Human NQO1 as a Selective Target for Anticancer Therapeutics and Tumor Imaging
Abstract
1. Introduction
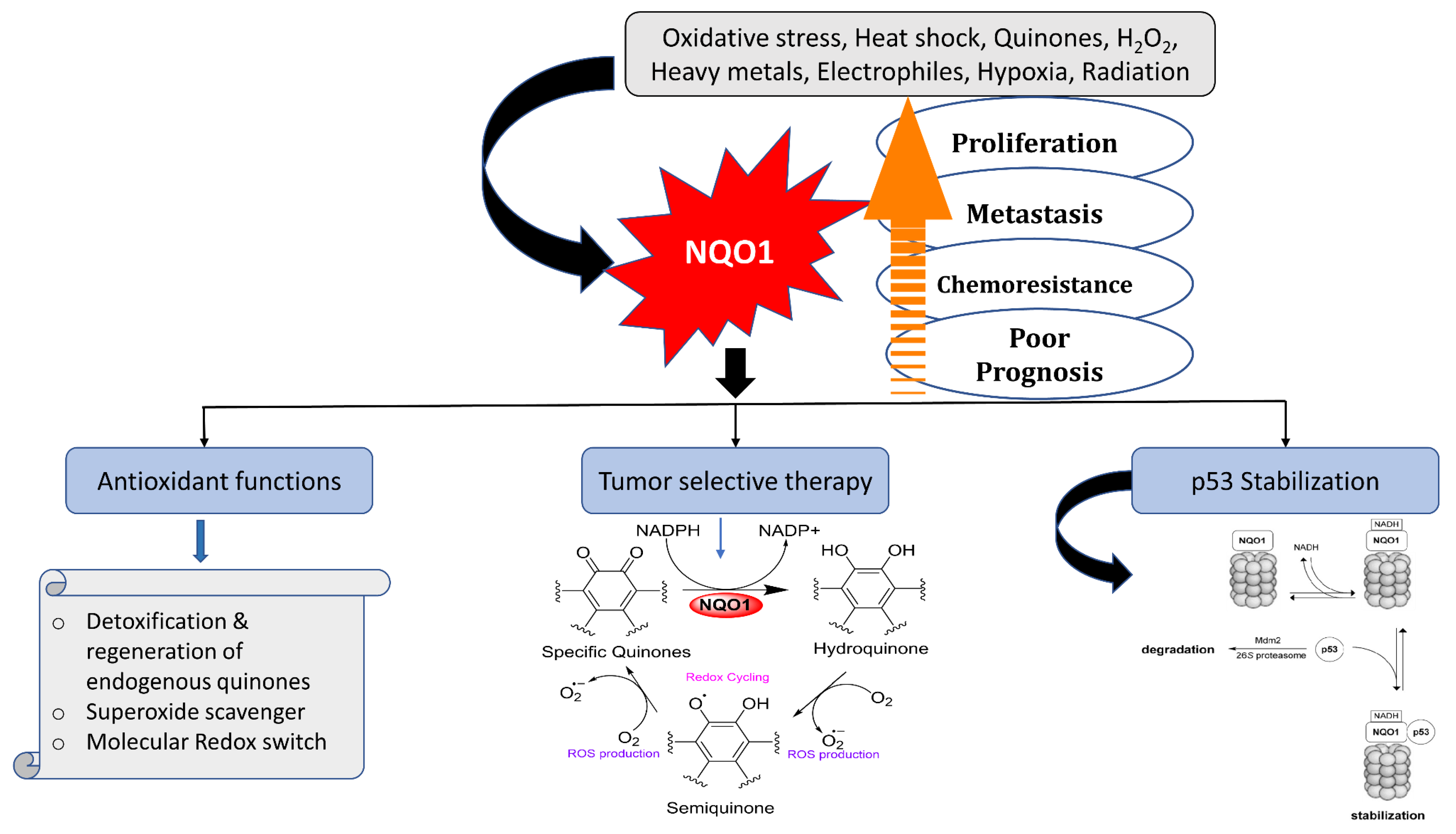
2. NQO1 Gene, Expression, Cellular Functions, and Anticancer Bioreductive Catalysis
2.1. Scavenging of Superoxide Radicals
2.2. Reduction and Activation of Quinone Compounds and Futile Redox Cycling
2.3. Preservation of Endogenous Antioxidants like Ubiquinone and α-Tocopherol
2.4. Stabilization of Tumor Suppressor Proteins p53 and p73
2.5. Cytotoxic Mechanisms of NQO1 Bioactivating Compounds
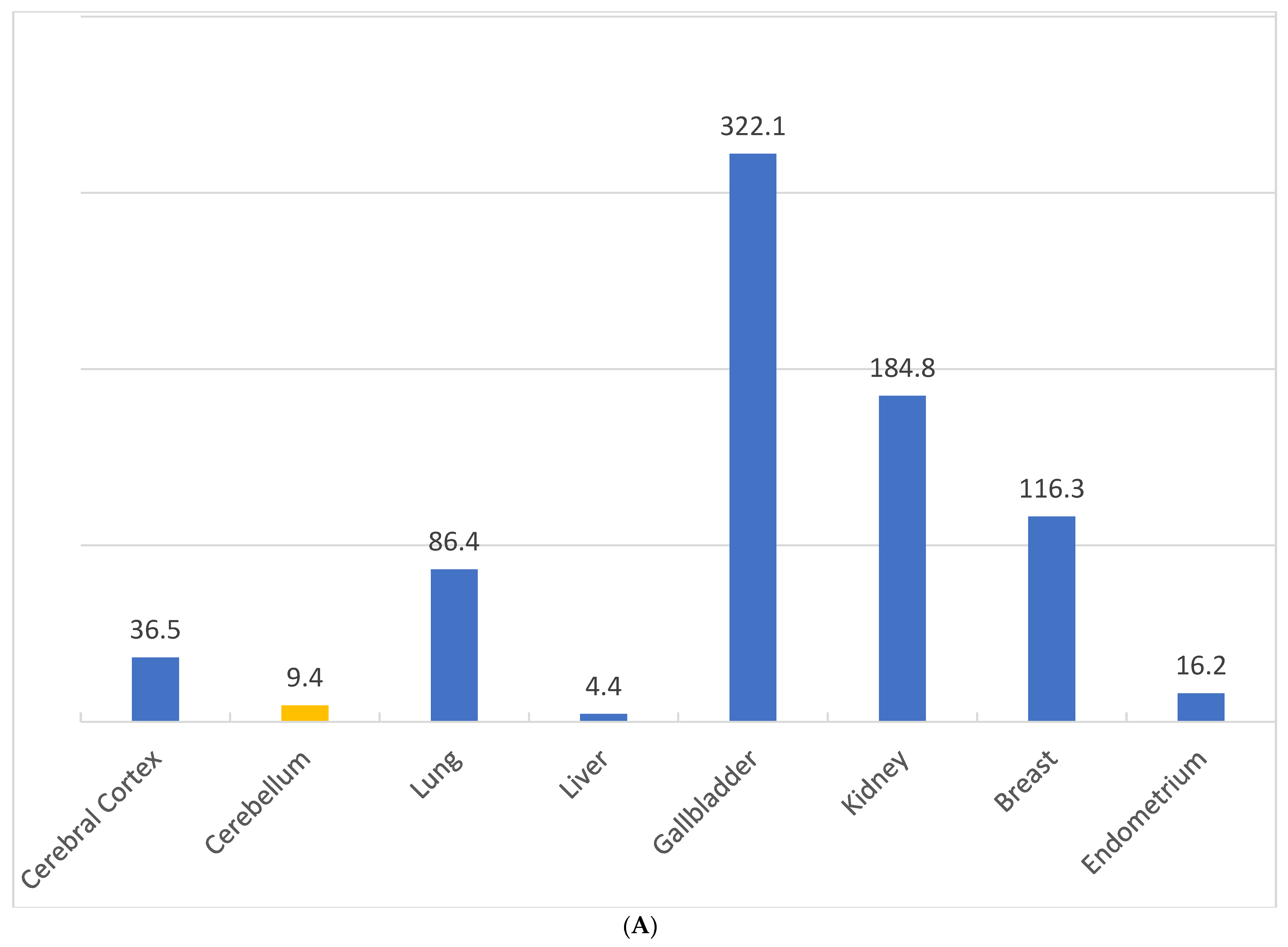
2.6. Differential Expression of NQO1 in Normal vs. Malignant Cells
2.7. NQO1 Bioreductive Quinone Substrates Eliciting Cytotoxicity
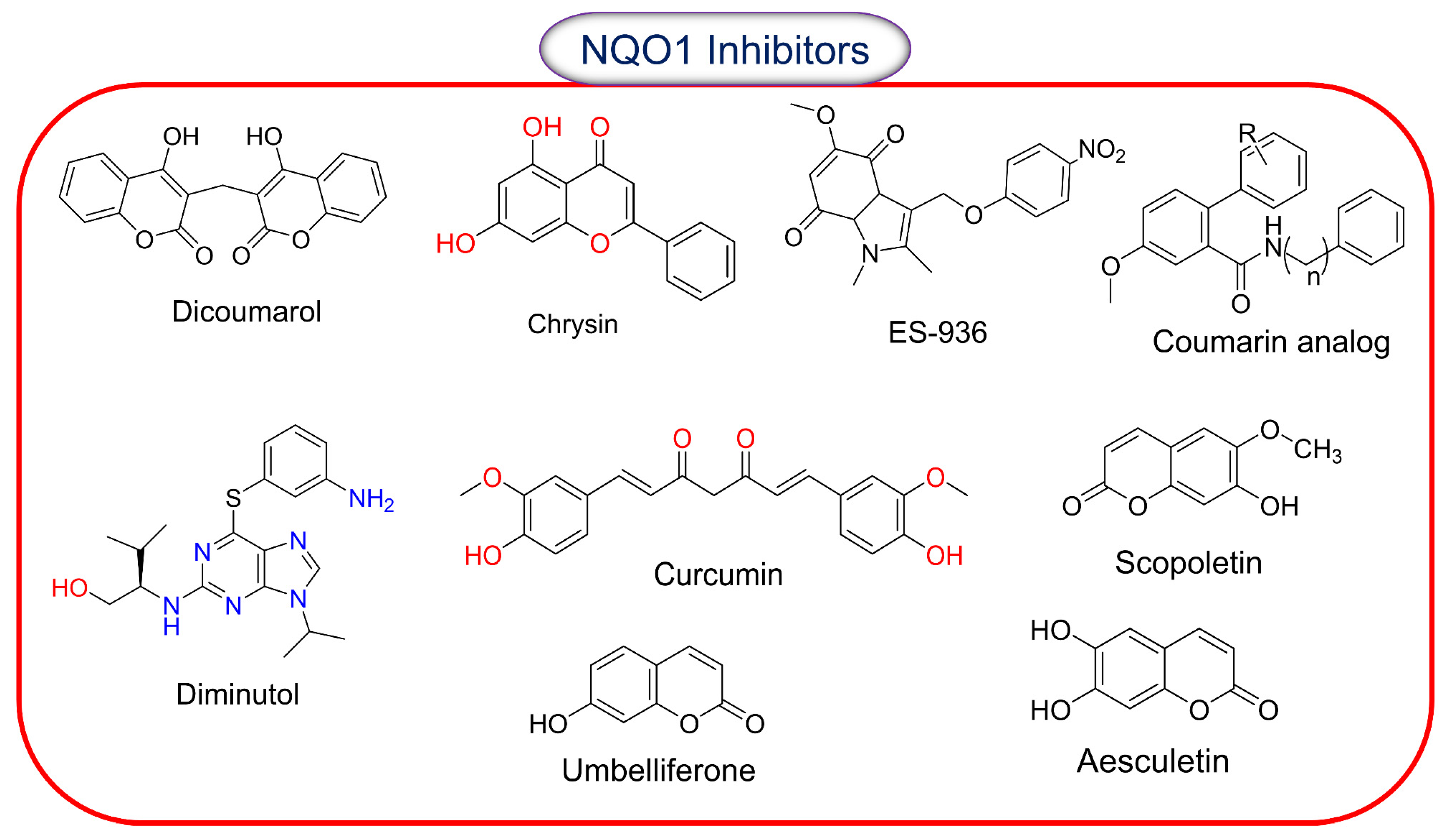
2.8. Inhibitors of NQO1 Catalytic Activity
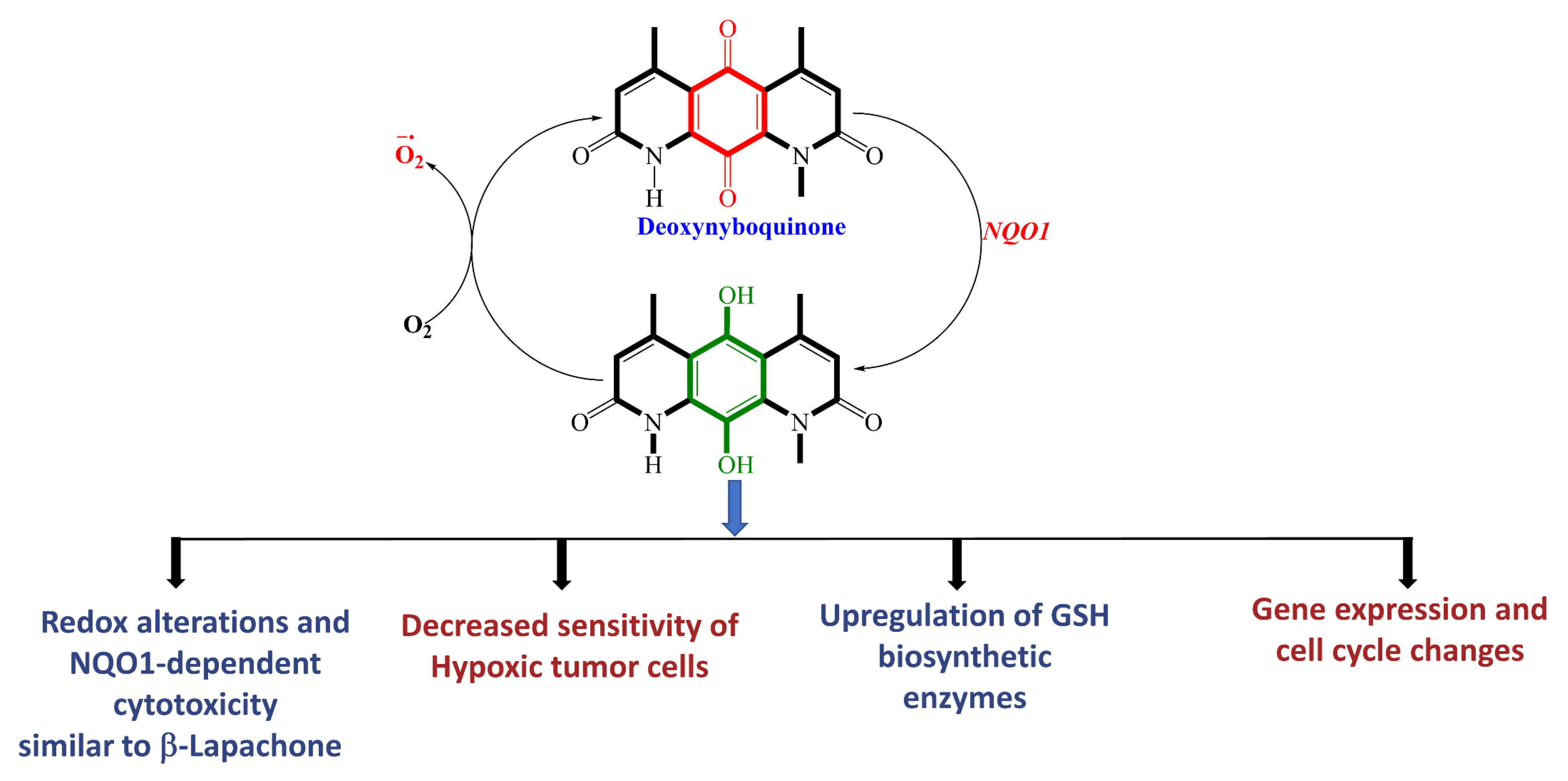
2.9. β-Lapachone and Deoxynyboquinone, Two Potent Futile Cycling Substrates of NQO1
2.10. Clinical Advances of β-Lapachone
3. Overview and Brief Descriptions of NQO1-Activatable Fluorescent Probes for Imaging Cancer Cells/Tissues
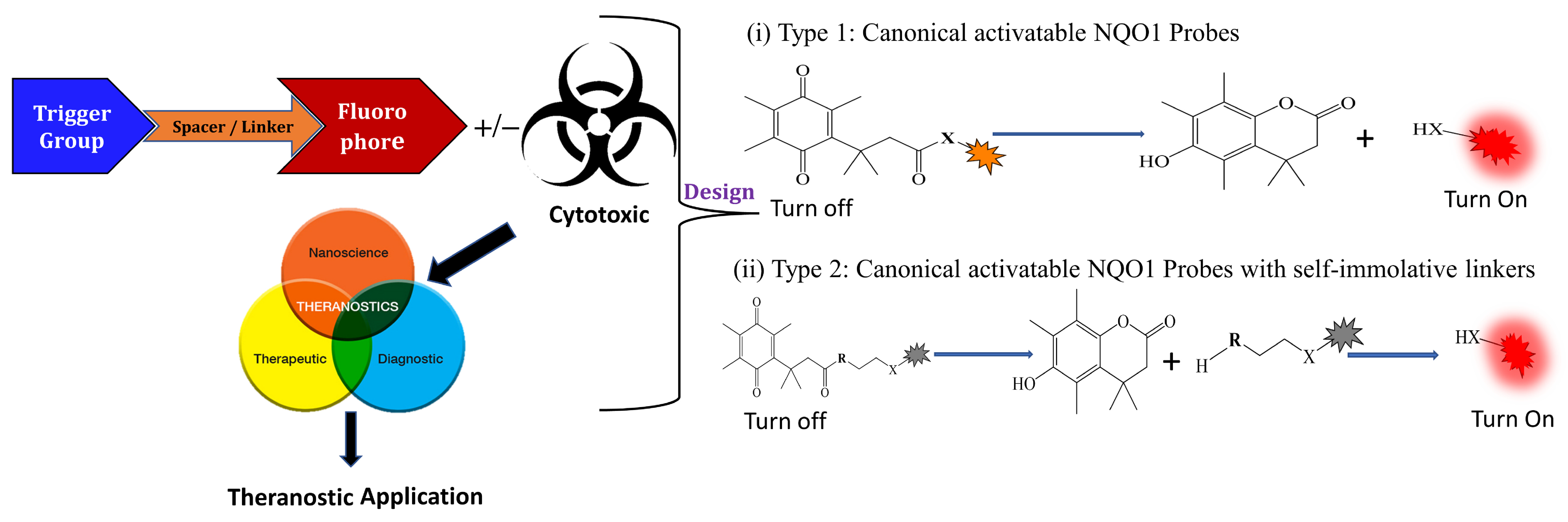
3.1. General Design and Approaches of Molecular Probes for NQO1 Sensing, Quantitation, and Imaging
3.2. NQO1 Fluorescent Probes with Emission Wavelengths in the Visible Spectrum
3.3. NIR/IR Probes for NQO1 Activity Imaging
3.4. Chemiluminescent Probes for NQO1
3.5. Bioluminescent Probes for NQO1 and NQO1-Deliverable Anticancer Drug Strategy
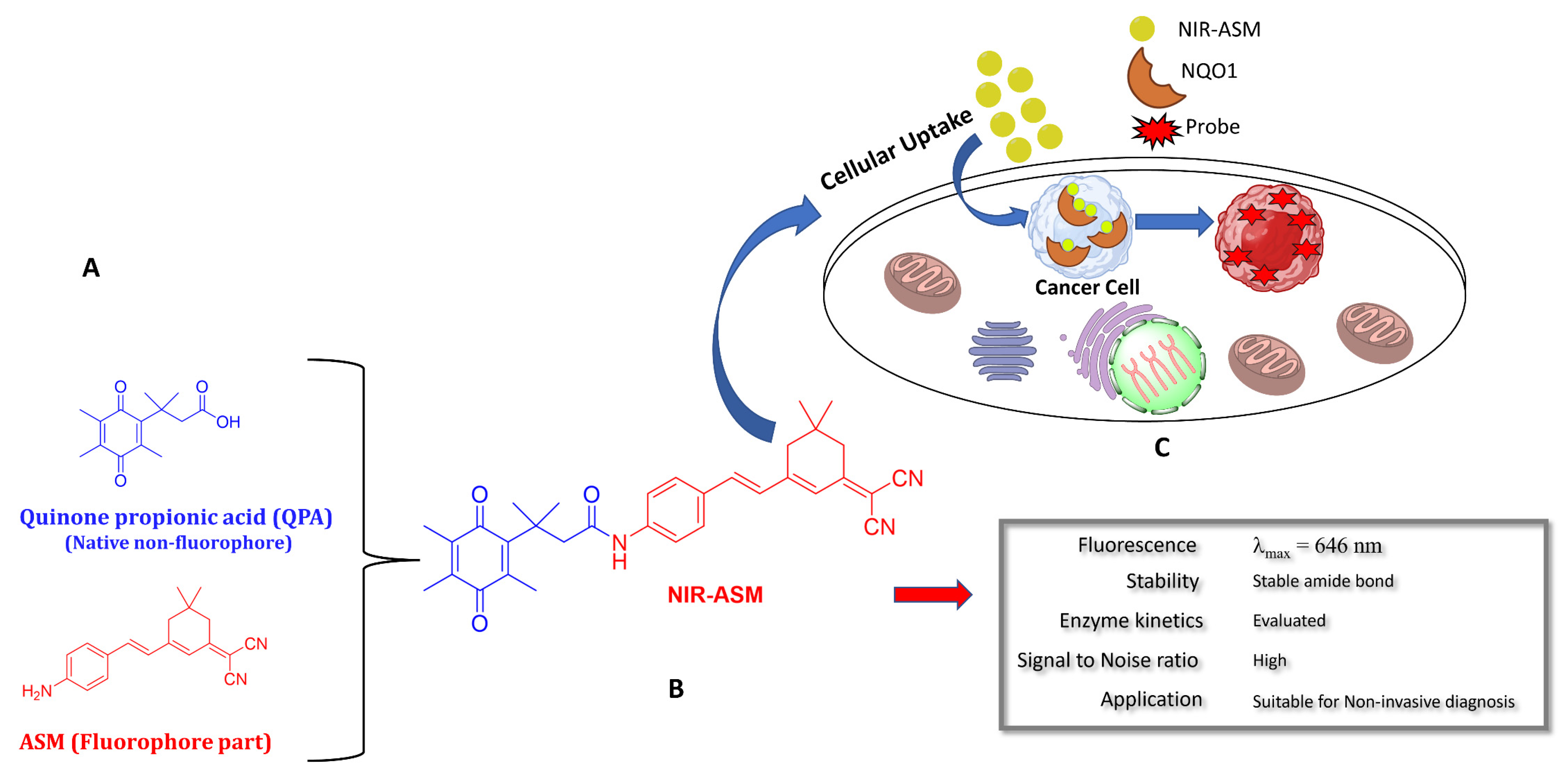
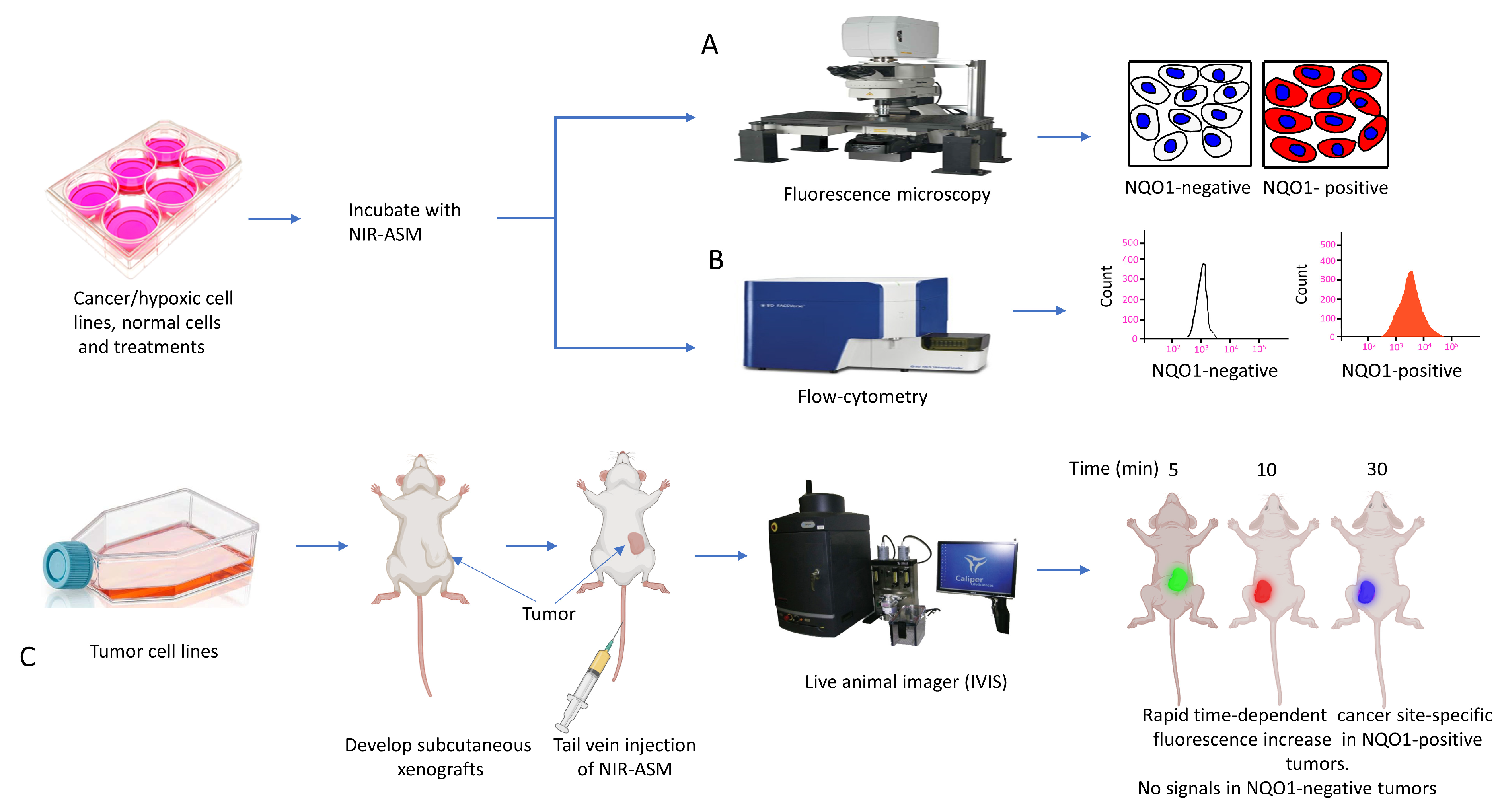
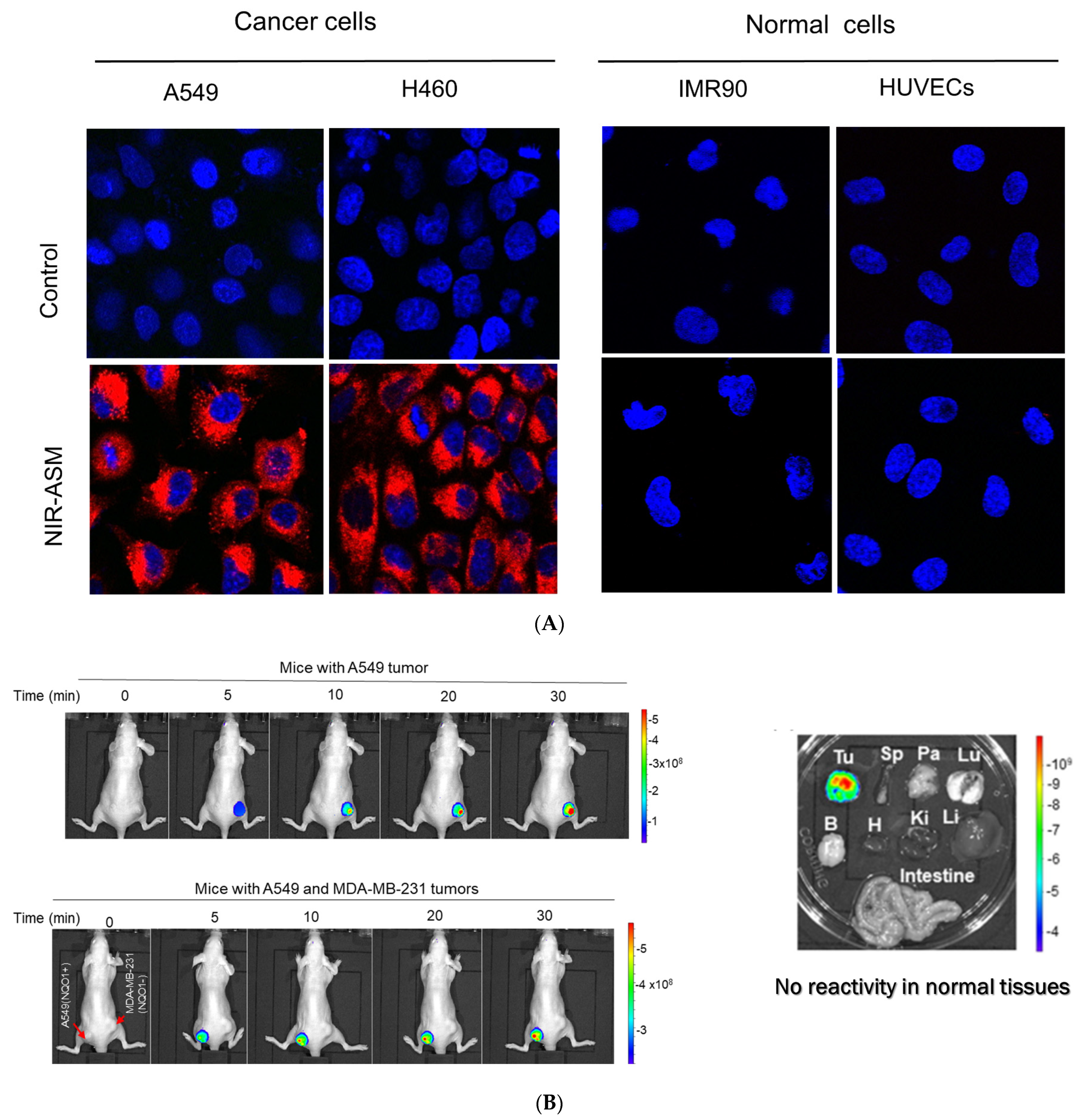
3.6. Biomedical Applications of NIR-ASM, a Superior Nontoxic NQO1-Imaging Agent
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zhu, H.; Li, Y. NAD(P)H: Quinone oxidoreductase 1 and its potential protective role in cardiovascular diseases and related conditions. Cardiovasc. Toxicol. 2012, 12, 39–45. [Google Scholar] [CrossRef]
- Ernster, L.; Lindberg, O. Animal mitochondria. Annu. Rev. Physiol. 1958, 20, 13–42. [Google Scholar] [CrossRef]
- Jaiswal, A.K.; Burnett, P.; Adesnik, M.; McBride, O.W. Nucleotide and deduced amino acid sequence of a human cDNA (NQO2) corresponding to a second member of the NAD(P)H:quinone oxidoreductase gene family. Extensive polymorphism at the NQO2 gene locus on chromosome 6. Biochemistry 1990, 29, 1899–1906. [Google Scholar] [CrossRef]
- Jaiswal, A.K. Human NAD(P)H:quinone oxidoreductase2. Gene structure, activity, and tissue-specific expression. J. Biol. Chem. 1994, 269, 14502–14508. [Google Scholar] [CrossRef]
- Dinkova-Kostova, A.T.; Talalay, P. NAD(P)H:quinone acceptor oxidoreductase 1 (NQO1), a multifunctional antioxidant enzyme and exceptionally versatile cytoprotector. Arch. Biochem. Biophys. 2010, 501, 116–123. [Google Scholar] [CrossRef] [PubMed]
- Foster, C.E.; Bianchet, M.A.; Talalay, P.; Zhao, Q.; Amzel, L.M. Crystal structure of human quinone reductase type 2, a metalloflavoprotein. Biochemistry 1999, 38, 9881–9886. [Google Scholar] [CrossRef] [PubMed]
- Skelly, J.V.; Sanderson, M.R.; Suter, D.A.; Baumann, U.; Read, M.A.; Gregory, D.S.; Bennett, M.; Hobbs, S.M.; Neidle, S. Crystal structure of human DT-diaphorase: A model for interaction with the cytotoxic prodrug 5-(aziridin-1-yl)-2,4-dinitrobenzamide (CB1954). J. Med. Chem. 1999, 42, 4325–4330. [Google Scholar] [CrossRef]
- Faig, M.; Bianchet, M.A.; Talalay, P.; Chen, S.; Winski, S.; Ross, D.; Amzel, L.M. Structures of recombinant human and mouse NAD(P)H:quinone oxidoreductases: Species comparison and structural changes with substrate binding and release. Proc. Natl. Acad. Sci. USA 2000, 97, 3177–3182. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Q.; Yang, X.L.; Holtzclaw, W.D.; Talalay, P. Unexpected genetic and structural relationships of a long-forgotten flavoenzyme to NAD(P)H:quinone reductase (DT-diaphorase). Proc. Natl. Acad. Sci. USA 1997, 94, 1669–1674. [Google Scholar] [CrossRef]
- Bianchet, M.A.; Faig, M.; Amzel, L.M. Structure and mechanism of NAD[P]H:quinone acceptor oxidoreductases (NQO). Methods Enzymol. 2004, 382, 144–174. [Google Scholar]
- Hassanpour, S.H.; Dehghani, M.A. Review of cancer from perspective of molecular. J. Cancer Res. Pract. 2017, 4, 127–129. [Google Scholar] [CrossRef]
- Falzone, L.; Salomone, S.; Libra, M. Evolution of Cancer Pharmacological Treatments at the Turn of the Third Millennium. Front. Pharmacol. 2018, 9, 1300. [Google Scholar] [CrossRef] [PubMed]
- Louie, T.M.; Yang, H.; Karnchanaphanurach, P.; Xie, X.S.; Xun, L. FAD is a preferred substrate and an inhibitor of Escherichia coli general NAD(P)H:flavin oxidoreductase. J. Biol. Chem. 2002, 277, 39450–39455. [Google Scholar] [CrossRef] [PubMed]
- Gorący, J.; Bogacz, A.; Uzar, I.; Wolek, M.; Łochyńska, M.; Ziętek, P.; Czerny, B.; Cymbaluk-Płoska, A.; Modliborski, P.; Kamiński, A. The Analysis of NADPH Quinone Reductase 1 (NQO1) Polymorphism in Polish Patients with Colorectal Cancer. Biomolecules 2021, 11, 1024. [Google Scholar] [CrossRef] [PubMed]
- Siegel, D.; Dehn, D.D.; Bokatzian, S.S.; Quinn, K.; Backos, D.S.; Di Francesco, A.; Bernier, M.; Reisdorph, N.; de Cabo, R.; Ross, D. Redox modulation of NQO1. PLoS ONE 2018, 13, e0190717. [Google Scholar] [CrossRef] [PubMed]
- Hayes, J.D.; Dinkova-Kostova, A.T.; Tew, K.D. Oxidative Stress in Cancer. Cancer Cell 2020, 38, 167–197. [Google Scholar] [CrossRef] [PubMed]
- Tie, J.K.; Jin, D.Y.; Straight, D.L.; Stafford, D.W. Functional study of the vitamin K cycle in mammalian cells. Blood 2011, 117, 2967–2974. [Google Scholar] [CrossRef] [PubMed]
- Criddle, D.N.; Gillies, S.; Baumgartner-Wilson, H.K.; Jaffar, M.; Chinje, E.C.; Passmore, S.; Chvanov, M.; Barrow, S.; Gerasimenko, O.V.; Tepikin, A.V.; et al. Menadione-induced reactive oxygen species generation via redox cycling promotes apoptosis of murine pancreatic acinar cells. J. Biol. Chem. 2006, 281, 40485–40492. [Google Scholar] [CrossRef] [PubMed]
- Nioi, P.; Hayes, J.D. Contribution of NAD(P)H:quinone oxidoreductase 1 to protection against carcinogenesis, and regulation of its gene by the Nrf2 basic-region leucine zipper and the arylhydrocarbon receptor basic helix-loop-helix transcription factors. Mutat. Res. 2004, 555, 149–171. [Google Scholar] [CrossRef]
- Lee, S.B.; Kim, C.Y.; Lee, H.J.; Yun, J.H.; Nho, C.W. Induction of the phase II detoxification enzyme NQO1 in hepatocarcinoma cells by lignans from the fruit of Schisandra chinensis through nuclear accumulation of Nrf2. Planta Med. 2009, 75, 1314–1318. [Google Scholar] [CrossRef]
- Nishiyama, T.; Izawa, T.; Usami, M.; Ohnuma, T.; Ogura, K.; Hiratsuka, A. Cooperation of NAD(P)H:quinone oxidoreductase 1 and UDP-glucuronosyltransferases reduces menadione cytotoxicity in HEK293 cells. Biochem. Biophys. Res. Commun. 2010, 394, 459–463. [Google Scholar] [CrossRef] [PubMed]
- Ross, D.; Kepa, J.K.; Winski, S.L.; Beall, H.D.; Anwar, A.; Siegel, D. NAD(P)H:quinone oxidoreductase 1 (NQO1): Chemoprotection, bioactivation, gene regulation and genetic polymorphisms. Chem. Biol. Interact. 2000, 129, 77–97. [Google Scholar] [CrossRef] [PubMed]
- Joseph, P.; Long, D.J.; 2nd Klein-Szanto, A.J.; Jaiswal, A.K. Role of NAD(P)H:quinone oxidoreductase 1 (DT diaphorase) in protection against quinone toxicity. Biochem. Pharmacol. 2000, 60, 207–214. [Google Scholar] [CrossRef] [PubMed]
- Joseph, P.; Jaiswal, A.K. NAD(P)H:quinone oxidoreductase1 (DT diaphorase) specifically prevents the formation of benzo[a]pyrene quinone-DNA adducts generated by cytochrome P4501A1 and P450 reductase. Proc. Natl. Acad. Sci. USA 1994, 91, 8413–8417. [Google Scholar] [CrossRef] [PubMed]
- Lajin, B.; Alachkar, A. The NQO1 polymorphism C609T (Pro187Ser) and cancer susceptibility: A comprehensive meta-analysis. Br. J. Cancer 2013, 109, 1325–1337. [Google Scholar] [CrossRef] [PubMed]
- Talalay, P.; Dinkova-Kostova, A.T. Role of nicotinamide quinone oxidoreductase 1 (NQO1) in protection against toxicity of electrophiles and reactive oxygen intermediates. Methods Enzymol. 2004, 382, 355–364. [Google Scholar] [PubMed]
- Nioi, P.; McMahon, M.; Itoh, K.; Yamamoto, M.; Hayes, J.D. Identification of a novel Nrf2-regulated antioxidant response element (ARE) in the mouse NAD(P)H:quinone oxidoreductase 1 gene: Reassessment of the ARE consensus sequence. Biochem. J. 2003, 374 Pt 2, 337–348. [Google Scholar] [CrossRef] [PubMed]
- Jaiswal, A.K. Regulation of genes encoding NAD(P)H:quinone oxidoreductases. Free Radic. Biol. Med. 2000, 29, 254–262. [Google Scholar] [CrossRef]
- Flicek, P.; Ahmed, I.; Amode, M.R.; Barrell, D.; Beal, K.; Brent, S.; Carvalho-Silva, D.; Clapham, P.; Coates, G.; Fairley, S.; et al. Ensembl 2013. Nucleic Acids Res. 2013, 41, D48–D55. [Google Scholar] [CrossRef]
- Cresteil, T.; Jaiswal, A.K. High levels of expression of the NAD(P)H:quinone oxidoreductase (NQO1) gene in tumor cells compared to normal cells of the same origin. Biochem. Pharmacol. 1991, 42, 1021–1027. [Google Scholar] [CrossRef]
- Oh, E.T.; Kim, J.W.; Kim, J.M.; Kim, S.J.; Lee, J.S.; Hong, S.S.; Goodwin, J.; Ruthenborg, R.J.; Jung, M.G.; Lee, H.J.; et al. NQO1 inhibits proteasome-mediated degradation of HIF-1α. Nat. Commun. 2016, 7, 13593. [Google Scholar] [CrossRef] [PubMed]
- Salido, E.; Timson, D.J.; Betancor-Fernández, I.; Palomino-Morales, R.; Anoz-Carbonell, E.; Pacheco-García, J.L.; Medina, M.; Pey, A.L. Targeting HIF-1α Function in Cancer through the Chaperone Action of NQO1: Implications of Genetic Diversity of NQO1. J. Pers. Med. 2022, 12, 747. [Google Scholar] [CrossRef] [PubMed]
- Siegel, D.; Ross, D. Immunodetection of NAD(P)H:quinone oxidoreductase 1 (NQO1) in human tissues. Free Radic. Biol. Med. 2000, 29, 246–253. [Google Scholar] [CrossRef] [PubMed]
- Siegel, D.; Gustafson, D.L.; Dehn, D.L.; Han, J.Y.; Boonchoong, P.; Berliner, L.J.; Ross, D. NAD(P)H:quinone oxidoreductase 1: Role as a superoxide scavenger. Mol. Pharmacol. 2004, 65, 1238–1247. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.; Jia, Z.; Mahaney, J.E.; Ross, D.; Misra, H.P.; Trush, M.A.; Li, Y. The highly expressed and inducible endogenous NAD(P)H:quinone oxidoreductase 1 in cardiovascular cells acts as a potential superoxide scavenger. Cardiovasc. Toxicol. 2007, 7, 202–211. [Google Scholar] [CrossRef] [PubMed]
- Cao, Z.; Li, Y. The chemical inducibility of mouse cardiac antioxidants and phase 2 enzymes in vivo. Biochem. Biophys. Res. Commun. 2004, 317, 1080–1088. [Google Scholar] [CrossRef] [PubMed]
- Boothman, D.A.; Meyers, M.; Fukunaga, N.; Lee, S.W. Isolation of x-ray-inducible transcripts from radioresistant human melanoma cells. Proc. Natl. Acad. Sci. USA 1993, 90, 7200–7204. [Google Scholar] [CrossRef]
- Ross, D.; Siegel, D. The diverse functionality of NQO1 and its roles in redox control. Redox Biol. 2021, 41, 101950. [Google Scholar] [CrossRef]
- Chesis, P.L.; Levin, D.E.; Smith, M.T.; Ernster, L.; Ames, B.N. Mutagenicity of quinones: Pathways of metabolic activation and detoxification. Proc. Natl. Acad. Sci. USA 1984, 81, 1696–1700. [Google Scholar] [CrossRef]
- Sekiguchi, M.; Seki, M.; Kawai, T.; Yoshida, K.; Yoshida, M.; Isobe, T.; Hoshino, N.; Shirai, R.; Tanaka, M.; Souzaki, R.; et al. Integrated multiomics analysis of hepatoblastoma unravels its heterogeneity and provides novel druggable targets. NPJ Precis. Oncol. 2020, 4, 20. [Google Scholar] [CrossRef]
- Wu, Y.L.; Wang, D.; Peng, X.E.; Chen, Y.L.; Zheng, D.L.; Chen, W.N.; Lin, X. Epigenetic silencing of NAD(P)H:quinone oxidoreductase 1 by hepatitis B virus X protein increases mitochondrial injury and cellular susceptibility to oxidative stress in hepatoma cells. Free Radic. Biol. Med. 2013, 65, 632–644. [Google Scholar] [CrossRef] [PubMed]
- Cadenas, E. Antioxidant and prooxidant functions of DT-diaphorase in quinone metabolism. Biochem. Pharmacol. 1995, 49, 127–140. [Google Scholar] [CrossRef] [PubMed]
- Shimokawa, M.; Yoshizumi, T.; Itoh, S.; Iseda, N.; Sakata, K.; Yugawa, K.; Toshima, T.; Harada, N.; Ikegami, T.; Mori, M. Modulation of Nqo1 activity intercepts anoikis resistance and reduces metastatic potential of hepatocellular carcinoma. Cancer Sci. 2020, 111, 1228–1240. [Google Scholar] [CrossRef] [PubMed]
- Pink, J.J.; Planchon, S.M.; Tagliarino, C.; Varnes, M.E.; Siegel, D.; Boothman, D.A. NAD(P)H:Quinone oxidoreductase activity is the principal determinant of beta-lapachone cytotoxicity. J. Biol. Chem. 2000, 275, 5416–5424. [Google Scholar] [CrossRef] [PubMed]
- Siegel, D.; Gibson, N.W.; Preusch, P.C.; Ross, D. Metabolism of mitomycin C by DT-diaphorase: Role in mitomycin C-induced DNA damage and cytotoxicity in human colon carcinoma cells. Cancer Res. 1990, 50, 7483–7489. [Google Scholar] [PubMed]
- Wang, J.; Su, X.; Jiang, L.; Boudreau, M.W.; Chatkewitz, L.E.; Kilgore, J.E.; Zahid, K.R.; Williams, N.S.; Chen, Y.; Liu, S.; et al. Augmented concentration of isopentyl-deoxynyboquinone in tumors selectively kills NAD(P)H quinone oxidoreductase 1-positive cancer Cells through programmed necrotic and apoptotic Mechanisms. Cancers 2023, 15, 5844. [Google Scholar] [CrossRef] [PubMed]
- Bailey, S.M.; Lewis, A.D.; Knox, R.J.; Patterson, L.H.; Fisher, G.R.; Workman, P. Reduction of the indoloquinone anticancer drug EO9 by purified DT-diaphorase: A detailed kinetic study and analysis of metabolites. Biochem. Pharmacol. 1998, 56, 613–621. [Google Scholar] [CrossRef] [PubMed]
- Choudry, G.A.; Stewart, P.A.; Double, J.A.; Krul, M.R.; Naylor, B.; Flannigan, G.M.; Shah, T.K.; Brown, J.E.; Phillips, R.M. A novel strategy for NQO1 (NAD(P)H:quinone oxidoreductase, EC 1.6.99.2) mediated therapy of bladder cancer based on the pharmacological properties of EO9. Br. J. Cancer 2001, 85, 1137–1146. [Google Scholar] [CrossRef]
- Guo, W.; Reigan, P.; Siegel, D.; Zirrolli, J.; Gustafson, D.; Ross, D. Formation of 17-allylamino-demethoxygeldanamycin (17-AAG) hydroquinone by NAD(P)H:quinone oxidoreductase 1: Role of 17-AAG hydroquinone in heat shock protein 90 inhibition. Cancer Res. 2005, 65, 10006–10015. [Google Scholar] [CrossRef]
- Siegel, D.; Gibson, N.W.; Preusch, P.C.; Ross, D. Metabolism of diaziquone by NAD(P)H:(quinone acceptor) oxidoreductase (DT-diaphorase): Role in diaziquone-induced DNA damage and cytotoxicity in human colon carcinoma cells. Cancer Res. 1990, 50, 7293–7300. [Google Scholar]
- Dehn, D.L.; Winski, S.L.; Ross, D. Development of a new isogenic cell-xenograft system for evaluation of NAD(P)H:quinone oxidoreductase-directed antitumor quinones: Evaluation of the activity of RH1. Clin. Cancer Res. 2004, 10, 3147–3155. [Google Scholar] [CrossRef] [PubMed]
- Dehn, D.L.; Inayat-Hussain, S.H.; Ross, D. RH1 induces cellular damage in an NAD(P)H:quinone oxidoreductase 1-dependent manner: Relationship between DNA cross-linking, cell cycle perturbations, and apoptosis. J. Pharmacol. Exp. Ther. 2005, 313, 771–779. [Google Scholar] [CrossRef] [PubMed]
- Awadallah, N.S.; Dehn, D.; Shah, R.J.; Russell Nash, S.; Chen, Y.K.; Ross, D.; Bentz, J.S.; Shroyer, K.R. NQO1 expression in pancreatic cancer and its potential use as a biomarker. Appl. Immunohistochem. Mol. Morphol. 2008, 16, 24–31. [Google Scholar] [CrossRef] [PubMed]
- Ross, D.; Siegel, D.; Beall, H.; Prakash, A.S.; Mulcahy, R.T.; Gibson, N.W. DT-diaphorase in activation and detoxification of quinones. Bioreductive activation of mitomycin C. Cancer Metastasis Rev. 1993, 12, 83–101. [Google Scholar] [CrossRef] [PubMed]
- Beyer, R.E.; Segura-Aguilar, J.; Di Bernardo, S.; Cavazzoni, M.; Fato, R.; Fiorentini, D.; Galli, M.C.; Setti, M.; Landi, L.; Lenaz, G. The role of DT-diaphorase in the maintenance of the reduced antioxidant form of coenzyme Q in membrane systems. Proc. Natl. Acad. Sci. USA 1996, 93, 2528–2532. [Google Scholar] [CrossRef] [PubMed]
- Ross, D.; Siegel, D. Functions of NQO1 in Cellular Protection and CoQ(10) Metabolism and its Potential Role as a Redox Sensitive Molecular Switch. Front. Physiol. 2017, 8, 595. [Google Scholar] [CrossRef] [PubMed]
- Asher, G.; Lotem, J.; Cohen, B.; Sachs, L.; Shaul, Y. Regulation of p53 stability and p53-dependent apoptosis by NADH quinone oxidoreductase 1. Proc. Natl. Acad. Sci. USA 2001, 98, 1188–1193. [Google Scholar] [CrossRef] [PubMed]
- Asher, G.; Lotem, J.; Kama, R.; Sachs, L.; Shaul, Y. NQO1 stabilizes p53 through a distinct pathway. Proc. Natl. Acad. Sci. USA 2002, 99, 3099–3104. [Google Scholar] [CrossRef] [PubMed]
- Fagerholm, R.; Hofstetter, B.; Tommiska, J.; Aaltonen, K.; Vrtel, R.; Syrjäkoski, K.; Kallioniemi, A.; Kilpivaara, O.; Mannermaa, A.; Kosma, V.M.; et al. NAD(P)H:quinone oxidoreductase 1 NQO1*2 genotype (P187S) is a strong prognostic and predictive factor in breast cancer. Nat. Genet. 2008, 40, 844–853. [Google Scholar] [CrossRef]
- Moscovitz, O.; Tsvetkov, P.; Hazan, N.; Michaelevski, I.; Keisar, H.; Ben-Nissan, G.; Shaul, Y.; Sharon, M. A mutually inhibitory feedback loop between the 20S proteasome and its regulator, NQO1. Mol. Cell 2012, 47, 76–86. [Google Scholar] [CrossRef]
- Hershkovitz Rokah, O.; Shpilberg, O.; Granot, G. NAD(P)H quinone oxidoreductase protects TAp63gamma from proteasomal degradation and regulates TAp63gamma-dependent growth arrest. PLoS ONE 2010, 5, e11401. [Google Scholar] [CrossRef] [PubMed]
- Adamovich, Y.; Shlomai, A.; Tsvetkov, P.; Umansky, K.B.; Reuven, N.; Estall, J.L.; Spiegelman, B.M.; Shaul, Y. The protein level of PGC-1α, a key metabolic regulator, is controlled by NADH-NQO1. Mol. Cell Biol. 2013, 33, 2603–2613. [Google Scholar] [CrossRef] [PubMed]
- Available online: https://www.proteinatlas.org/ENSG00000181019-NQO1/tissue (accessed on 24 July 2024).
- Blaza, J.N.; Bridges, H.R.; Aragão, D.; Dunn, E.A.; Heikal, A.; Cook, G.M.; Nakatani, Y.; Hirst, J. The mechanism of catalysis by type-II NADH:quinone oxidoreductases. Sci. Rep. 2017, 7, 40165. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, M.C.; Morais, J.A.V.; Ganassin, R.; Oliveira, G.R.T.; Costa, F.C.; Morais, A.A.C.; Silveira, A.P.; Silva, V.C.M.; Longo, J.P.F.; Muehlmann, L.A. An Overview on Immunogenic Cell Death in Cancer Biology and Therapy. Pharmaceutics 2022, 14, 1564. [Google Scholar] [CrossRef] [PubMed]
- Available online: https://www.targetedonc.com/view/fda-rejects-apaziquone-application-for-bladder-cancer (accessed on 27 July 2024).
- Siegel, D.; Yan, C.; Ross, D. NAD(P)H:quinone oxidoreductase 1 (NQO1) in the sensitivity and resistance to antitumor quinones. Biochem. Pharmacol. 2012, 83, 1033–1040. [Google Scholar] [CrossRef] [PubMed]
- Siegel, D.; Beall, H.; Senekowitsch, C.; Kasai, M.; Arai, H.; Gibson, N.W.; Ross, D. Bioreductive activation of mitomycin C by DT-diaphorase. Biochemistry 1992, 31, 7879–7885. [Google Scholar] [CrossRef] [PubMed]
- Gerber, D.E.; Beg, M.S.; Fattah, F.; Frankel, A.E.; Fatunde, O.; Arriaga, Y.; Dowell, J.E.; Bisen, A.; Leff, R.D.; Meek, C.C.; et al. Phase 1 study of ARQ 761, a β-lapachone analogue that promotes NQO1-mediated programmed cancer cell necrosis. Br. J. Cancer 2018, 119, 928–936. [Google Scholar] [CrossRef] [PubMed]
- Marín, A.; López de Cerain, A.; Hamilton, E.; Lewis, A.D.; Martinez-Peñuela, J.M.; Idoate, M.A.; Bello, J. DT-diaphorase and cytochrome B5 reductase in human lung and breast tumours. Br. J. Cancer 1997, 76, 923–929. [Google Scholar] [CrossRef] [PubMed]
- Belinsky, M.; Jaiswal, A.K. NAD(P)H:quinone oxidoreductase1 (DT-diaphorase) expression in normal and tumor tissues. Cancer Metastasis Rev. 1993, 12, 103–117. [Google Scholar] [CrossRef]
- Ough, M.; Lewis, A.; Bey, E.A.; Gao, J.; Ritchie, J.M.; Bornmann, W.; Boothman, D.A.; Oberley, L.W.; Cullen, J.J. Efficacy of beta-lapachone in pancreatic cancer treatment: Exploiting the novel, therapeutic target NQO1. Cancer Biol. Ther. 2005, 4, 95–102. [Google Scholar] [CrossRef]
- Dong, Y.; Chin, S.F.; Blanco, E.; Bey, E.A.; Kabbani, W.; Xie, X.J.; Bornmann, W.G.; Boothman, D.A.; Gao, J. Intratumoral delivery of beta-lapachone via polymer implants for prostate cancer therapy. Clin. Cancer Res. 2009, 15, 131–139. [Google Scholar] [CrossRef]
- Li, Y.; Feng, M.; Guo, T.; Wang, Z.; Zhao, Y. Tailored Beta-Lapachone Nanomedicines for Cancer-Specific Therapy. Adv. Healthc. Mater. 2023, 12, e2300349. [Google Scholar] [CrossRef] [PubMed]
- Bey, E.A.; Bentle, M.S.; Reinicke, K.E.; Dong, Y.; Yang, C.R.; Girard, L.; Minna, J.D.; Bornmann, W.G.; Gao, J.; Boothman, D.A. An NQO1- and PARP-1-mediated cell death pathway induced in non-small-cell lung cancer cells by beta-lapachone. Proc. Natl. Acad. Sci. USA 2007, 104, 11832–11837. [Google Scholar] [CrossRef] [PubMed]
- Blanco, E.; Bey, E.A.; Khemtong, C.; Yang, S.G.; Setti-Guthi, J.; Chen, H.; Kessinger, C.W.; Carnevale, K.A.; Bornmann, W.G.; Boothman, D.A.; et al. Beta-lapachone micellar nanotherapeutics for non-small cell lung cancer therapy. Cancer Res. 2010, 70, 3896–3904. [Google Scholar] [CrossRef]
- Li, C.J.; Li, Y.Z.; Pinto, A.V.; Pardee, A.B. Potent inhibition of tumor survival in vivo by beta-lapachone plus taxol: Combining drugs imposes different artificial checkpoints. Proc. Natl. Acad. Sci. USA 1999, 96, 13369–13374. [Google Scholar] [CrossRef] [PubMed]
- Park, H.J.; Ahn, K.J.; Ahn, S.D.; Choi, E.; Lee, S.W.; Williams, B.; Kim, E.J.; Griffin, R.; Bey, E.A.; Bornmann, W.G.; et al. Susceptibility of cancer cells to beta-lapachone is enhanced by ionizing radiation. Int. J. Radiat. Oncol. Biol. Phys. 2005, 61, 212–219. [Google Scholar] [CrossRef]
- Punganuru, S.R.; Madala, H.R.; Arutla, V.; Srivenugopal, K.S. Abstract 1952: Design and characterization of an NQO1-activated spiroisoindolinone derivative for glioma treatment. Cancer Res. 2018, 78 (Suppl. S13), 1952. [Google Scholar] [CrossRef]
- Silvers, M.A.; Deja, S.; Singh, N.; Egnatchik, R.A.; Sudderth, J.; Luo, X.; Beg, M.S.; Burgess, S.C.; DeBerardinis, R.J.; Boothman, D.A.; et al. The NQO1 bioactivatable drug, β-lapachone, alters the redox state of NQO1+ pancreatic cancer cells, causing perturbation in central carbon metabolism. J. Biol. Chem. 2017, 292, 18203–18216. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Motea, E.A.; Moore, Z.R.; Yao, J.; Dong, Y.; Chakrabarti, G.; Kilgore, J.A.; Silvers, M.A.; Patidar, P.L.; Cholka, A.; et al. Leveraging an NQO1 Bioactivatable Drug for Tumor-Selective Use of Poly(ADP-ribose) Polymerase Inhibitors. Cancer Cell 2016, 30, 940–952. [Google Scholar] [CrossRef] [PubMed]
- Zhao, W.; Jiang, L.; Fang, T.; Fang, F.; Liu, Y.; Zhao, Y.; You, Y.; Zhou, H.; Su, X.; Wang, J.; et al. β-Lapachone selectively kills hepatocellular carcinoma cells by targeting NQO1 to induce extensive DNA damage and PARP1 hyperactivation. Front. Oncol. 2021, 11, 747282. [Google Scholar] [CrossRef]
- Huang, X.; Dong, Y.; Bey, E.A.; Kilgore, J.A.; Bair, J.S.; Li, L.S.; Patel, M.; Parkinson, E.I.; Wang, Y.; Williams, N.S.; et al. An NQO1 substrate with potent antitumor activity that selectively kills by PARP1-induced programmed necrosis. Cancer Res. 2012, 72, 3038–3047. [Google Scholar] [CrossRef] [PubMed]
- Parkinson, E.I.; Hergenrother, P.J. Deoxynyboquinones as NQO1-Activated Cancer Therapeutics. Acc. Chem. Res. 2015, 48, 2715–2723. [Google Scholar] [CrossRef]
- Wondrak, G.T. NQO1-activated phenothiazinium redox cyclers for the targeted bioreductive induction of cancer cell apoptosis. Free Radic. Biol. Med. 2007, 43, 178–190. [Google Scholar] [CrossRef]
- Digby, T.; Leith, M.K.; Thliveris, J.A.; Begleiter, A. Effect of NQO1 induction on the antitumor activity of RH1 in human tumors in vitro and in vivo. Cancer Chemother. Pharmacol. 2005, 56, 307–316. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Qu, Y.; Ma, X.; Li, M.; Li, S.; Li, Y.; Wu, L. NQO1-selective activated prodrugs of combretastatin A-4: Synthesis and biological evaluation. Bioorg. Chem. 2020, 103, 104200. [Google Scholar] [CrossRef]
- Qu, Y.; Zhang, C.; Ma, X.; Gao, Y.; Liu, J.; Wu, L. Synthesis and biological evaluation of NQO1-activated prodrugs of podophyllotoxin as antitumor agents. Bioorg. Med. Chem. 2020, 28, 115821. [Google Scholar] [CrossRef]
- Beall, H.D.; Liu, Y.; Siegel, D.; Bolton, E.M.; Gibson, N.W.; Ross, D. Role of NAD(P)H:quinone oxidoreductase (DT-diaphorase) in cytotoxicity and induction of DNA damage by streptonigrin. Biochem. Pharmacol. 1996, 51, 645–652. [Google Scholar] [CrossRef] [PubMed]
- Gustafson, D.L.; Beall, H.D.; Bolton, E.M.; Ross, D.; Waldren, C.A. Expression of human NAD(P)H: Quinone oxidoreductase (DT-diaphorase) in Chinese hamster ovary cells: Effect on the toxicity of antitumor quinones. Mol. Pharmacol. 1996, 50, 728–735. [Google Scholar] [PubMed]
- Fitzsimmons, S.A.; Workman, P.; Grever, M.; Paull, K.; Camalier, R.; Lewis, A.D. Reductase enzyme expression across the National Cancer Institute Tumor cell line panel: Correlation with sensitivity to mitomycin C and EO9. J. Natl. Cancer. Inst. 1996, 88, 259–269. [Google Scholar] [CrossRef]
- Keyes, S.R.; Fracasso, P.M.; Heimbrook, D.C.; Rockwell, S.; Sligar, S.G.; Sartorelli, A.C. Role of NADPH:cytochrome c reductase and DT-diaphorase in the biotransformation of mitomycin C1. Cancer Res. 1984, 44 Pt 1, 5638–5643. [Google Scholar]
- Plumb, J.A.; Gerritsen, M.; Workman, P. DT-diaphorase protects cells from the hypoxic cytotoxicity of indoloquinone EO9. Br. J. Cancer 1994, 70, 1136–1143. [Google Scholar] [CrossRef][Green Version]
- Plumb, J.A.; Workman, P. Unusually marked hypoxic sensitization to indoloquinone EO9 and mitomycin C in a human colon-tumour cell line that lacks DT-diaphorase activity. Int. J. Cancer 1994, 56, 134–139. [Google Scholar] [CrossRef]
- Phillips, R.M.; Hendriks, H.R.; Peters, G.J. EO9 (Apaziquone): From the clinic to the laboratory and back again. Br. J. Pharmacol. 2013, 168, 11–18. [Google Scholar] [CrossRef]
- Zhang, X.; Li, X.; Li, Z.; Wu, X.; Wu, Y.; You, Q.; Zhang, X. An NAD(P)H:Quinone Oxidoreductase 1 Responsive and Self-Immolative Prodrug of 5-Fluorouracil for Safe and Effective Cancer Therapy. Org. Lett. 2018, 20, 3635–3638. [Google Scholar] [CrossRef]
- Danson, S.J.; Johnson, P.; Ward, T.H.; Dawson, M.; Denneny, O.; Dickinson, G.; Aarons, L.; Watson, A.; Jowle, D.; Cummings, J.; et al. Phase I pharmacokinetic and pharmacodynamic study of the bioreductive drug RH1. Ann. Oncol. 2011, 22, 1653–1660. [Google Scholar] [CrossRef]
- Yan, C.; Kepa, J.K.; Siegel, D.; Stratford, I.J.; Ross, D. Dissecting the Role of Multiple Reductases in Bioactivation and Cytotoxicity of the Antitumor Agent 2,5-Diaziridinyl-3-(hydroxymethyl)-6-methyl-1,4-benzoquinone (RH1). Mol. Pharmacol. 2008, 74, 1657–1665. [Google Scholar] [CrossRef]
- Yang, X.; Duan, J.; Wu, L. Research advances in NQO1-responsive prodrugs and nanocarriers for cancer treatment. Future Med. Chem. 2022, 14, 363–383. [Google Scholar] [CrossRef]
- Lewis, A.M.; Ough, M.; Hinkhouse, M.M.; Tsao, M.S.; Oberley, L.W.; Cullen, J.J. Targeting NAD(P)H:quinone oxidoreductase (NQO1) in pancreatic cancer. Mol. Carcinog. 2005, 43, 215–224. [Google Scholar] [CrossRef]
- Danson, S.; Ward, T.H.; Butler, J.; Ranson, M. DT-diaphorase: A target for new anticancer drugs. Cancer Treat. Rev. 2004, 30, 437–449. [Google Scholar] [CrossRef]
- Ross, D.; Beall, H.D.; Siegel, D.; Traver, R.D.; Gustafson, D.L. Enzymology of bioreductive drug activation. Br. J. Cancer Suppl. 1996, 27, S1–S8. [Google Scholar]
- Tudor, G.; Alley, M.; Nelson, C.M.; Huang, R.; Covell, D.G.; Gutierrez, P.; Sausville, E.A. Cytotoxicity of RH1: NAD(P)H:quinone acceptor oxidoreductase (NQO1)-independent oxidative stress and apoptosis induction. Anticancer Drugs 2005, 16, 381–391. [Google Scholar] [CrossRef]
- Loadman, P.M.; Bibby, M.C.; Phillips, R.M. Pharmacological approach towards the development of indolequinone bioreductive drugs based on the clinically inactive agent EO9. Br. J. Pharmacol. 2002, 137, 701–709. [Google Scholar] [CrossRef][Green Version]
- Miao, X.S.; Song, P.; Savage, R.E.; Zhong, C.; Yang, R.Y.; Kizer, D.; Wu, H.; Volckova, E.; Ashwell, M.A.; Supko, J.G.; et al. Identification of the in vitro metabolites of 3,4-dihydro-2,2-dimethyl-2H-naphthol[1,2-b]pyran-5,6-dione (ARQ 501; beta-lapachone) in whole blood. Drug Metab. Dispos. 2008, 36, 641–648. [Google Scholar] [CrossRef]
- Nolan, K.A.; Scott, K.A.; Barnes, J.; Doncaster, J.; Whitehead, R.C.; Stratford, I.J. Pharmacological inhibitors of NAD(P)H quinone oxidoreductase, NQO1: Structure/activity relationships and functional activity in tumour cells. Biochem. Pharmacol. 2010, 80, 977–981. [Google Scholar] [CrossRef]
- Khunluck, T.; Kukongviriyapan, V.; Senggunprai, L.; Duangarsong, W.; Prawan, A. The Inhibition Kinetics and Potential Anti-Migration Activity of NQO1 Inhibitory Coumarins on Cholangiocarcinoma Cells. Integr. Cancer Ther. 2019, 18, 1534735418820444. [Google Scholar] [CrossRef]
- Available online: https://www.scbt.com/p/diminutol-361431-33-6 (accessed on 27 July 2024).
- Winski, S.L.; Faig, M.; Bianchet, M.A.; Siegel, D.; Swann, E.; Fung, K.; Duncan, M.W.; Moody, C.J.; Amzel, L.M.; Ross, D. Characterization of a mechanism-based inhibitor of NAD(P)H:quinone oxidoreductase 1 by biochemical, X-ray crystallographic, and mass spectrometric approaches. Biochemistry 2001, 40, 15135–15142. [Google Scholar] [CrossRef]
- Lei, K.; Gu, X.; Alvarado, A.G.; Du, Y.; Luo, S.; Ahn, E.H.; Kang, S.S.; Ji, B.; Liu, X.; Mao, H.; et al. Discovery of a dual inhibitor of NQO1 and GSTP1 for treating glioblastoma. J. Hematol. Oncol. 2020, 13, 141. [Google Scholar] [CrossRef]
- Gong, Q.; Hu, J.; Wang, P.; Li, X.; Zhang, X. A comprehensive review on β-lapachone: Mechanisms, structural modifications, and therapeutic potentials. Eur. J. Med. Chem. 2021, 210, 112962. [Google Scholar] [CrossRef]
- Bey, E.A.; Reinicke, K.E.; Srougi, M.C.; Varnes, M.; Anderson, V.E.; Pink, J.J.; Li, L.S.; Patel, M.; Cao, L.; Moore, Z.; et al. Catalase Abrogates β-Lapachone–Induced PARP1 Hyperactivation–Directed Programmed Necrosis in NQO1-Positive Breast Cancers. Mol. Cancer Ther. 2013, 12, 2110–2120. [Google Scholar] [CrossRef]
- Moore, Z.; Chakrabarti, G.; Luo, X.; Ali, A.; Hu, Z.; Fattah, F.J.; Vemireddy, R.; DeBerardinis, R.J.; Brekken, R.A.; Boothman, D.A. NAMPT inhibition sensitizes pancreatic adenocarcinoma cells to tumor-selective, PAR-independent metabolic catastrophe and cell death induced by β-lapachone. Cell Death Dis. 2015, 6, e1599. [Google Scholar] [CrossRef]
- Musaogullari, A.; Chai, Y.C. Redox Regulation by Protein S-Glutathionylation: From Molecular Mechanisms to Implications in Health and Disease. Int. J. Mol. Sci. 2020, 21, 8113. [Google Scholar] [CrossRef] [PubMed]
- Krishnan, P.; Bastow, K.F. Novel mechanisms of DNA topoisomerase II inhibition by pyranonaphthoquinone derivatives-eleutherin, alpha lapachone, and beta lapachone. Biochem. Pharmacol. 2000, 60, 1367–1379. [Google Scholar] [CrossRef] [PubMed]
- Li, C.J.; Averboukh, L.; Pardee, A.B. Beta-Lapachone, a novel DNA topoisomerase I inhibitor with a mode of action different from camptothecin. J. Biol. Chem. 1993, 268, 22463–22468. [Google Scholar] [CrossRef] [PubMed]
- Park, M.T.; Song, M.J.; Lee, H.; Oh, E.T.; Choi, B.H.; Jeong, S.Y.; Choi, E.K.; Park, H.J. β-lapachone significantly increases the effect of ionizing radiation to cause mitochondrial apoptosis via JNK activation in cancer cells. PLoS ONE 2011, 6, e25976. [Google Scholar] [CrossRef] [PubMed]
- Bair, J.S.; Palchaudhuri, R.; Hergenrother, P.J. Chemistry and Biology of Deoxynyboquinone, a potent inducer of cancer cell death. J. Am. Chem. Soc. 2010, 132, 5469–5478. [Google Scholar] [CrossRef] [PubMed]
- Garland, M.; Yim, J.J.; Bogyo, M. A Bright Future for Precision Medicine: Advances in Fluorescent Chemical Probe Design and Their Clinical Application. Cell Chem. Biol. 2016, 23, 122–136. [Google Scholar] [CrossRef]
- Luo, Z.; Feng, L.; An, R.; Duan, G.; Yan, R.; Shi, H.; He, J.; Zhou, Z.; Ji, C.; Chen, H.Y.; et al. Activatable Near-Infrared Probe for Fluorescence Imaging of γ-Glutamyl Transpeptidase in Tumor Cells and In Vivo. Chemistry 2017, 23, 14778–14785. [Google Scholar] [CrossRef]
- Tung, C.-H.; Zeng, Q.; Shah, K.; Kim, D.-E.; Schellingerhout, D.; Weissleder, R. In Vivo Imaging of β-Galactosidase Activity Using Far Red Fluorescent Switch. Cancer Res. 2004, 64, 1579–1583. [Google Scholar] [CrossRef] [PubMed]
- Guo, T.; Cui, L.; Shen, J.; Zhu, W.; Xu, Y.; Qian, X. A highly sensitive long-wavelength fluorescence probe for nitroreductase and hypoxia: Selective detection and quantification. Chem. Commun. 2013, 49, 10820–10822. [Google Scholar] [CrossRef]
- Li, H.; Li, Y.; Yao, Q.; Fan, J.; Sun, W.; Long, S.; Shao, K.; Du, J.; Wang, J.; Peng, X. In situ imaging of aminopeptidase N activity in hepatocellular carcinoma: A migration model for tumour using an activatable two-photon NIR fluorescent probe. Chem. Sci. 2019, 10, 1619–1625. [Google Scholar] [CrossRef]
- Gao, X.; Ma, G.; Jiang, C.; Zeng, L.; Jiang, S.; Huang, P.; Lin, J. In Vivo Near-Infrared Fluorescence and Photoacoustic Dual-Modal Imaging of Endogenous Alkaline Phosphatase. Anal. Chem. 2019, 91, 7112–7117. [Google Scholar] [CrossRef] [PubMed]
- Tian, Y.; Li, Y.; Jiang, W.L.; Zhou, D.Y.; Fei, J.; Li, C.Y. In-Situ Imaging of Azoreductase Activity in the Acute and Chronic Ulcerative Colitis Mice by a Near-Infrared Fluorescent Probe. Anal. Chem. 2019, 91, 10901–10907. [Google Scholar] [CrossRef] [PubMed]
- Zhan, C.; Cheng, J.; Li, B.; Huang, S.; Zeng, F.; Wu, S. A Fluorescent Probe for Early Detection of Melanoma and Its Metastasis by Specifically Imaging Tyrosinase Activity in a Mouse Model. Anal. Chem. 2018, 90, 8807–8815. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Lee, D.; Yu, S.; Kim, G.; Lee, S.; Cho, Y.; Jeong, H.; Nam, K.T.; Yoon, J. In vivo near-infrared imaging and phototherapy of tumors using a cathepsin B-activated fluorescent probe. Biomaterials 2017, 122, 130–140. [Google Scholar] [CrossRef]
- Neijenhuis, L.K.A.; de Myunck, L.; Bijlstra, O.D.; Kuppen, P.J.K.; Hilling, D.E.; Borm, F.J.; Cohen, D.; Mieog, J.S.D.; Steup, W.H.; Braun, J.; et al. Near-Infrared Fluorescence Tumor-Targeted Imaging in Lung Cancer: A Systematic Review. Life 2022, 12, 446. [Google Scholar] [CrossRef] [PubMed]
- Scott, J.I.; Deng, Q.; Vendrell, M. Near-Infrared Fluorescent Probes for the Detection of Cancer-Associated Proteases. ACS Chem. Biol. 2021, 16, 1304–1317. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Kim, D.; Yao, Q.; Ge, H.; Chung, J.; Fan, J.; Wang, J.; Peng, X.; Yoon, J. Activity-Based NIR Enzyme Fluorescent Probes for the Diagnosis of Tumors and Image-Guided Surgery. Angew. Chem. Int. Ed. Engl. 2021, 60, 17268–17289. [Google Scholar] [CrossRef] [PubMed]
- Available online: https://www.cancernetwork.com/view/fda-approves-pegulicianine-based-imaging-for-breast-cancer-surgery (accessed on 27 July 2024).
- Smith, B.L.; Hunt, K.K.; Carr, D.; Blumencranz, P.W.; Hwang, E.S.; Gadd, M.A.; Stone, K.; Dyess, D.L.; Dodge, D.; Valente, S.; et al. Intraoperative Fluorescence Guidance for Breast Cancer Lumpectomy Surgery. NEJM Evid. 2023, 2, EVIDoa2200333. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Zhang, Y.; Wu, Q.; Cui, X.; Lin, Z.; Liu, S.; Chen, L. Clinical implications of high NQO1 expression in breast cancers. J. Exp. Clin. Cancer Res. 2014, 33, 14. [Google Scholar] [CrossRef]
- Cui, X.; Jin, T.; Wang, X.; Jin, G.; Li, Z.; Lin, L. NAD(P)H:quinone oxidoreductase-1 overexpression predicts poor prognosis in small cell lung cancer. Oncol. Rep. 2014, 32, 2589–2595. [Google Scholar] [CrossRef]
- Dong, Y.; Bey, E.A.; Li, L.S.; Kabbani, W.; Yan, J.; Xie, X.J.; Hsieh, J.T.; Gao, J.; Boothman, D.A. Prostate cancer radiosensitization through poly(ADP-Ribose) polymerase-1 hyperactivation. Cancer Res. 2010, 70, 8088–8096. [Google Scholar] [CrossRef] [PubMed]
- Ji, L.; Wei, Y.; Jiang, T.; Wang, S. Correlation of Nrf2, NQO1, MRP1, cmyc and p53 in colorectal cancer and their relationships to clinicopathologic features and survival. Int. J. Clin. Exp. Pathol. 2014, 7, 1124–1131. [Google Scholar] [PubMed]
- Okamura, T.; Kurisu, K.; Yamamoto, W.; Takano, H.; Nishiyama, M. NADPH/quinone oxidoreductase is a priority target of glioblastoma chemotherapy. Int. J. Oncol. 2000, 16, 295–303. [Google Scholar] [CrossRef] [PubMed]
- Silvers, W.C.; Prasai, B.; Burk, D.H.; Brown, M.L.; McCarley, R.L. Profluorogenic reductase substrate for rapid, selective, and sensitive visualization and detection of human cancer cells that overexpress NQO1. J. Am. Chem. Soc. 2013, 135, 309–314. [Google Scholar] [CrossRef] [PubMed]
- Gong, Q.; Yang, F.; Hu, J.; Li, T.; Wang, P.; Li, X.; Zhang, X. Rational designed highly sensitive NQO1-activated near-infrared fluorescent probe combined with NQO1 substrates in vivo: An innovative strategy for NQO1-overexpressing cancer theranostics. Eur. J. Med. Chem. 2021, 224, 113707. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.X.S.; Song, Z.; Li, K. Trimethyl Lock Quinone-Based Organic Molecular Probes for NQO1 Sensing and Imaging. Chemosensors 2023, 11, 221. [Google Scholar] [CrossRef]
- Su, D.; Chen, X.; Zhang, Y.; Gao, X. Activatable imaging probes for cancer-linked NAD (P) H: Quinone oxidoreductase-1 (NQO1): Advances and future prospects. Trends Analyt. Chem. 2020, 133, 116112. [Google Scholar] [CrossRef]
- Srivenugopal, K.S.; Arutla, V.; Punganuru, S.R.; Khan, A.E.M.A. Application of a Specific and Sensitive NQO1 Turn-On Near-Infrared Fluorescence Probe for Live Cancer Cell and Xenografted Tumor Imaging in Nude Mice. In Hypoxia: Methods and Protocols; Gilkes, D.M., Ed.; Springer: New York, NY, USA, 2024; pp. 63–74. [Google Scholar]
- Zhang, X.; Yu, F.; Wang, Z.; Jiang, T.; Song, X.; Yu, F. Fluorescence probes for lung carcinoma diagnosis and clinical application. Sens. Diagn. 2023, 2, 1077–1096. [Google Scholar] [CrossRef]
- Cao, C.; Li, J.; Zhang, X.; Zhang, X.; Gong, X.; Wang, S. NQO1-activated multifunctional theranostic probe for imaging-guided mitochondria-targeted photodynamic therapy and boosting immunogenic cell death. Talanta 2024, 272, 125786. [Google Scholar] [CrossRef]
- Mendoza, M.F.; Hollabaugh, N.M.; Hettiarachchi, S.U.; McCarley, R.L. Human NAD(P)H:quinone oxidoreductase type I (hNQO1) activation of quinone propionic acid trigger groups. Biochemistry 2012, 51, 8014–8026. [Google Scholar] [CrossRef]
- Hettiarachchi, S.U.; Prasai, B.; McCarley, R.L. Detection and cellular imaging of human cancer enzyme using a turn-on, wavelength-shiftable, self-immolative profluorophore. J. Am. Chem. Soc. 2014, 136, 7575–7578. [Google Scholar] [CrossRef] [PubMed]
- Prasai, B.; Silvers, W.C.; McCarley, R.L. Oxidoreductase-Facilitated Visualization and Detection of Human Cancer Cells. Anal. Chem. 2015, 87, 6411–6418. [Google Scholar] [CrossRef] [PubMed]
- Silvers, W.C.; Payne, A.S.; McCarley, R.L. Shedding light by cancer redox—Human NAD(P)H:quinone oxidoreductase 1 activation of a cloaked fluorescent dye. Chem. Commun. 2011, 47, 11264–11266. [Google Scholar] [CrossRef] [PubMed]
- Best, Q.A.; Johnson, A.E.; Prasai, B.; Rouillere, A.; McCarley, R.L. Environmentally Robust Rhodamine Reporters for Probe-based Cellular Detection of the Cancer-linked Oxidoreductase hNQO1. ACS Chem. Biol. 2016, 11, 231–240. [Google Scholar] [CrossRef] [PubMed]
- Fei, Q.; Zhou, L.; Wang, F.; Shi, B.; Li, C.; Wang, R.; Zhao, C. Rational Construction of Probes Rendering Ratiometric Response to the Cancer-Specific Enzyme NQO1. Dye. Pigment. 2017, 136, 846–851. [Google Scholar] [CrossRef]
- Cuff, S.; Lewis, R.D.; Chinje, E.; Jaffar, M.; Knox, R.; Weeks, I. An improved cell-permeable fluorogenic substrate as the basis for a highly sensitive test for NAD(P)H quinone oxidoreductase 1 (NQO1) in living cells. Free Radic. Biol. Med. 2018, 116, 141–148. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Z.; Xu, M.; Wu, T.; Zhang, X.; Shen, Y.; Ernest, U.; Gui, L.; Wang, F.; He, Q.; Chen, H. Design and synthesis of NQO1 responsive fluorescence probe and its application in bio-imaging for cancer diagnosis. Talanta 2019, 198, 323–329. [Google Scholar] [CrossRef] [PubMed]
- Kwon, N.; Cho, M.K.; Park, S.J.; Kim, D.; Nam, S.-J.; Cui, L.; Kim, H.M.; Yoon, J. An efficient two-photon fluorescent probe for human NAD(P)H:quinone oxidoreductase (hNQO1) detection and imaging in tumor cells. Chem. Commun. 2017, 53, 525–528. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.P.; Qi, F.J.; Qian, Y.P.; Bao, X.Z.; Zhang, H.C.; Ma, B.; Dai, F.; Zhang, S.X.; Zhou, B. Developing Push-Pull Hydroxylphenylpolyenylpyridinium Chromophores as Ratiometric Two-Photon Fluorescent Probes for Cellular and Intravital Imaging of Mitochondrial NQO1. Anal. Chem. 2021, 93, 2385–2393. [Google Scholar] [CrossRef]
- Shin, W.S.; Han, J.; Verwilst, P.; Kumar, R.; Kim, J.H.; Kim, J.S. Cancer Targeted Enzymatic Theranostic Prodrug: Precise Diagnosis and Chemotherapy. Bioconjug. Chem. 2016, 27, 1419–1426. [Google Scholar] [CrossRef]
- Punganuru, S.R.; Madala, H.R.; Arutla, V.; Srivenugopal, K.S. Cancer-Specific Biomarker hNQO1-Activatable Fluorescent Probe for Imaging Cancer Cells In Vitro and In Vivo. Cancers 2018, 10, 470. [Google Scholar] [CrossRef] [PubMed]
- Shin, W.S.; Lee, M.-G.; Verwilst, P.; Lee, J.H.; Chi, S.-G.; Kim, J.S. Mitochondria-targeted aggregation induced emission theranostics: Crucial importance of in situ activation. Chem. Sci. 2016, 7, 6050–6059. [Google Scholar] [CrossRef] [PubMed]
- Best, Q.A.; Prasai, B.; Rouillere, A.; Johnson, A.E.; McCarley, R.L. Efficacious fluorescence turn-on probe for high-contrast imaging of human cells overexpressing quinone reductase activity. Chem. Commun. 2017, 53, 783–786. [Google Scholar] [CrossRef] [PubMed]
- Pan, D.; Luo, F.; Liu, X.; Liu, W.; Chen, W.; Liu, F.; Kuang, Y.-Q.; Jiang, J.-H. A novel two-photon fluorescent probe with a long Stokes shift and a high signal-to-background ratio for human NAD(P)H:quinone oxidoreductase 1 (hNQO1) detection and imaging in living cells and tissues. Analyst 2017, 142, 2624–2630. [Google Scholar] [CrossRef] [PubMed]
- Digby, E.M.; Sadovski, O.; Beharry, A.A. An Activatable Photosensitizer Targeting Human NAD(P)H: Quinone Oxidoreductase 1. Chemistry 2020, 26, 2713–2718. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Liu, P.; Yan, D.; Zeng, F.; Wu, S. A self-immolative and DT-diaphorase-activatable prodrug for drug-release tracking and therapy. J. Mater. Chem. B 2017, 5, 2635–2643. [Google Scholar] [CrossRef] [PubMed]
- Dai, M.; Song, C.W.; Yang, Y.J.; Kim, H.R.; Reo, Y.J.; Ahn, K.H. Toward Ratiometric Detection of NAD(P)H Quinone Oxidoreductase-1: Benzocoumarin-Based Fluorescent Probes. Sens. Actuators B Chem. 2021, 330, 129277. [Google Scholar] [CrossRef]
- Zhang, C.; Zhai, B.-B.; Peng, T.; Zhong, Z.; Xu, L.; Zhang, Q.-Z.; Li, L.-Y.; Yi, L.; Xi, Z. Design and synthesis of near-infrared fluorescence-enhancement probes for the cancer-specific enzyme hNQO1. Dye. Pigment. 2017, 143, 245–251. [Google Scholar] [CrossRef]
- Shen, Z.; Prasai, B.; Nakamura, Y.; Kobayashi, H.; Jackson, M.S.; McCarley, R.L. A Near-Infrared, Wavelength-Shiftable, Turn-on Fluorescent Probe for the Detection and Imaging of Cancer Tumor Cells. ACS Chem. Biol. 2017, 12, 1121–1132. [Google Scholar] [CrossRef]
- Punganuru, S.R.; Madala, H.R.; Arutla, V.; Zhang, R.; Srivenugopal, K.S. Characterization of a highly specific NQO1-activated near-infrared fluorescent probe and its application for in vivo tumor imaging. Sci. Rep. 2019, 9, 8577. [Google Scholar] [CrossRef]
- Zheng, Y.; Pan, D.; Zhang, Y.; Zhang, Y.; Shen, Y. Hemicyanine-based near-infrared fluorescent probe for the ultrasensitive detection of hNQO1 activity and discrimination of human cancer cells. Anal. Chim. Acta 2019, 1090, 125–132. [Google Scholar] [CrossRef]
- Wu, W.; Li, X.; Zhao, L.; Li, S.; Han, J.; Zhang, Y.; Zhao, Z. Design and synthesis of a deep tissue penetrating near-infrared two-photon fluorescence probe for the specific detection of NQO1. Chem. Commun. 2022, 58, 5634–5637. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Chen, J.; Gao, J.; Chen, J.-A.; Xu, G.; Zhu, T.; Gu, X.; Guo, Z.; Zhu, W.-H.; Zhao, C. A molecular design strategy toward enzyme-activated probes with near-infrared I and II fluorescence for targeted cancer imaging. Chem. Sci. 2019, 10, 7222–7227. [Google Scholar] [CrossRef]
- Zhang, X.; Jiang, K.; Jiang, S.; Zhao, F.; Chen, P.; Huang, P.; Lin, J. In Vivo Near-Infrared Fluorescence/Ratiometric Photoacoustic Duplex Imaging of Lung Cancer-Specific hNQO1. Anal. Chem. 2022, 94, 13770–13776. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Jiang, Y.; Li, J.; Huang, J.; Pu, K. Molecular Chemiluminescent Probes with a Very Long Near-Infrared Emission Wavelength for in Vivo Imaging. Angew. Chem. Int. Ed. Engl. 2021, 60, 3999–4003. [Google Scholar] [CrossRef] [PubMed]
- Ye, S.; Hananya, N.; Green, O.; Chen, H.; Zhao, A.Q.; Shen, J.; Shabat, D.; Yang, D. A Highly Selective and Sensitive Chemiluminescent Probe for Real-Time Monitoring of Hydrogen Peroxide in Cells and Animals. Angew. Chem. Int. Ed. Engl. 2020, 59, 14326–14330. [Google Scholar] [CrossRef]
- Bruemmer, K.J.; Green, O.; Su, T.A.; Shabat, D.; Chang, C.J. Chemiluminescent Probes for Activity-Based Sensing of Formaldehyde Released from Folate Degradation in Living Mice. Angew. Chem. Int. Ed. Engl. 2018, 57, 7508–7512. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; Huang, J.; Fan, J.; Du, J.; Pu, K.; Peng, X. Chemiluminescence for bioimaging and therapeutics: Recent advances and challenges. Chem. Soc. Rev. 2020, 49, 6800–6815. [Google Scholar] [CrossRef] [PubMed]
- Hananya, N.; Reid, J.P.; Green, O.; Sigman, M.S.; Shabat, D. Rapid chemiexcitation of phenoxy-dioxetane luminophores yields ultrasensitive chemiluminescence assays. Chem. Sci. 2019, 10, 1380–1385. [Google Scholar] [CrossRef]
- Liu, J.; Chen, Z.; Huo, H.; Chen, L.; Wu, Y.; Zhang, X.; Su, L.; Li, Q.; Song, J. An Activatable Near-Infrared Molecular Chemiluminescence Probe for Visualization of NQO1 Activity In Vivo. Chin. J. Chem. 2022, 40, 2400–2406. [Google Scholar] [CrossRef]
- Son, S.; Won, M.; Green, O.; Hananya, N.; Sharma, A.; Jeon, Y.; Kwak, J.H.; Sessler, J.L.; Shabat, D.; Kim, J.S. Chemiluminescent Probe for the In Vitro and In Vivo Imaging of Cancers Over-Expressing NQO1. Angew. Chem. Int. Ed. Engl. 2019, 58, 1739–1743. [Google Scholar] [CrossRef] [PubMed]
- Luo, Y.; Wang, W.; Zeng, Y.; Wang, S.; Guo, X.; Hu, R.; Yang, G. A bioluminescent probe for NQO1 overexpressing cancer cell imaging in vitro and in vivo. Analyst 2022, 147, 5264–5268. [Google Scholar] [CrossRef] [PubMed]
- Dai, Y.; Qian, M.; Li, Y. Structural Modification Endows Small-Molecular SN38 Derivatives with Multifaceted Functions. Molecules 2023, 28, 4931. [Google Scholar] [CrossRef]
- Sadikot, R.T.; Blackwell, T.S. Bioluminescence imaging. Proc. Am. Thorac. Soc. 2005, 2, 511–532, 537–540. [Google Scholar] [CrossRef] [PubMed]
- Ozawa, T.; Yoshimura, H.; Kim, S.B. Advances in fluorescence and bioluminescence imaging. Anal. Chem. 2013, 85, 590–609. [Google Scholar] [CrossRef] [PubMed]
- Welsh, D.K.; Kay, S.A. Bioluminescence imaging in living organisms. Curr. Opin. Biotechnol. 2005, 16, 73–78. [Google Scholar] [CrossRef] [PubMed]
- Cohen, A.S.; Dubikovskaya, E.A.; Rush, J.S.; Bertozzi, C.R. Real-time bioluminescence imaging of glycans on live cells. J. Am. Chem. Soc. 2010, 132, 8563–8565. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; Tang, J.; Xu, Y.; Dai, Z. Bioluminescence Imaging of Inflammation in Vivo Based on Bioluminescence and Fluorescence Resonance Energy Transfer Using Nanobubble Ultrasound Contrast Agent. ACS Nano 2019, 13, 5124–5132. [Google Scholar] [CrossRef]
- Lu, L.; Li, B.; Ding, S.; Fan, Y.; Wang, S.; Sun, C.; Zhao, M.; Zhao, C.X.; Zhang, F. NIR-II bioluminescence for in vivo high contrast imaging and in situ ATP-mediated metastases tracing. Nat. Commun. 2020, 11, 4192. [Google Scholar] [CrossRef]
- Gross, S.; Gammon, S.T.; Moss, B.L.; Rauch, D.; Harding, J.; Heinecke, J.W.; Ratner, L.; Piwnica-Worms, D. Bioluminescence imaging of myeloperoxidase activity in vivo. Nat. Med. 2009, 15, 455–461. [Google Scholar] [CrossRef]
- Zhou, W.; Leippe, D.; Duellman, S.; Sobol, M.; Vidugiriene, J.; O’Brien, M.; Shultz, J.W.; Kimball, J.J.; DiBernardo, C.; Moothart, L.; et al. Self-immolative bioluminogenic quinone luciferins for NAD(P)H assays and reducing capacity-based cell viability assays. Chembiochem 2014, 15, 670–675. [Google Scholar] [CrossRef]
- Si, J.; Zhao, X.; Gao, S.; Huang, D.; Sui, M. Advances in delivery of Irinotecan (CPT-11) active metabolite 7-ethyl-10-hydroxycamptothecin. Int. J. Pharm. 2019, 568, 118499. [Google Scholar] [CrossRef]
- Zhu, S.; Zhang, T.; Gao, H.; Jin, G.; Yang, J.; He, X.; Guo, H.; Xu, F. Combination therapy of Lox inhibitor and stimuli-responsive drug for mechanochemically synergistic breast cancer treatment. Adv. Healthc. Mater. 2023, 12, e2300103. [Google Scholar] [CrossRef] [PubMed]
- BioTracker NIR-ASM NQO1 Live Cell Dye. Available online: https://www.emdmillipore.com/US/en/product/BioTracker-NIR-ASM-NQO1-Live-Cell-Dye,MM_NF-SCT072 (accessed on 27 July 2024).
- Rashid, M.H.; Babu, D.; Siraki, A.G. Interactions of the antioxidant enzymes NAD(P)H: Quinone oxidoreductase 1 (NQO1) and NRH: Quinone oxidoreductase 2 (NQO2) with pharmacological agents, endogenous biochemicals and environmental contaminants. Chem. Biol. Interact. 2021, 345, 109574. [Google Scholar] [CrossRef] [PubMed]





| Compound | Structure | Properties/Limitations to Therapy | Reference(s) |
|---|---|---|---|
| β-Lapachone |  | Well-characterized, excellent futile substrate, complex structure, solubility problems, potent efficacy in vitro and in preclinical cancer models; however, anemia was a prominent adverse effect in a single clinical trial. | [44,69,75,80,81,82] |
| Deoxynyboquinone | 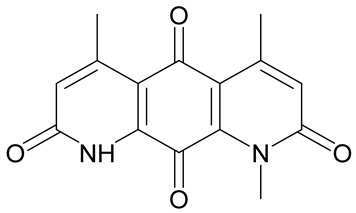 | Natural product, has also been synthesized, more potent than β-lapachone as a futile cycle substrate and for induction of tumor cell killing, insoluble, limited data on anticancer efficacy. | [83,84] |
| GNQ-9 |  | Synthetic quinone substrate, BBB-permeable, strong cytotoxicity in cancer cell lines, eliminated glioblastoma in orthotopic xenografts. Some evidence indicates immunogenic cell death may play a part. | [79] |
| Phenothiazinium redox cyclers |  | Robust induction of oxidative stress, and cell death in NQO1 stably transfected MCF-7 cells was demonstrated. | [85] |
| RH1 | 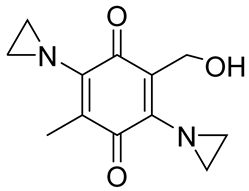 | RH1 is similar to mitomycin, activation by NQO1 and similar enzymes leads to the formation of cytotoxic species, which then alkylate and crosslink the DNA. Phase I clinical trial indicated bone marrow suppression in patients. | [52,86] |
| Combretastatin A-4 prodrug | 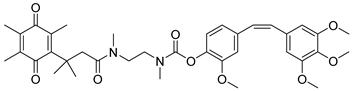 | CA-4 is an anti-microtubule and anti-angiogenesis drug. NQO1-mediated release of CA-4 was reported in a prodrug approach. Selective killing of NQO1-rich cells was indicated. | [87] |
| Podophyllotoxin prodrug |  | Podophyllotoxin derivatives inhibit topoisomerase II. Prodrug 3 of the study induced greater cell kill in NQO1-positive cells and suppressed the growth of HepG2 xenografts. | [88] |
| Steptonigrin (STN) | 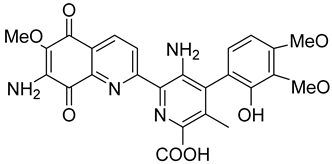 | A quinone antibiotic, has been reported to induce NQO1-dependent and dicumarol-sensitive cell death. | [89,90] |
| Geldanamycin AAG |  | The hydroquinone resulting from NQO1 action on Gelda-AAG inactivates HSP-90, leading to apoptosis. Thus, these cytotoxic quinones exhibit synthetic lethality with NRF2 inducers. | [39] |
| Mitomycin C (MMC) | 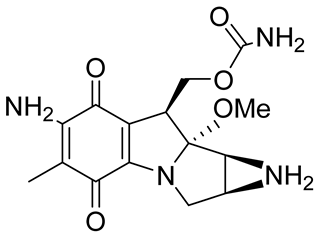 | Mitomycin C (MMC), an established clinically used antitumor antibiotic, can be activated by NQO1 to alkylate DNA and generate crosslinks. Other enzymes have also been implicated in MMC conversion to semiquinone species. | [67,68,91,92] |
| EO9 | 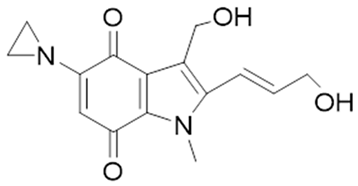 | EO9 was engineered as an NQO1-targeted drug; however, it penetrates the cancers poorly and is rapidly eliminated, thus hindering its antitumor efficacy. | [66,93,94,95] |
| 5 FU prodrug | 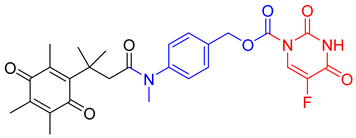 | A tripartite prodrug composed of a trigger group, a self-immolative linker, and 5-FU was synthesized. The prodrug had a cytotoxicity similar to 5-FU and may have a better safety profile. | [96] |
| Reagent Number | Original Designation | Structure | Reference |
|---|---|---|---|
| 1 | NMPABA |  | [138,146] |
| 2 | Q3NI | 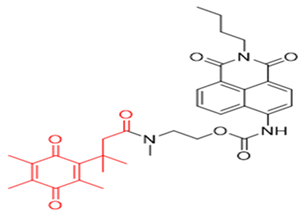 | [146] |
| 3 | Q3PA |  | [146] |
| 4 | QMeNN | 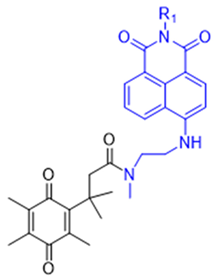 | [147] |
| 5 | Q3PA Rhodamine fluorophore |  | [148] |
| 6 | Q3MJSNR | 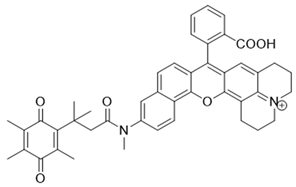 | [149] |
| 7 | 6-Hydroxyl phenolprobe-13 | 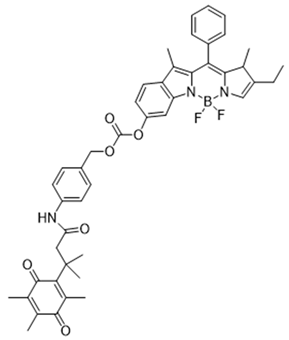 | [150] |
| 8 | 4-Methylumbelliferone (4-MU), probe-14 |  | [151] |
| 9 | 7-Nitro-2,1,3-benzoxadiazole, probe-16 |  | [152] |
| 10 | Aminoacetyl-naphthalene, probe,18-TPQ | 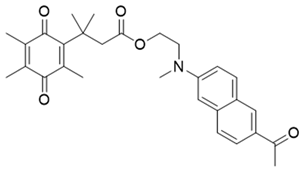 | [153] |
| 11 | Hydroxylphenylpolyenyl pyridinium, probe 19-QBMP | 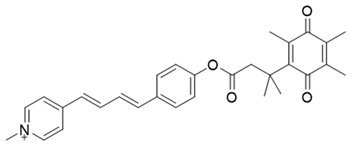 | [154] |
| 12 | 7-Ethyl-10-hydroxycamptothecinprobe-24 |  | [155] |
| 13 | NQ-DCM |  | [156] |
| 14 | Triphenylphosphoniumprobe-21 | 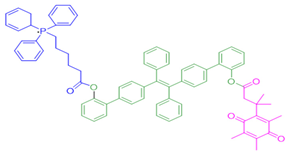 | [157] |
| Reagent Number | Designation | Structure | Reference |
|---|---|---|---|
| 15 | Q3STCY |  | [164] |
| 16 | NIR-ASM |  | [165] |
| 17 | HCYSN |  | [166] |
| 18 | LET-10 | 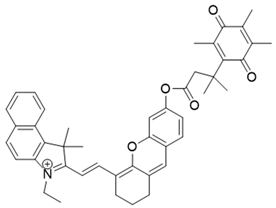 | [167] |
| 19 | NQO-Iml | 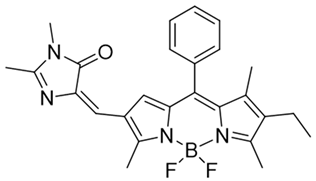 | [168] |
| Reagent Number | Chemiluminescent Probes for NQO1 | Structure | Reference |
|---|---|---|---|
| 20 | NIR CL probe |  | [175] |
| 21 | LET-10 |  | [176] |
| 22 | Luciferin probe 1 (Bioluminescent) | 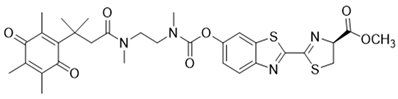 | [177] |
| 23 | Luciferin probe 2 (Bioluminescent) | 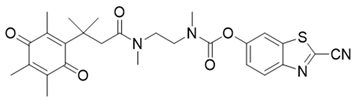 | [177] |
| 24 | SN-38 conjugate | 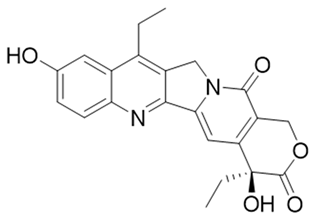 | [178] |
| 25 | Phenalenone conjugate | 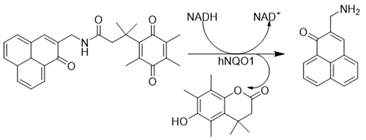 | [165] |
| 26 | Prodrug for detecting DT-diaphorase |  | [161] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Khan, A.E.M.A.; Arutla, V.; Srivenugopal, K.S. Human NQO1 as a Selective Target for Anticancer Therapeutics and Tumor Imaging. Cells 2024, 13, 1272. https://doi.org/10.3390/cells13151272
Khan AEMA, Arutla V, Srivenugopal KS. Human NQO1 as a Selective Target for Anticancer Therapeutics and Tumor Imaging. Cells. 2024; 13(15):1272. https://doi.org/10.3390/cells13151272
Chicago/Turabian StyleKhan, A. E. M. Adnan, Viswanath Arutla, and Kalkunte S. Srivenugopal. 2024. "Human NQO1 as a Selective Target for Anticancer Therapeutics and Tumor Imaging" Cells 13, no. 15: 1272. https://doi.org/10.3390/cells13151272
APA StyleKhan, A. E. M. A., Arutla, V., & Srivenugopal, K. S. (2024). Human NQO1 as a Selective Target for Anticancer Therapeutics and Tumor Imaging. Cells, 13(15), 1272. https://doi.org/10.3390/cells13151272






