miRNA-Driven Regulation of Endothelial-to-Mesenchymal Transition Differs among Thoracic Aortic Aneurysms
Abstract
:1. Introduction
2. Materials and Methods
2.1. Tissue and Serum Sample Collection
2.2. Immunohistochemical Analysis
2.3. Gene Expression Analysis on Tissue Samples
2.4. Gene Expression Analysis on Serum Samples
2.5. Statistical Analysis
3. Results
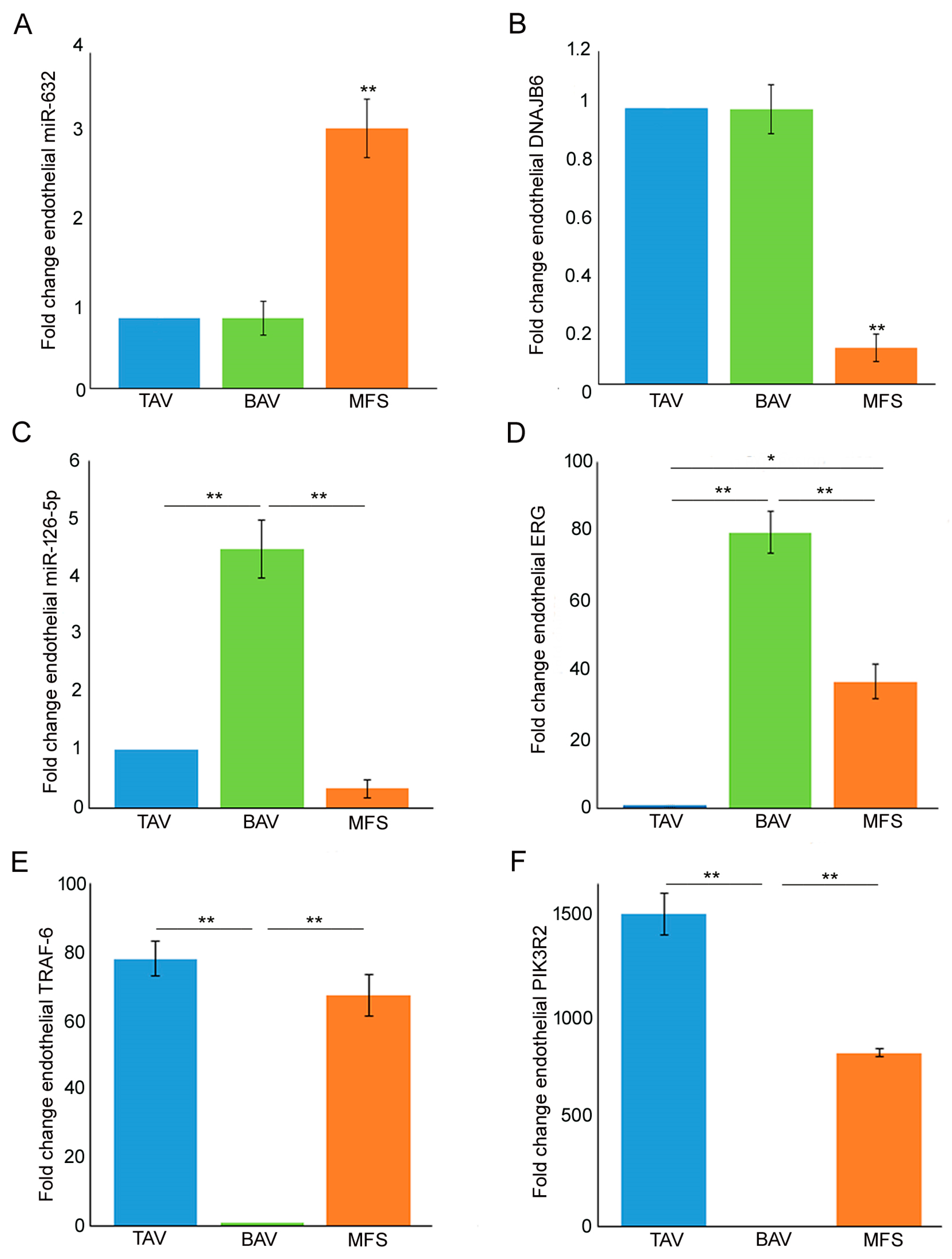
3.1. Different miRNA Deregulation Characterizes the Endothelium of Thoracic Aortic Aneurysms
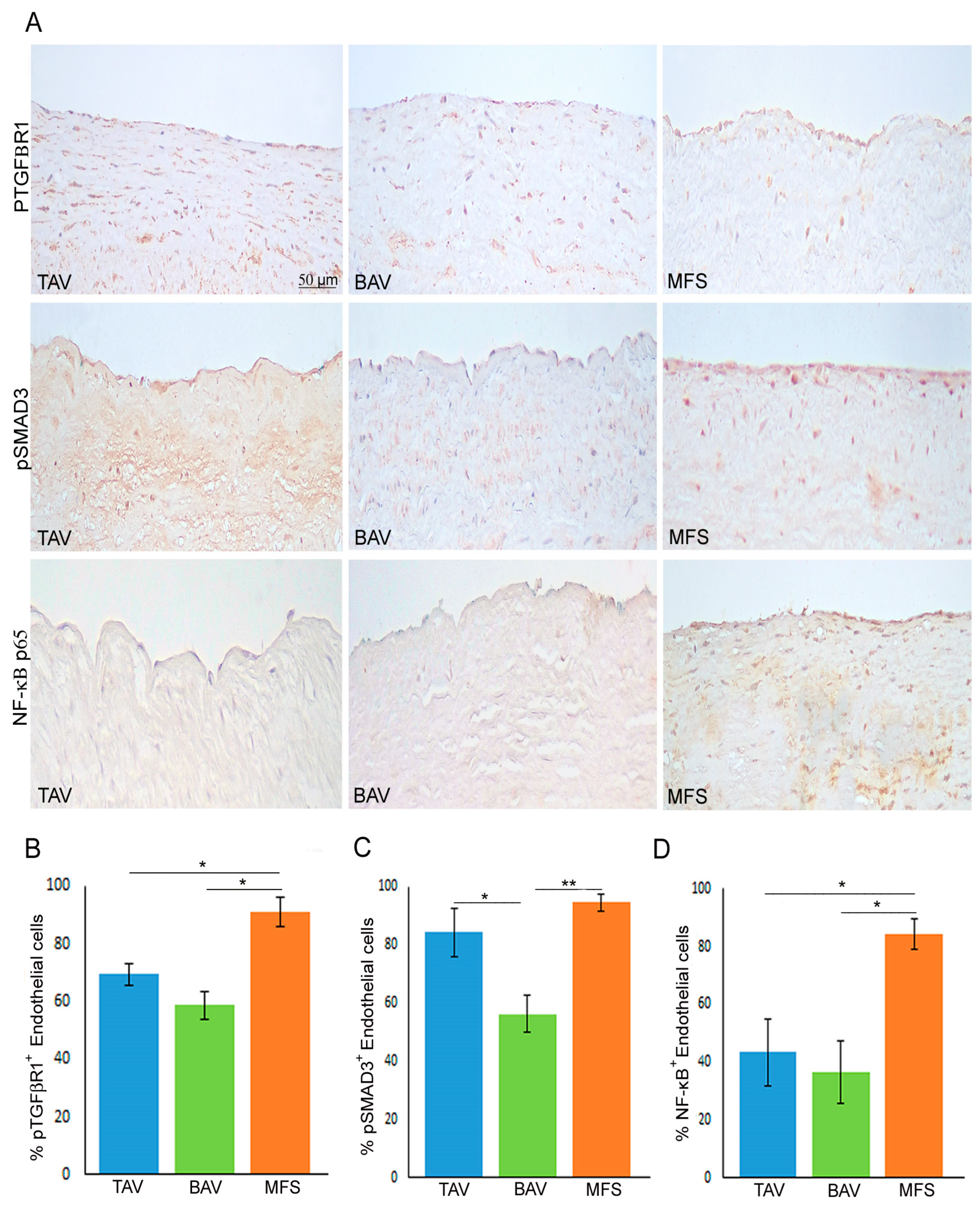
3.2. End–MT Is Exacerbated in MFS Endothelium Compared with TAV and BAV
3.3. Non-Canonical TGF-β Signaling Is Hyper-Activated in MFS Aorta
3.4. MFS Patients’ Serum Showed the Same miR-632 Deregulation Observed in Aortic Endothelium
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Salmasi, M.Y.; Alwis, S.; Cyclewala, S.; Jarral, O.A.; Mohamed, H.; Mozalbat, D.; Nienaber, C.A.; Athanasiou, T.; Morris-Rosendahl, D. The genetic basis of thoracic aortic disease: The future of aneurysm classification? Hell. J. Cardiol. 2023, 69, 41–50. [Google Scholar] [CrossRef]
- Pyeritz, R.E. The Marfan syndrome. Annu. Rev. Med. 2000, 51, 481–510. [Google Scholar] [CrossRef]
- Dawson, A.; Li, Y.; Li, Y.; Ren, P.; Vasquez, H.G.; Zhang, C.; Rebello, K.R.; Ageedi, W.; Azares, A.R.; Mattar, A.B.; et al. Single-Cell Analysis of Aneurysmal Aortic Tissue in Patients with Marfan Syndrome Reveals Dysfunctional TGF-β Signaling. Genes 2021, 13, 95. [Google Scholar] [CrossRef]
- Yassine, N.M.; Shahram, J.T.; Body, S.C. Pathogenic Mechanisms of Bicuspid Aortic Valve Aortopathy. Front. Physiol. 2017, 8, 687. [Google Scholar] [CrossRef]
- Gillis, E.; Kumar, A.A.; Luyckx, I.; Preuss, C.; Cannaerts, E.; van de Beek, G.; Wieschendorf, B.; Alaerts, M.; Bolar, N.; Vandeweyer, G.; et al. Candidate Gene Resequencing in a Large Bicuspid Aortic Valve-Associated Thoracic Aortic Aneurysm Cohort: SMAD6 as an Important Contributor. Front. Physiol. 2017, 8, 400. [Google Scholar] [CrossRef]
- Ma, M.; Li, Z.; Mohamed, M.A.; Liu, L.; Wei, X. Aortic root aortopathy in bicuspid aortic valve associated with high genetic risk. BMC Cardiovasc. Disord. 2021, 21, 413. [Google Scholar] [CrossRef]
- Malashicheva, A.; Kostina, D.; Kostina, A.; Irtyuga, O.; Voronkina, I.; Smagina, L.; Ignatieva, E.; Gavriliuk, N.; Uspensky, V.; Moiseeva, O.; et al. Phenotypic and Functional Changes of Endothelial and Smooth Muscle Cells in Thoracic Aortic Aneurysms. Int. J. Vasc. Med. 2016, 2016, 3107879. [Google Scholar] [CrossRef]
- Souilhol, C.; Harmsen, M.C.; Evans, P.C.; Krenning, G. Endothelial-mesenchymal transition in atherosclerosis. Cardiovasc. Res. 2018, 114, 565–577. [Google Scholar] [CrossRef]
- Terriaca, S.; Scioli, M.G.; Pisano, C.; Ruvolo, G.; Ferlosio, A.; Orlandi, A. miR-632 Induces DNAJB6 Inhibition Stimulating Endothelial-to-Mesenchymal Transition and Fibrosis in Marfan Syndrome Aortopathy. Int. J. Mol. Sci. 2023, 24, 5133. [Google Scholar] [CrossRef]
- Eisenberg, L.M.; Markwald, R.R. Molecular regulation of atrioventricular valvuloseptal morphogenesis. Circ. Res. 1995, 77, 1–6. [Google Scholar] [CrossRef]
- Kovacic, J.C.; Mercader, N.; Torres, M.; Boehm, M.; Fuster, V. Epithelial-to-mesenchymal and endothelial-to-mesenchymal transition: From cardiovascular development to disease. Circulation 2012, 125, 1795–1808. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Duffhues, G.; García de Vinuesa, A.; Ten Dijke, P. Endothelial-to-mesenchymal transition in cardiovascular diseases: Developmental signaling pathways gone awry. Dev. Dyn. 2018, 247, 492–508. [Google Scholar] [CrossRef] [PubMed]
- Muylaert, D.E.P.; de Jong, O.G.; Slaats, G.G.G.; Nieuweboer, F.E.; Fledderus, J.O.; Goumans, M.J.; Hierck, B.P.; Verhaar, M.C. Environmental Influences on Endothelial to Mesenchymal Transition in Developing Implanted Cardiovascular Tissue-Engineered Grafts. Tissue Eng. Part B Rev. 2016, 22, 58–67. [Google Scholar] [CrossRef] [PubMed]
- Zhong, A.; Mirzaei, Z.; Simmons, C.A. The Roles of Matrix Stiffness and ß-Catenin Signaling in Endothelial-to-Mesenchymal Transition of Aortic Valve Endothelial Cells. Cardiovasc. Eng. Technol. 2018, 9, 158–167. [Google Scholar] [CrossRef]
- Abu-Halima, M.; Kahraman, M.; Henn, D.; Rädle-Hurst, T.; Keller, A.; Abdul-Khaliq, H.; Meese, E. Deregulated microRNA and mRNA expression profiles in the peripheral blood of patients with Marfan syndrome. J. Transl. Med. 2018, 16, 60. [Google Scholar] [CrossRef] [PubMed]
- Kim, R.; Lee, S.; Lee, J.; Kim, M.; Kim, W.J.; Lee, H.W.; Lee, M.Y.; Kim, J.; Chang, W. Exosomes derived from microRNA-584 transfected mesenchymal stem cells: Novel alternative therapeutic vehicles for cancer therapy. BMB Rep. 2018, 51, 406–411. [Google Scholar] [CrossRef] [PubMed]
- Gunaratne, A.; Benchabane, H.; Di Guglielmo, G.M. Regulation of TGFβ receptor trafficking and signaling by atypical protein kinase C. Cell. Signal. 2012, 24, 119–130. [Google Scholar] [CrossRef]
- Clayton, S.W.; Ban, G.I.; Liu, C.; Serra, R. Canonical and noncanonical TGF-β signaling regulate fibrous tissue differentiation in the axial skeleton. Sci. Rep. 2020, 10, 21364. [Google Scholar] [CrossRef]
- Wang, S.; Aurora, A.B.; Johnson, B.A.; Qi, X.; McAnally, J.; Hill, J.A.; Richardson, J.A.; Bassel-Duby, R.; Olson, E.N. The endothelial-specific microRNA miR-126 governs vascular integrity and angiogenesis. Dev. Cell 2008, 15, 261–271. [Google Scholar] [CrossRef]
- Pisano, C.; Terriaca, S.; Scioli, M.G.; Nardi, P.; Altieri, C.; Orlandi, A.; Ruvolo, G.; Balistreri, C.R. The Endothelial Transcription Factor ERG Mediates a Differential Role in the Aneurysmatic Ascending Aorta with Bicuspid or Tricuspid Aorta Valve: A Preliminary Study. Int. J. Mol. Sci. 2022, 23, 0848. [Google Scholar] [CrossRef]
- Harris, T.A.; Yamakuchi, M.; Kondo, M.; Oettgen, P.; Lowenstein, C.J. Ets-1 and Ets-2 regulate the expression of microRNA-126 in endothelial cells. Arterioscler. Thromb. Vasc. Biol. 2010, 30, 1990–1997. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Sun, J.; Kahaleh, B. Epigenetic down-regulation of microRNA-126 in scleroderma endothelial cells is associated with impaired responses to VEGF and defective angiogenesis. J. Cell. Mol. Med. 2021, 25, 7078–7088. [Google Scholar] [CrossRef] [PubMed]
- Fu, R.; Tong, J.S. miR-126 reduces trastuzumab resistance by targeting PIK3R2 and regulating AKT/mTOR pathway in breast cancer cells. J. Cell. Mol. Med. 2020, 24, 7600–7608. [Google Scholar] [CrossRef] [PubMed]
- Mao, X.; Wu, Y.; Xu, W. miR-126-5p expression in the plasma of patients with sepsis-induced acute lung injury and its correlation with inflammation and immune function. Clin. Respir. J. 2023, 17, 629–637. [Google Scholar] [CrossRef] [PubMed]
- Kostina, A.S.; Uspensky, V.; Irtyuga, O.B.; Ignatieva, E.V.; Freylikhman, O.; Gavriliuk, N.D.; Moiseeva, O.M.; Zhuk, S.; Tomilin, A.; Kostareva, A.; et al. Notch-dependent EMT is attenuated in patients with aortic aneurysm and bicuspid aortic valve. Biochim. Biophys. Acta 2016, 1862, 733–740. [Google Scholar] [CrossRef] [PubMed]
- Timmerman, L.A.; Grego-Bessa, J.; Raya, A.; Bertrán, E.; Pérez-Pomares, J.M.; Díez, J.; Aranda, S.; Palomo, S.; McCormick, F.; Izpisúa-Belmonte, J.C.; et al. Notch promotes epithelial-mesenchymal transition during cardiac development and oncogenic transformation. Genes Dev. 2004, 18, 99–115. [Google Scholar] [CrossRef] [PubMed]
- Gould, R.A.; Aziz, H.; Woods, C.E.; Seman-Senderos, M.A.; Sparks, E.; Preuss, C.; Wünnemann, F.; Bedja, D.; Moats, C.R.; McClymont, S.A.; et al. ROBO4 variants predispose individuals to bicuspid aortic valve and thoracic aortic aneurysm. Nat. Genet. 2019, 51, 42–50. [Google Scholar] [CrossRef] [PubMed]
- Maleki, S.; Kjellqvist, S.; Paloschi, V.; Magné, J.; Branca, R.M.; Du, L.; Hultenby, K.; Petrini, J.; Fuxe, J.; Lehtiö, J.; et al. Mesenchymal state of intimal cells may explain higher propensity to ascending aortic aneurysm in bicuspid aortic valves. Sci. Rep. 2016, 6, 35712. [Google Scholar] [CrossRef] [PubMed]
- Loeys, B.L.; Dietz, H.C.; Braverman, A.C.; Callewaert, B.L.; De Backer, J.; Devereux, R.B.; Hilhorst-Hofstee, Y.; Jondeau, G.; Faivre, L.; Milewicz, D.M.; et al. The revised Ghent nosology for the Marfan syndrome. J. Med. Genet. 2010, 47, 476–485. [Google Scholar] [CrossRef]
- Evangelista, A.; Flachskampf, F.A.; Erbel, R.; Antonini-Canterin, F.; Vlachopoulos, C.; Rocchi, G.; Sicari, R.; Nihoyannopoulos, P.; Zamorano, J.; Pepi, M.; et al. Echocardiography in aortic diseases: EAE recommendations for clinical practice. Eur. J. Echocardiogr. 2010, 11, 645–658. [Google Scholar] [CrossRef]
- Kouchoukos, N.T.; Wareing, T.H.; Murphy, S.F.; Perrillo, J.B. Sixteen-year experience with aortic root replacement. Results of 172 operations. Ann. Surg. 1991, 214, 308–318; discussion 318–320. [Google Scholar] [CrossRef] [PubMed]
- Orlandi, A.; Ferlosio, A.; Gabbiani, G.; Spagnoli, L.G.; Ehrlich, P.H. Phenotypic heterogeneity influences the behavior of rat aortic smooth muscle cells in collagen lattice. Exp. Cell Res. 2005, 311, 317–327. [Google Scholar] [CrossRef]
- D’Amico, F.; Doldo, E.; Pisano, C.; Scioli, M.G.; Centofanti, F.; Proietti, G.; Falconi, M.; Sangiuolo, F.; Ferlosio, A.; Ruvolo, G.; et al. Specific miRNA and Gene Deregulation Characterize the Increased Angiogenic Remodeling of Thoracic Aneurysmatic Aortopathy in Marfan Syndrome. Int. J. Mol. Sci. 2020, 21, 6886. [Google Scholar] [CrossRef] [PubMed]
- Green, M.R.; Sambrook, J. Quantifying and Storing RNA. Cold Spring Harb. Protoc. 2020, 2020, 101709. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Want, A.; Staniak, K.; Grabowska-Pyrzewicz, W.; Fesiuk, A.; Barczak, A.; Gabryelewicz, T.; Kulczyńska-Przybik, A.; Mroczko, B.; Wojda, U. Optimized RT-qPCR and a novel normalization method for validating circulating miRNA biomarkers in ageing-related diseases. Sci. Rep. 2023, 13, 20869. [Google Scholar] [CrossRef] [PubMed]
- Caporarello, N.; Lee, J.; Pham, T.X.; Jones, D.L.; Guan, J.; Link, P.A.; Meridew, J.A.; Marden, G.; Yamashita, T.; Osborne, C.A.; et al. Dysfunctional ERG signaling drives pulmonary vascular aging and persistent fibrosis. Nat. Commun. 2022, 13, 4170. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Terriaca, S.; Scioli, M.G.; Bertoldo, F.; Pisano, C.; Nardi, P.; Balistreri, C.R.; Magro, D.; Belmonte, B.; Savino, L.; Ferlosio, A.; et al. miRNA-Driven Regulation of Endothelial-to-Mesenchymal Transition Differs among Thoracic Aortic Aneurysms. Cells 2024, 13, 1252. https://doi.org/10.3390/cells13151252
Terriaca S, Scioli MG, Bertoldo F, Pisano C, Nardi P, Balistreri CR, Magro D, Belmonte B, Savino L, Ferlosio A, et al. miRNA-Driven Regulation of Endothelial-to-Mesenchymal Transition Differs among Thoracic Aortic Aneurysms. Cells. 2024; 13(15):1252. https://doi.org/10.3390/cells13151252
Chicago/Turabian StyleTerriaca, Sonia, Maria Giovanna Scioli, Fabio Bertoldo, Calogera Pisano, Paolo Nardi, Carmela Rita Balistreri, Daniele Magro, Beatrice Belmonte, Luca Savino, Amedeo Ferlosio, and et al. 2024. "miRNA-Driven Regulation of Endothelial-to-Mesenchymal Transition Differs among Thoracic Aortic Aneurysms" Cells 13, no. 15: 1252. https://doi.org/10.3390/cells13151252





