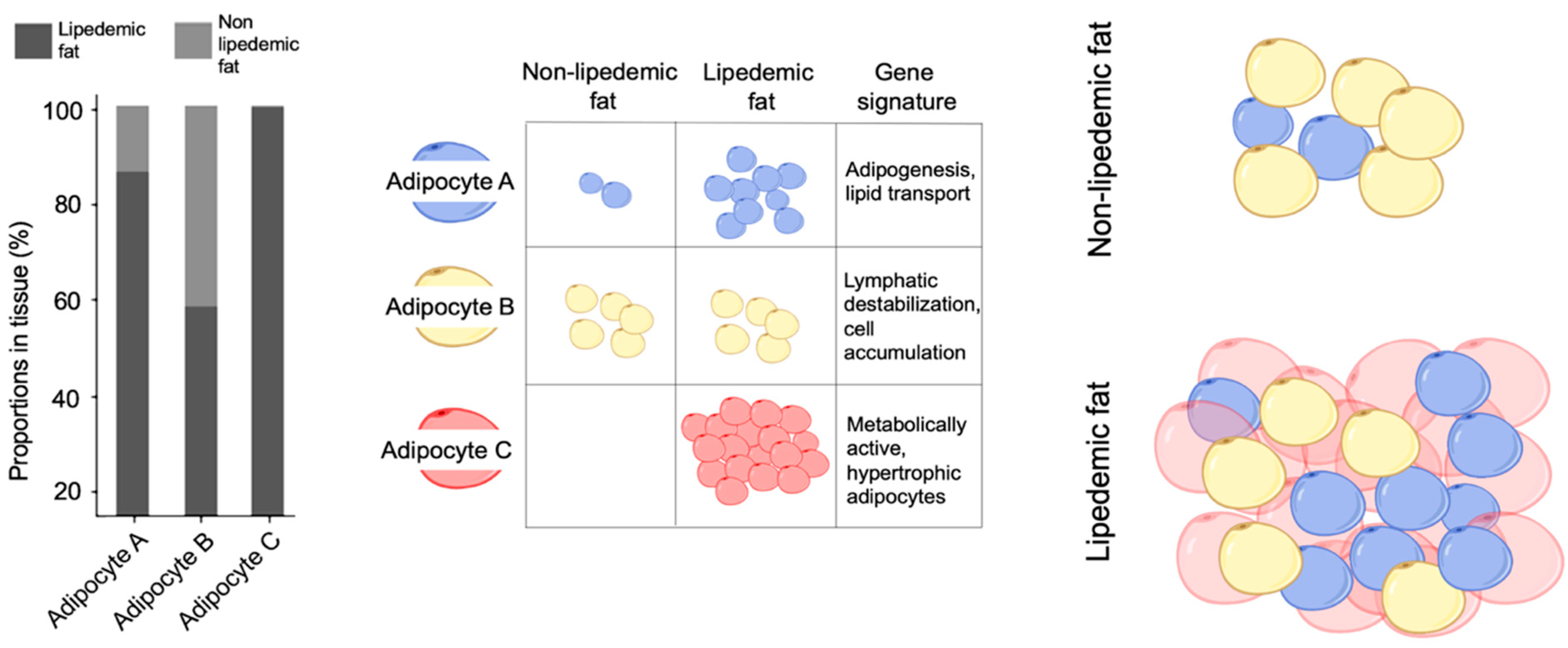
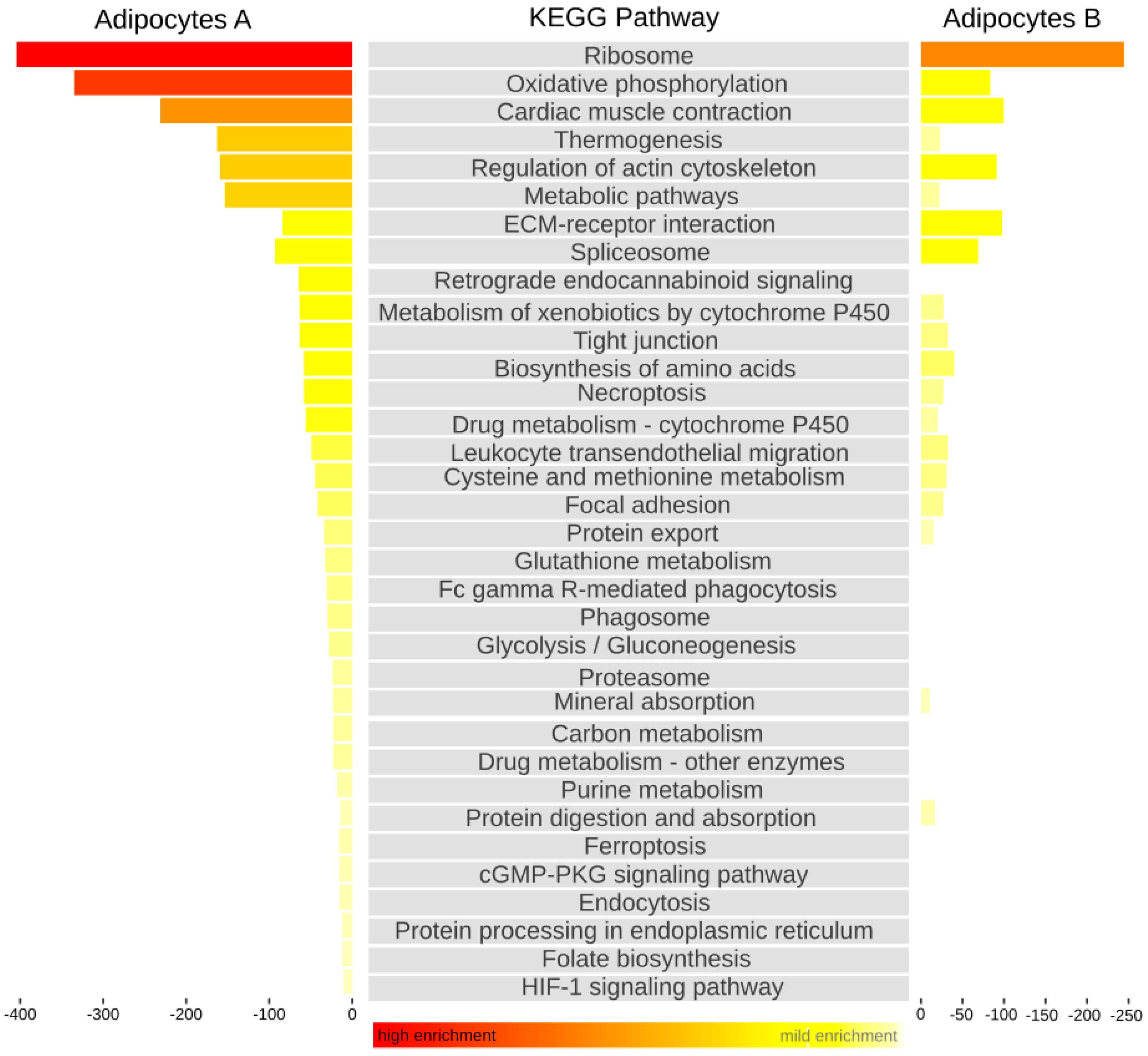
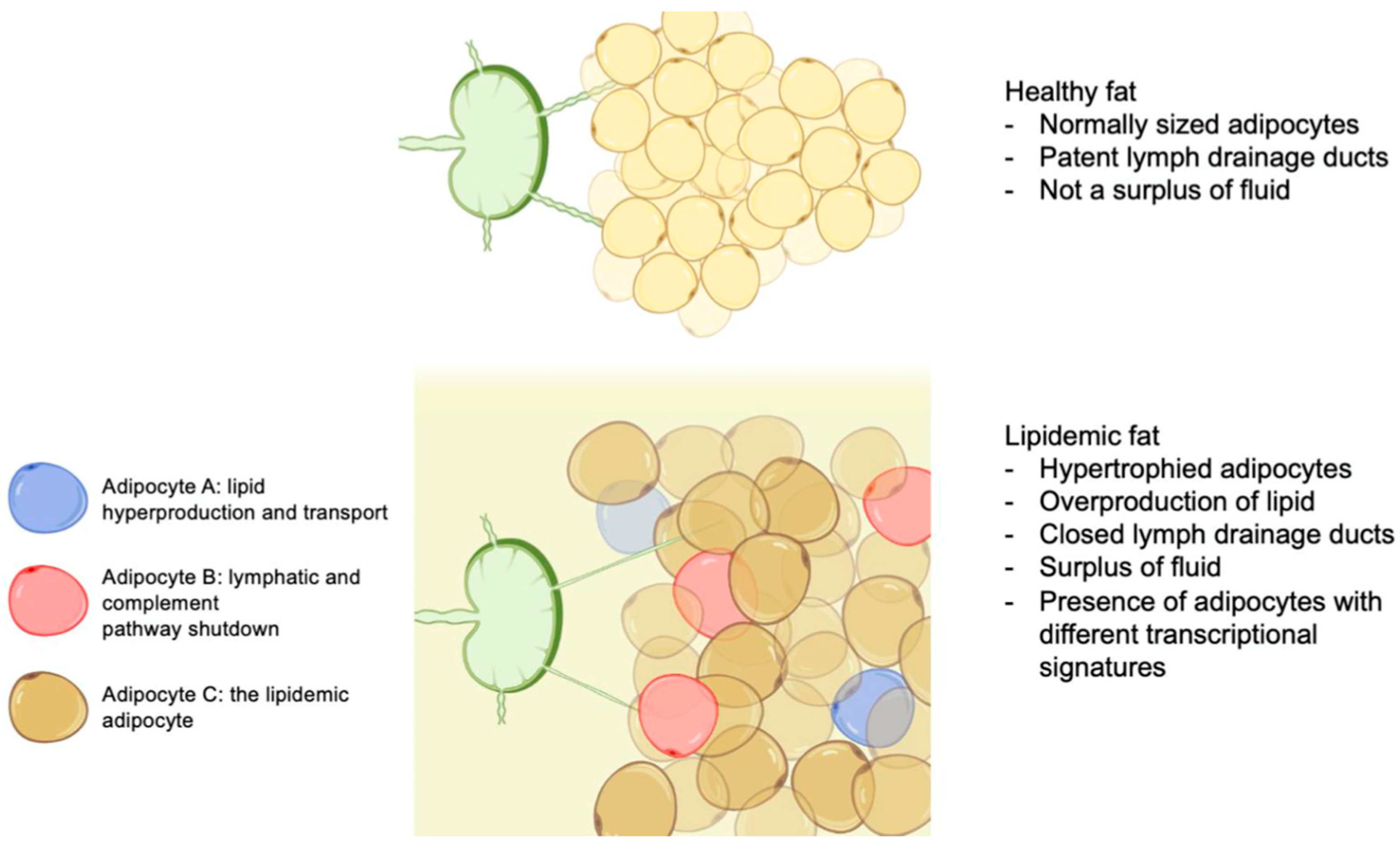
3. Results and Discussion
4. Summary and Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kruppa, P.; Georgiou, I.; Biermann, N.; Prantl, L.; Klein-Weigel, P.; Ghods, M. Lipedema-Pathogenesis, Diagnosis, and Treatment Options. Dtsch. Aerzteblatt Online 2020, 117, 396–403. [Google Scholar] [CrossRef] [PubMed]
- Wold, L.E.; Hines, E.A., Jr.; Allen, E.V. Lipedema of the legs; a syndrome characterized by fat legs and edema. Ann. Intern. Med. 1951, 34, 1243–1250. [Google Scholar] [CrossRef] [PubMed]
- Peled, A.A.W.; Kappos, E. Lipedema: Diagnostic and management challenges. Int. J. Women’s Health 2016, 8, 389–395. [Google Scholar] [CrossRef] [PubMed]
- Dudek, J.E.; Białaszek, W.; Ostaszewski, P.; Smidt, T. Depression and appearance-related distress in functioning with lipedema. Psychol. Health Med. 2018, 23, 846–853. [Google Scholar] [CrossRef] [PubMed]
- Ghods, M.; Georgiou, I.; Schmidt, J.; Kruppa, P. Disease progression and comorbidities in lipedema patients: A 10-year retrospective analysis. Dermatol. Ther. 2020, 33, e14534. [Google Scholar] [CrossRef]
- Buck, D.W.; Herbst, K.L. Lipedema: A Relatively Common Disease with Extremely Common Misconceptions. Plast. Reconstr. Surg.-Glob. Open 2016, 4, e1043. [Google Scholar] [CrossRef]
- Vyas, A.; Adnan, G. Lipedema. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2024. Available online: http://www.ncbi.nlm.nih.gov/books/NBK573066/ (accessed on 11 March 2024).
- Al-Ghadban, S.; Bunnell, B.A. Adipose Tissue-Derived Stem Cells: Immunomodulatory Effects and Therapeutic Potential. Physiology 2020, 35, 125–133. [Google Scholar] [CrossRef] [PubMed]
- Szolnoky, G.; Ifeoluwa, A.; Tuczai, M.; Varga, E.; Varga, M.; Dosa-Racz, E.; Kemeny, L. Measurement of capillary fragility: A useful tool to differentiate lipedema from obesity? Lymphology 2017, 50, 203–209. [Google Scholar]
- Kempa, S.; Buechler, C.; Föh, B.; Felthaus, O.; Prantl, L.; Günther, U.L.; Müller, M.; Derer-Petersen, S.; Sina, C.; Schmelter, F.; et al. Serum Metabolomic Profiling of Patients with Lipedema. Int. J. Mol. Sci. 2023, 24, 17437. [Google Scholar] [CrossRef]
- Felmerer, G.; Stylianaki, A.; Hollmén, M.; Ströbel, P.; Stepniewski, A.; Wang, A.; Frueh, F.S.; Kim, B.-S.; Giovanoli, P.; Lindenblatt, N.; et al. Increased levels of VEGF-C and macrophage infiltration in lipedema patients without changes in lymphatic vascular morphology. Sci. Rep. 2020, 10, 10947. [Google Scholar] [CrossRef]
- Fernandes, H.J.; Patikas, N.; Foskolou, S.; Field, S.F.; Park, J.-E.; Byrne, M.L.; Bassett, A.R.; Metzakopian, E. Single-Cell Transcriptomics of Parkinson’s Disease Human In Vitro Models Reveals Dopamine Neuron-Specific Stress Responses. Cell Rep. 2020, 33, 108263. [Google Scholar] [CrossRef] [PubMed]
- Strohmeier, K.; Hofmann, M.; Jacak, J.; Narzt, M.-S.; Wahlmueller, M.; Mairhofer, M.; Schaedl, B.; Holnthoner, W.; Barsch, M.; Sandhofer, M.; et al. Multi-Level Analysis of Adipose Tissue Reveals the Relevance of Perivascular Subpopulations and an Increased Endothelial Permeability in Early-Stage Lipedema. Biomedicines 2022, 10, 1163. [Google Scholar] [CrossRef] [PubMed]
- Veniaminova, N.A.; Jia, Y.Y.; Hartigan, A.M.; Huyge, T.J.; Tsai, S.-Y.; Grachtchouk, M.; Nakagawa, S.; Dlugosz, A.A.; Atwood, S.X.; Wong, S.Y. Distinct mechanisms for sebaceous gland self-renewal and regeneration provide durability in response to injury. Cell Rep. 2023, 42, 113121. [Google Scholar] [CrossRef] [PubMed]
- Izar, B.; Tirosh, I.; Stover, E.H.; Wakiro, I.; Cuoco, M.S.; Alter, I.; Rodman, C.; Leeson, R.; Su, M.-J.; Shah, P.; et al. A single-cell landscape of high-grade serous ovarian cancer. Nat. Med. 2020, 26, 1271–1279. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Chang, H.-W.; Huang, Z.-M.; Nakamura, M.; Sekhon, S.; Ahn, R.; Munoz-Sandoval, P.; Bhattarai, S.; Beck, K.M.; Sanchez, I.M.; et al. Single-cell RNA sequencing of psoriatic skin identifies pathogenic Tc17 cell subsets and reveals distinctions between CD8+ T cells in autoimmunity and cancer. J. Allergy Clin. Immunol. 2021, 147, 2370–2380. [Google Scholar] [CrossRef] [PubMed]
- Stephenson, W.; Donlin, L.T.; Butler, A.; Rozo, C.; Bracken, B.; Rashidfarrokhi, A.; Goodman, S.M.; Ivashkiv, L.B.; Bykerk, V.P.; Orange, D.E.; et al. Single-cell RNA-seq of rheumatoid arthritis synovial tissue using low-cost microfluidic instrumentation. Nat. Commun. 2018, 9, 791. [Google Scholar] [CrossRef] [PubMed]
- Muffat, J.; Walker, D.W. Apolipoprotein D: An overview of its role in aging and age-related diseases. Cell Cycle 2010, 9, 269–273. [Google Scholar] [CrossRef] [PubMed]
- Buso, G.; Depairon, M.; Tomson, D.; Raffoul, W.; Vettor, R.; Mazzolai, L. Lipedema: A Call to Action! Obesity 2019, 27, 1567–1576. [Google Scholar] [CrossRef] [PubMed]
- Ruan, H.-B. Developmental and functional heterogeneity of thermogenic adipose tissue. J. Mol. Cell Biol. 2020, 12, 775–784. [Google Scholar] [CrossRef]
- Varlamov, O.; Chu, M.; Cornea, A.; Sampath, H.; Roberts, C.T., Jr. Cell-Autonomous Heterogeneity of Nutrient Uptake in White Adipose Tissue of Rhesus Macaques. Endocrinology 2015, 156, 80–89. [Google Scholar] [CrossRef]
- Luong, Q.; Huang, J.; Lee, K.Y. Deciphering White Adipose Tissue Heterogeneity. Biology 2019, 8, 23. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Li, J.; He, F.; Li, K.; Li, X.; Zhang, Y. Matrix Gla protein regulates adipogenesis and is serum marker of visceral adiposity. Adipocyte 2020, 9, 68–76. [Google Scholar] [CrossRef] [PubMed]
- Hu, L.; Yang, G.; Hägg, D.; Sun, G.; Jiang, N.; Ricupero, C.L.; Wu, J.; Rodhe, C.H.; Ascherman, J.A.; Chen, L.; et al. IGF1 Promotes Adipogenesis by a Lineage Bias of Endogenous Adipose Stem/Progenitor Cells. Stem Cells 2015, 33, 2483–2495. [Google Scholar] [CrossRef] [PubMed]
- Charkoftaki, G.; Thompson, D.C.; Golla, J.P.; Garcia-Milian, R.; Lam, T.T.; Engel, J.; Vasiliou, V. Integrated multi-omics approach reveals a role of ALDH1A1 in lipid metabolism in human colon cancer cells. Chem. Interact. 2019, 304, 88–96. [Google Scholar] [CrossRef] [PubMed]
- Hara, T.; Nakayama, Y. Chapter 5 CXCL14 and Insulin Action. In Vitamins & Hormones; Elsevier: Amsterdam, The Netherlands, 2009; Volume 80, pp. 107–123. [Google Scholar] [CrossRef]
- Dho, S.H.; Lim, J.C.; Kim, L.K. Beyond the Role of CD55 as a Complement Component. Immune Netw. 2018, 18, e11. [Google Scholar] [CrossRef] [PubMed]
- Festy, F.; Hoareau, L.; Bes-Houtmann, S.; Péquin, A.-M.; Gonthier, M.-P.; Munstun, A.; Hoarau, J.J.; Césari, M.; Roche, R. Surface protein expression between human adipose tissue-derived stromal cells and mature adipocytes. Histochem. Cell Biol. 2005, 124, 113–121. [Google Scholar] [CrossRef] [PubMed]
- Nam, J.S.; Ahn, C.W.; Park, H.J.; Kim, Y.S. Semaphorin 3C is a Novel Adipokine Representing Exercise-Induced Improvements of Metabolism in Metabolically Healthy Obese Young Males. Sci. Rep. 2020, 10, 10005. [Google Scholar] [CrossRef] [PubMed]
- Mumblat, Y.; Kessler, O.; Ilan, N.; Neufeld, G. Full-Length Semaphorin-3C Is an Inhibitor of Tumor Lymphangiogenesis and Metastasis. Cancer Res. 2015, 75, 2177–2186. [Google Scholar] [CrossRef]
- Amann-Vesti, B.R.; Franzeck, U.K.; Bollinger, A. Microlymphatic aneurysms in patients with lipedema. Lymphology 2001, 34, 170–175. [Google Scholar]
- Han, C.Y.; Kang, I.; Harten, I.A.; Gebe, J.A.; Chan, C.K.; Omer, M.; Alonge, K.M.; Hartigh, L.J.D.; Kjerulf, D.G.; Goodspeed, L.; et al. Adipocyte-Derived Versican and Macrophage-Derived Biglycan Control Adipose Tissue Inflammation in Obesity. Cell Rep. 2020, 31, 107818. [Google Scholar] [CrossRef]
- Han, J.; Chu, W.; Lin, K.; Wang, X.; Gao, Y. β-catenin activation in Gli-1+ stem cells leads to reprograming of the hair follicle. Eur. J. Dermatol. 2023, 33, 710–712. [Google Scholar] [CrossRef] [PubMed]
- Gao, H.; Volat, F.; Sandhow, L.; Galitzky, J.; Nguyen, T.; Esteve, D.; Åström, G.; Mejhert, N.; Ledoux, S.; Thalamas, C.; et al. CD36 Is a Marker of Human Adipocyte Progenitors with Pronounced Adipogenic and Triglyceride Accumulation Potential. Stem Cells 2017, 35, 1799–1814. [Google Scholar] [CrossRef] [PubMed]
- Park, J.; Park, J.; Nahm, S.-S.; Choi, I.; Kim, J. Identification of anti-adipogenic proteins in adult bovine serum suppressing 3T3-L1 preadipocyte differentiation. BMB Rep. 2013, 46, 582–587. [Google Scholar] [CrossRef] [PubMed]
- Furuhashi, M.; Saitoh, S.; Shimamoto, K.; Miura, T. Fatty Acid-Binding Protein 4 (FABP4): Pathophysiological Insights and Potent Clinical Biomarker of Metabolic and Cardiovascular Diseases. Clin. Med. Insights Cardiol. 2014, 8 (Suppl. S3), 22–33. [Google Scholar] [CrossRef]
- Jin, W.; Jin, W.; Pan, D. Ifi27 is indispensable for mitochondrial function and browning in adipocytes. Biochem. Biophys. Res. Commun. 2018, 501, 273–279. [Google Scholar] [CrossRef]
| LIPEDEMA CLASSIFICATION: STAGES (I–III) AND MORPHOLOGY (I–V) | |||||
|---|---|---|---|---|---|
| STAGES | Stage I Thickened but soft subcutaneous tissue with small palpable nodules; smooth skin surface | Stage II Thickened subcutaneous tissue, soft; some larger nodules; uneven skin surface. | Stage III Thickened subcutaneous tissue; indurated, large nodules and formation of folds | ||
| MORPHOLOGY | Type I: Buttock | Type II: Thigh | Type III: Entire leg | Type IV: Arm (often associated withType II/III) | Type V: Lower Leg |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pagani, A.; Duscher, D.; Kempa, S.; Ghods, M.; Prantl, L. Preliminary Single-Cell RNA-Sequencing Analysis Uncovers Adipocyte Heterogeneity in Lipedema. Cells 2024, 13, 1028. https://doi.org/10.3390/cells13121028
Pagani A, Duscher D, Kempa S, Ghods M, Prantl L. Preliminary Single-Cell RNA-Sequencing Analysis Uncovers Adipocyte Heterogeneity in Lipedema. Cells. 2024; 13(12):1028. https://doi.org/10.3390/cells13121028
Chicago/Turabian StylePagani, Andrea, Dominik Duscher, Sally Kempa, Mojtaba Ghods, and Lukas Prantl. 2024. "Preliminary Single-Cell RNA-Sequencing Analysis Uncovers Adipocyte Heterogeneity in Lipedema" Cells 13, no. 12: 1028. https://doi.org/10.3390/cells13121028
APA StylePagani, A., Duscher, D., Kempa, S., Ghods, M., & Prantl, L. (2024). Preliminary Single-Cell RNA-Sequencing Analysis Uncovers Adipocyte Heterogeneity in Lipedema. Cells, 13(12), 1028. https://doi.org/10.3390/cells13121028








