A Review of Nanotechnology in microRNA Detection and Drug Delivery
Abstract
1. Introduction
2. MicroRNA
3. Nanotechnology in microRNA Detection
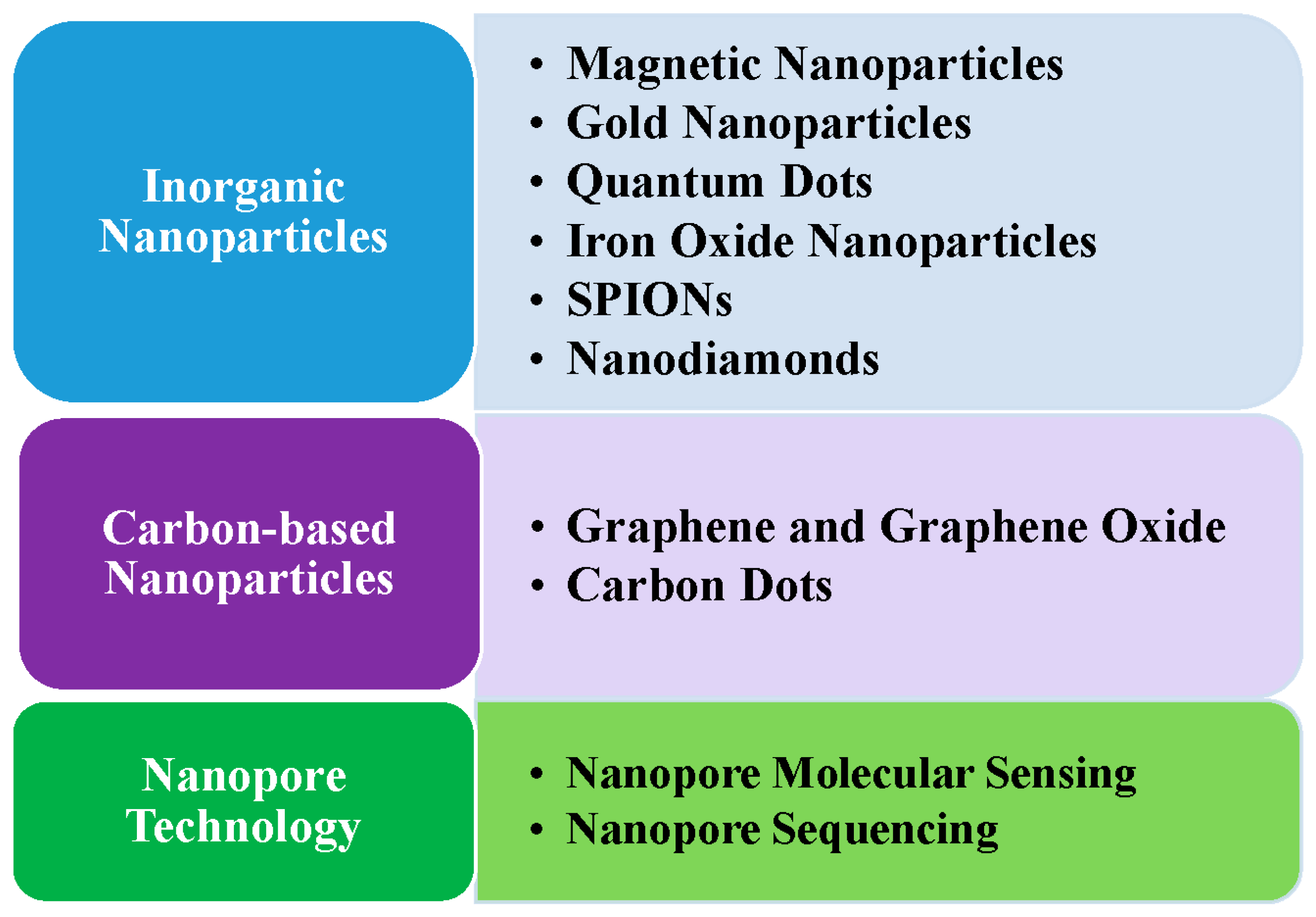
3.1. Inorganic Nanoparticles
3.1.1. Magnetic Nanoparticles
3.1.2. Gold Nanoparticles
3.1.3. Quantum Dots
3.2. Carbon-Based Nanoparticles
3.3. Nanopore Technology
4. Drug Delivery
4.1. Inorganic Nanoparticles
4.2. Polymeric Nanoparticles
4.3. Lipid Nanoparticles
4.4. Carbon-Based Nanomaterials
5. Discussion
6. Conclusions
Funding
Conflicts of Interest
Abbreviations
| CaCO3 | calcium carbonate |
| Cdots | carbon dots |
| CNTs | carbon nanotubes |
| CQDs | carbon quantum dots |
| chNPs | chitosan nanoparticles |
| C18P | C18-peptide |
| EGFR | epidermal growth factor receptor |
| GO/IO | graphene-oxide-loaded iron oxide |
| GQDs | graphene quantum dots |
| GNPs | gold nanoparticles |
| HCC | hepatocellular carcinoma |
| IAVs | influenza A viruses |
| LNA | locked nucleic acid |
| KSHV | Kaposi’s sarcoma-associated herpesvirus |
| MNPs | magnetic nanoparticles |
| MSCs | mesenchymal stromal cells |
| miRNAs | microRNAs |
| f-GQDs | modified GQDs |
| NDs | nanodiamonds |
| ncRNA | non-coding RNAs |
| NIR | near-infrared |
| P-L-Arg | poly-L-arginine |
| PCR | polymerase chain reaction |
| PLGA | poly (lactic-co-glycolic acid) |
| PLA | poly(l-lactide) |
| PEG | polyethylene glycol |
| PEL | primary effusion lymphoma |
| qPCR | quantitative PCR |
| RT-qPCR | quantitative polymerase chain reaction |
| QDs | quantum dots |
| RT-PCR | reverse transcription polymerase chain reaction |
| SPCE | screen-printed carbon electrodes |
| SPIONs | superparamagnetic iron-oxide-based nanoparticles |
References
- Bhushan, B. Introduction to nanotechnology. In Springer Handbook of Nanotechnology; Springer: Berlin/Heidelberg, Germany, 2017; pp. 1–19. [Google Scholar]
- Abou Neel, E.A.; Bozec, L.; Perez, R.A.; Kim, H.-W.; Knowles, J.C. Nanotechnology in dentistry: Prevention, diagnosis, and therapy. Int. J. Nanomed. 2015, 10, 6371–6394. [Google Scholar] [CrossRef]
- McNeil, S.E. Nanotechnology for the biologist. J. Leukoc. Biol. 2005, 78, 585–594. [Google Scholar] [CrossRef] [PubMed]
- Pané, S.; Puigmartí-Luis, J.; Bergeles, C.; Chen, X.Z.; Pellicer, E.; Sort, J.; Pocepcova, V.; Ferreira, A.; Nelson, B.J. Imaging Technologies for Biomedical Micro- and Nanoswimmers. Adv. Mater. Technol.-US 2019, 4, 1800575. [Google Scholar] [CrossRef]
- Rajendran, A.K.; Kim, H.D.; Kim, J.-W.; Bae, J.W.; Hwang, N.S. Nanotechnology in tissue engineering and regenerative medicine. Korean J. Chem. Eng. 2023, 40, 286–301. [Google Scholar] [CrossRef]
- Wang, H. MicroRNAs, Parkinson’s disease, and diabetes mellitus. Int. J. Mol. Sci. 2021, 22, 2953. [Google Scholar] [CrossRef] [PubMed]
- Wang, H. MicroRNAs and apoptosis in colorectal cancer. Int. J. Mol. Sci. 2020, 21, 5353. [Google Scholar] [CrossRef] [PubMed]
- Lewis, B.P.; Burge, C.B.; Bartel, D.P. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 2005, 120, 15–20. [Google Scholar] [CrossRef] [PubMed]
- Wang, H. Predicting cancer-related MiRNAs using expression profiles in tumor tissue. Curr. Pharm. Biotechnol. 2014, 15, 438–444. [Google Scholar] [CrossRef]
- Wang, H. Predicting microRNA biomarkers for cancer using phylogenetic tree and microarray analysis. Int. J. Mol. Sci. 2016, 17, 773. [Google Scholar] [CrossRef]
- Wang, H. microRNA Pathological Mechanisms between Parkinson’s Disease, Alzheimer’s Disease, Glaucoma, and Macular Degeneration. Expert Rev. Mol. Med. 2023, 25, e24. [Google Scholar] [CrossRef]
- Wang, H.; Taguchi, Y.; Liu, X. miRNAs and neurological diseases. Front. Neurol. 2021, 12, 662373. [Google Scholar]
- Gentile, G.; Morello, G.; La Cognata, V.; Guarnaccia, M.; Conforti, F.L.; Cavallaro, S. Dysregulated miRNAs as biomarkers and therapeutical targets in neurodegenerative diseases. J. Pers. Med. 2022, 12, 770. [Google Scholar] [CrossRef] [PubMed]
- Jung, H.J.; Lee, K.-P.; Kwon, K.-S.; Suh, Y. MicroRNAs in skeletal muscle aging: Current issues and perspectives. J. Gerontol. Ser. A 2019, 74, 1008–1014. [Google Scholar] [CrossRef] [PubMed]
- So, J.B.Y.; Kapoor, R.; Zhu, F.; Koh, C.; Zhou, L.; Zou, R.; Tang, Y.C.; Goo, P.C.; Rha, S.Y.; Chung, H.C. Development and validation of a serum microRNA biomarker panel for detecting gastric cancer in a high-risk population. Gut 2021, 70, 829–837. [Google Scholar] [CrossRef] [PubMed]
- Wang, N.; Wang, L.; Yang, Y.; Gong, L.; Xiao, B.; Liu, X. A serum exosomal microRNA panel as a potential biomarker test for gastric cancer. Biochem. Biophys. Res. Commun. 2017, 493, 1322–1328. [Google Scholar] [CrossRef] [PubMed]
- Wang, H. Phylogenetic analysis to explore the association between anti-NMDA receptor encephalitis and tumors based on microRNA biomarkers. Biomolecules 2019, 9, 572. [Google Scholar] [CrossRef] [PubMed]
- Wang, H. MicroRNAs, Multiple Sclerosis, and Depression. Int. J. Mol. Sci. 2021, 22, 7802. [Google Scholar] [CrossRef] [PubMed]
- Wang, H. MicroRNA, Diabetes Mellitus and Colorectal Cancer. Biomedicines 2020, 8, 530. [Google Scholar] [CrossRef]
- Ouyang, T.; Liu, Z.; Han, Z.; Ge, Q. MicroRNA detection specificity: Recent advances and future perspective. Anal. Chem. 2019, 91, 3179–3186. [Google Scholar] [CrossRef]
- Git, A.; Dvinge, H.; Salmon-Divon, M.; Osborne, M.; Kutter, C.; Hadfield, J.; Bertone, P.; Caldas, C. Systematic comparison of microarray profiling, real-time PCR, and next-generation sequencing technologies for measuring differential microRNA expression. RNA 2010, 16, 991–1006. [Google Scholar] [CrossRef]
- Shabaninejad, Z.; Yousefi, F.; Movahedpour, A.; Ghasemi, Y.; Dokanehiifard, S.; Rezaei, S.; Aryan, R.; Savardashtaki, A.; Mirzaei, H. Electrochemical-based biosensors for microRNA detection: Nanotechnology comes into view. Anal. Biochem. 2019, 581, 113349. [Google Scholar] [CrossRef] [PubMed]
- Alam, M.K. Nanocarrier-Based Drug Delivery Systems using Microfluidic-Assisted Techniques. Adv. NanoBiomed. Res. 2023, 3, 2300041. [Google Scholar] [CrossRef]
- Chandrasekaran, A.R.; Punnoose, J.A.; Zhou, L.; Dey, P.; Dey, B.K.; Halvorsen, K. DNA nanotechnology approaches for microRNA detection and diagnosis. Nucleic Acids Res. 2019, 47, 10489–10505. [Google Scholar] [CrossRef] [PubMed]
- Scheideler, M.; Vidakovic, I.; Prassl, R. Lipid nanocarriers for microRNA delivery. Chem. Phys. Lipids 2020, 226, 104837. [Google Scholar] [CrossRef] [PubMed]
- Sukocheva, O.A.; Liu, J.; Neganova, M.E.; Beeraka, N.M.; Aleksandrova, Y.R.; Manogaran, P.; Grigorevskikh, E.M.; Chubarev, V.N.; Fan, R. Perspectives of using microRNA-loaded nanocarriers for epigenetic reprogramming of drug resistant colorectal cancers. In Seminars in Cancer Biology; Elsevier: Amsterdam, The Netherlands, 2022; pp. 358–375. [Google Scholar]
- Dasgupta, I.; Chatterjee, A. Recent advances in miRNA delivery systems. Methods Protoc. 2021, 4, 10. [Google Scholar] [CrossRef] [PubMed]
- Awasthi, R.; Rathbone, M.J.; Hansbro, P.M.; Bebawy, M.; Dua, K. Therapeutic prospects of microRNAs in cancer treatment through nanotechnology. Drug Deliv. Transl. Res. 2018, 8, 97–110. [Google Scholar] [CrossRef]
- Lee, R.C.; Feinbaum, R.L.; Ambros, V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 1993, 75, 843–854. [Google Scholar] [CrossRef] [PubMed]
- Hammond, S.M. An overview of microRNAs. Adv. Drug Deliv. Rev. 2015, 87, 3–14. [Google Scholar] [CrossRef]
- Kozomara, A.; Birgaoanu, M.; Griffiths-Jones, S. miRBase: From microRNA sequences to function. Nucleic Acids Res. 2019, 47, D155–D162. [Google Scholar] [CrossRef]
- Fromm, B.; Domanska, D.; Høye, E.; Ovchinnikov, V.; Kang, W.; Aparicio-Puerta, E.; Johansen, M.; Flatmark, K.; Mathelier, A.; Hovig, E. MirGeneDB 2.0: The metazoan microRNA complement. Nucleic Acids Res. 2020, 48, D132–D141. [Google Scholar] [CrossRef]
- Condrat, C.E.; Thompson, D.C.; Barbu, M.G.; Bugnar, O.L.; Boboc, A.; Cretoiu, D.; Suciu, N.; Cretoiu, S.M.; Voinea, S.C. miRNAs as biomarkers in disease: Latest findings regarding their role in diagnosis and prognosis. Cells 2020, 9, 276. [Google Scholar] [CrossRef] [PubMed]
- Huang, W. MicroRNAs: Biomarkers, diagnostics, and therapeutics. In Bioinformatics in MicroRNA Research; Springer: Berlin/Heidelberg, Germany, 2017; pp. 57–67. [Google Scholar]
- Guay, C.; Regazzi, R. Circulating microRNAs as novel biomarkers for diabetes mellitus. Nat. Rev. Endocrinol. 2013, 9, 513–521. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.-H.; Wang, H. The association between depression and gastroesophageal reflux based on phylogenetic analysis of miRNA biomarkers. Curr. Med. Chem. 2020, 27, 6536–6547. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.-H.; Wang, H. The association between migraine and depression based on miRNA biomarkers and cohort studies. Curr. Med. Chem. 2021, 28, 5648–5656. [Google Scholar] [CrossRef] [PubMed]
- Wang, H. The distance distribution of human microRNAs in MirGeneDB database. Sci. Rep. 2022, 12, 17696. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Ho, C. The Human Pre-miRNA Distance Distribution for Exploring Disease Association. Int. J. Mol. Sci. 2023, 24, 1009. [Google Scholar] [CrossRef] [PubMed]
- Liu, K.; Tong, H.; Li, T.; Wang, X.; Chen, Y. Research progress in molecular biology related quantitative methods of MicroRNA. Am. J. Transl. Res. 2020, 12, 3198. [Google Scholar] [PubMed]
- Hunt, E.A.; Broyles, D.; Head, T.; Deo, S.K. MicroRNA detection: Current technology and research strategies. Annu. Rev. Anal. Chem. 2015, 8, 217–237. [Google Scholar]
- Li, W.; Ruan, K. MicroRNA detection by microarray. Anal. Bioanal. Chem. 2009, 394, 1117–1124. [Google Scholar] [CrossRef]
- Costa, V.; Angelini, C.; De Feis, I.; Ciccodicola, A. Uncovering the complexity of transcriptomes with RNA-Seq. BioMed Res. Int. 2010, 2010, 853916. [Google Scholar] [CrossRef]
- Hu, Y.; Lan, W.; Miller, D. Next-generation sequencing for MicroRNA expression profile. Bioinform. MicroRNA Res. 2017, 1617, 169–177. [Google Scholar]
- Willenbrock, H.; Salomon, J.; Søkilde, R.; Barken, K.B.; Hansen, T.N.; Nielsen, F.C.; Møller, S.; Litman, T. Quantitative miRNA expression analysis: Comparing microarrays with next-generation sequencing. RNA 2009, 15, 2028–2034. [Google Scholar] [CrossRef] [PubMed]
- Hurd, P.J.; Nelson, C.J. Advantages of next-generation sequencing versus the microarray in epigenetic research. Brief Funct. Genom. Proteomic 2009, 8, 174–183. [Google Scholar] [CrossRef] [PubMed]
- De la Rosa, C.; Reyes, J.L. Northern blot analysis of microRNAs and other small RNAs in plants. Plant MicroRNAs Methods Protoc. 2019, 1932, 121–129. [Google Scholar]
- Várallyay, E.; Burgyán, J.; Havelda, Z. MicroRNA detection by northern blotting using locked nucleic acid probes. Nat. Protoc. 2008, 3, 190–196. [Google Scholar] [CrossRef] [PubMed]
- Válóczi, A.; Hornyik, C.; Varga, N.; Burgyán, J.; Kauppinen, S.; Havelda, Z. Sensitive and specific detection of microRNAs by northern blot analysis using LNA-modified oligonucleotide probes. Nucleic Acids Res. 2004, 32, e175. [Google Scholar] [CrossRef] [PubMed]
- Várallyay, E.; Burgyán, J.; Havelda, Z. Detection of microRNAs by Northern blot analyses using LNA probes. Methods 2007, 43, 140–145. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.; Zhang, H.; Xu, Y.; Laššáková, S.; Korabečná, M.; Neužil, P. PCR past, present and future. Biotechniques 2020, 69, 317–325. [Google Scholar] [CrossRef]
- Schmittgen, T.D.; Lee, E.J.; Jiang, J.; Sarkar, A.; Yang, L.; Elton, T.S.; Chen, C. Real-time PCR quantification of precursor and mature microRNA. Methods 2008, 44, 31–38. [Google Scholar] [CrossRef]
- Dymond, J.S. Explanatory chapter: Quantitative PCR. In Methods in Enzymology; Elsevier: Amsterdam, The Netherlands, 2013; Volume 529, pp. 279–289. [Google Scholar]
- Bustin, S.; Benes, V.; Nolan, T.; Pfaffl, M. Quantitative real-time RT-PCR–a perspective. J. Mol. Endocrinol. 2005, 34, 597–601. [Google Scholar]
- Adams, G. A beginner’s guide to RT-PCR, qPCR and RT-qPCR. Biochemist 2020, 42, 48–53. [Google Scholar] [CrossRef]
- Norouzi, M.; Yasamineh, S.; Montazeri, M.; Dadashpour, M.; Sheervalilou, R.; Abasi, M.; Pilehvar-Soltanahmadi, Y. Recent advances on nanomaterials-based fluorimetric approaches for microRNAs detection. Mater. Sci. Eng. C 2019, 104, 110007. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Cheng, Z.; Wang, Y.; Han, T. The risks of miRNA therapeutics: In a drug target perspective. Drug Des. Dev. Ther. 2021, 15, 721–733. [Google Scholar] [CrossRef] [PubMed]
- Asakiya, C.; Zhu, L.; Yuhan, J.; Zhu, L.; Huang, K.; Xu, W. Current progress of miRNA-derivative nucleotide drugs: Modifications, delivery systems, applications. Expert Opin. Drug Deliv. 2022, 19, 435–450. [Google Scholar] [CrossRef] [PubMed]
- Gessner, I.; Fries, J.W.; Brune, V.; Mathur, S. Magnetic nanoparticle-based amplification of microRNA detection in body fluids for early disease diagnosis. J. Mater. Chem. B 2021, 9, 9–22. [Google Scholar] [CrossRef] [PubMed]
- Xu, S.; Hossaini Nasr, S.; Chen, D.; Zhang, X.; Sun, L.; Huang, X.; Qian, C. MiRNA extraction from cell-free biofluid using protein corona formed around carboxyl magnetic nanoparticles. ACS Biomater. Sci. Eng. 2018, 4, 654–662. [Google Scholar] [CrossRef] [PubMed]
- Balaban Hanoglu, S.; Harmanci, D.; Ucar, N.; Evran, S.; Timur, S. Recent Approaches in Magnetic Nanoparticle-Based Biosensors of miRNA Detection. Magnetochemistry 2023, 9, 23. [Google Scholar] [CrossRef]
- Tavallaie, R.; McCarroll, J.; Le Grand, M.; Ariotti, N.; Schuhmann, W.; Bakker, E.; Tilley, R.D.; Hibbert, D.B.; Kavallaris, M.; Gooding, J.J. Nucleic acid hybridization on an electrically reconfigurable network of gold-coated magnetic nanoparticles enables microRNA detection in blood. Nat. Nanotechnol. 2018, 13, 1066–1071. [Google Scholar] [CrossRef]
- Yuan, Y.-H.; Wu, Y.-D.; Chi, B.-Z.; Wen, S.-H.; Liang, R.-P.; Qiu, J.-D. Simultaneously electrochemical detection of microRNAs based on multifunctional magnetic nanoparticles probe coupling with hybridization chain reaction. Biosens. Bioelectron. 2017, 97, 325–331. [Google Scholar] [CrossRef]
- Ali, A.; Zafar, H.; Zia, M.; ul Haq, I.; Phull, A.R.; Ali, J.S.; Hussain, A. Synthesis, characterization, applications, and challenges of iron oxide nanoparticles. Nanotechnol. Sci. Appl. 2016, 9, 49–67. [Google Scholar] [CrossRef]
- Islam, M.N.; Gorgannezhad, L.; Masud, M.K.; Tanaka, S.; Hossain, M.S.A.; Yamauchi, Y.; Nguyen, N.T.; Shiddiky, M.J. Graphene-oxide-loaded superparamagnetic iron oxide nanoparticles for ultrasensitive electrocatalytic detection of MicroRNA. ChemElectroChem 2018, 5, 2488–2495. [Google Scholar] [CrossRef]
- Dreaden, E.C.; Alkilany, A.M.; Huang, X.; Murphy, C.J.; El-Sayed, M.A. The golden age: Gold nanoparticles for biomedicine. Chem. Soc. Rev. 2012, 41, 2740–2779. [Google Scholar] [CrossRef] [PubMed]
- Sztandera, K.; Gorzkiewicz, M.; Klajnert-Maculewicz, B. Gold nanoparticles in cancer treatment. Mol. Pharm. 2018, 16, 1–23. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Wang, Y.; Yang, G.; Xu, J.-J.; Chen, H.-Y. MicroRNA-mediated signal amplification coupled with GNP/dendrimers on a mass-sensitive biosensor and its applications in intracellular microRNA quantification. Biosens. Bioelectron. 2016, 85, 897–902. [Google Scholar] [CrossRef]
- Mohammadniaei, M.; Yoon, J.; Lee, T.; Choi, J.-W. Spectroelectrochemical detection of microRNA-155 based on functional RNA immobilization onto ITO/GNP nanopattern. J. Biotechnol. 2018, 274, 40–46. [Google Scholar] [CrossRef] [PubMed]
- Coutinho, C.; Somoza, Á. MicroRNA sensors based on gold nanoparticles. Anal. Bioanal. Chem. 2019, 411, 1807–1824. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Yang, Y.; Zhang, C.-y. Toward biocompatible semiconductor quantum dots: From biosynthesis and bioconjugation to biomedical application. Chem. Rev. 2015, 115, 11669–11717. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Xu, D.; Gao, J.; Chen, X.; Duo, Y.; Zhang, H. Semiconducting quantum dots: Modification and applications in biomedical science. Sci. China Mater. 2020, 63, 1631–1650. [Google Scholar] [CrossRef]
- Goryacheva, O.A.; Novikova, A.S.; Drozd, D.D.; Pidenko, P.S.; Ponomaryeva, T.S.; Bakal, A.A.; Mishra, P.K.; Beloglazova, N.V.; Goryacheva, I.Y. Water-dispersed luminescent quantum dots for miRNA detection. TrAC Trends Anal. Chem. 2019, 111, 197–205. [Google Scholar] [CrossRef]
- Goryacheva, O.; Mishra, P.; Goryacheva, I.Y. Luminescent quantum dots for miRNA detection. Talanta 2018, 179, 456–465. [Google Scholar] [CrossRef]
- Hu, O.; Li, Z.; Tong, Y.; Wang, Q.; Chen, Z. DNA functionalized double quantum dots-based fluorescence biosensor for one-step simultaneous detection of multiple microRNAs. Talanta 2021, 235, 122763. [Google Scholar] [CrossRef] [PubMed]
- Martins, C.S.; LaGrow, A.P.; Prior, J.o.A. Quantum dots for cancer-related miRNA monitoring. ACS Sens. 2022, 7, 1269–1299. [Google Scholar] [CrossRef]
- Ajgaonkar, R.; Lee, B.; Valimukhametova, A.; Nguyen, S.; Gonzalez-Rodriguez, R.; Coffer, J.; Akkaraju, G.R.; Naumov, A.V. Detection of pancreatic cancer miRNA with biocompatible nitrogen-doped graphene quantum dots. Materials 2022, 15, 5760. [Google Scholar] [CrossRef] [PubMed]
- Borghei, Y.S.; Hosseini, M.; Ganjali, M.R. A label-free luminescent light switching system for miRNA detection based on two color quantum dots. J. Photochem. Photobiol. A Chem. 2020, 391, 112351. [Google Scholar] [CrossRef]
- Ma, Y.; Zhang, Y.; William, W.Y. Near infrared emitting quantum dots: Synthesis, luminescence properties and applications. J. Mater. Chem. C 2019, 7, 13662–13679. [Google Scholar] [CrossRef]
- Ratre, P.; Nazeer, N.; Kumari, R.; Thareja, S.; Jain, B.; Tiwari, R.; Kamthan, A.; Srivastava, R.K.; Mishra, P.K. Carbon-based fluorescent nano-biosensors for the detection of cell-free circulating MicroRNAs. Biosensors 2023, 13, 226. [Google Scholar] [CrossRef]
- Hosseini, S.M.; Mohammadnejad, J.; Najafi-Taher, R.; Zadeh, Z.B.; Tanhaei, M.; Ramakrishna, S. Multifunctional carbon-based nanoparticles: Theranostic applications in cancer therapy and diagnosis. ACS Appl. Bio Mater. 2023, 6, 1323–1338. [Google Scholar] [CrossRef] [PubMed]
- Eivazzadeh-Keihan, R.; Noruzi, E.B.; Chidar, E.; Jafari, M.; Davoodi, F.; Kashtiaray, A.; Gorab, M.G.; Hashemi, S.M.; Javanshir, S.; Cohan, R.A. Applications of carbon-based conductive nanomaterials in biosensors. Chem. Eng. J. 2022, 442, 136183. [Google Scholar] [CrossRef]
- Wang, J.; Wen, J.; Yan, H. Recent applications of carbon nanomaterials for microRNA electrochemical sensing. Chem.–Asian J. 2021, 16, 114–128. [Google Scholar] [CrossRef]
- Gutiérrez-Gálvez, L.; García-Mendiola, T.; Gutiérrez-Sánchez, C.; Guerrero-Esteban, T.; García-Diego, C.; Buendía, I.; García-Bermejo, M.L.; Pariente, F.; Lorenzo, E. Carbon nanodot–based electrogenerated chemiluminescence biosensor for miRNA-21 detection. Microchim. Acta 2021, 188, 1–12. [Google Scholar] [CrossRef]
- Su, J.; Liu, W.; Chen, S.; Deng, W.; Dou, Y.; Zhao, Z.; Li, J.; Li, Z.; Yin, H.; Ding, X. A carbon-based DNA framework nano–bio interface for biosensing with high sensitivity and a high signal-to-noise ratio. ACS Sens. 2020, 5, 3979–3987. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Miao, P.; Sun, M.; Yan, M.; Liu, H. Progress in miRNA detection using graphene material–based biosensors. Small 2019, 15, 1901867. [Google Scholar] [CrossRef]
- Shuai, H.-L.; Huang, K.-J.; Xing, L.-L.; Chen, Y.-X. Ultrasensitive electrochemical sensing platform for microRNA based on tungsten oxide-graphene composites coupling with catalyzed hairpin assembly target recycling and enzyme signal amplification. Biosens. Bioelectron. 2016, 86, 337–345. [Google Scholar] [CrossRef] [PubMed]
- Shuai, H.-L.; Huang, K.-J.; Zhang, W.-J.; Cao, X.; Jia, M.-P. Sandwich-type microRNA biosensor based on magnesium oxide nanoflower and graphene oxide–gold nanoparticles hybrids coupling with enzyme signal amplification. Sens. Actuators B Chem. 2017, 243, 403–411. [Google Scholar] [CrossRef]
- Lin, B.; Hui, J.; Mao, H. Nanopore technology and its applications in gene sequencing. Biosensors 2021, 11, 214. [Google Scholar] [CrossRef]
- Ying, Y.-L.; Hu, Z.-L.; Zhang, S.; Qing, Y.; Fragasso, A.; Maglia, G.; Meller, A.; Bayley, H.; Dekker, C.; Long, Y.-T. Nanopore-based technologies beyond DNA sequencing. Nat. Nanotechnol. 2022, 17, 1136–1146. [Google Scholar] [CrossRef]
- Takeuchi, N.; Hiratani, M.; Kawano, R. Pattern recognition of microRNA expression in body fluids using nanopore decoding at subfemtomolar concentrations. JACS Au 2022, 2, 1829–1838. [Google Scholar] [CrossRef]
- Wanunu, M.; Dadosh, T.; Ray, V.; Jin, J.; McReynolds, L.; Drndić, M. Rapid electronic detection of probe-specific microRNAs using thin nanopore sensors. Nat. Nanotechnol. 2010, 5, 807–814. [Google Scholar] [CrossRef]
- Koch, C.; Reilly-O’Donnell, B.; Gutierrez, R.; Lucarelli, C.; Ng, F.S.; Gorelik, J.; Ivanov, A.P.; Edel, J.B. Nanopore sequencing of DNA-barcoded probes for highly multiplexed detection of microRNA, proteins and small biomarkers. Nat. Nanotechnol. 2023, 18, 1483–1491. [Google Scholar] [CrossRef]
- Huang, H.-C.; Barua, S.; Sharma, G.; Dey, S.K.; Rege, K. Inorganic nanoparticles for cancer imaging and therapy. J. Control. Release 2011, 155, 344–357. [Google Scholar] [CrossRef]
- Boca, S.; Gulei, D.; Zimta, A.-A.; Onaciu, A.; Magdo, L.; Tigu, A.B.; Ionescu, C.; Irimie, A.; Buiga, R.; Berindan-Neagoe, I. Nanoscale delivery systems for microRNAs in cancer therapy. Cell. Mol. Life Sci. 2020, 77, 1059–1086. [Google Scholar] [CrossRef] [PubMed]
- Yu, M.; Lei, B.; Gao, C.; Yan, J.; Ma, P.X. Optimizing surface-engineered ultra-small gold nanoparticles for highly efficient miRNA delivery to enhance osteogenic differentiation of bone mesenchymal stromal cells. Nano Res. 2017, 10, 49–63. [Google Scholar] [CrossRef]
- Schade, A.; Delyagina, E.; Scharfenberg, D.; Skorska, A.; Lux, C.; David, R.; Steinhoff, G. Innovative strategy for microRNA delivery in human mesenchymal stem cells via magnetic nanoparticles. Int. J. Mol. Sci. 2013, 14, 10710–10726. [Google Scholar] [CrossRef]
- Assali, A.; Akhavan, O.; Adeli, M.; Razzazan, S.; Dinarvand, R.; Zanganeh, S.; Soleimani, M.; Dinarvand, M.; Atyabi, F. Multifunctional core-shell nanoplatforms (gold@ graphene oxide) with mediated NIR thermal therapy to promote miRNA delivery. Nanomed. Nanotechnol. Biol. Med. 2018, 14, 1891–1903. [Google Scholar] [CrossRef] [PubMed]
- Kröger, N.; Poulsen, N. Diatoms—From cell wall biogenesis to nanotechnology. Annu. Rev. Genet. 2008, 42, 83–107. [Google Scholar] [CrossRef] [PubMed]
- Haddick, L.; Zhang, W.; Reinhard, S.; Möller, K.; Engelke, H.; Wagner, E.; Bein, T. Particle-size-dependent delivery of antitumoral miRNA using targeted mesoporous silica nanoparticles. Pharmaceutics 2020, 12, 505. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Xie, Y.; Kilchrist, K.V.; Li, J.; Duvall, C.L.; Oupický, D. Endosomolytic and tumor-penetrating mesoporous silica nanoparticles for siRNA/miRNA combination cancer therapy. ACS Appl. Mater. Interfaces 2020, 12, 4308–4322. [Google Scholar] [CrossRef] [PubMed]
- Yang, T.; Ao, Y.; Feng, J.; Wang, C.; Zhang, J. Biomineralization inspired synthesis of CaCO3-based DDS for pH-responsive release of anticancer drug. Mater. Today Commun. 2021, 27, 102256. [Google Scholar] [CrossRef]
- Roy, B.; Ghose, S.; Biswas, S. Therapeutic strategies for miRNA delivery to reduce hepatocellular carcinoma. In Seminars in Cell & Developmental Biology; Elsevier: Amsterdam, The Netherlands, 2022; pp. 134–144. [Google Scholar]
- Zhao, P.; Li, M.; Wang, Y.; Chen, Y.; He, C.; Zhang, X.; Yang, T.; Lu, Y.; You, J.; Lee, R.J. Enhancing anti-tumor efficiency in hepatocellular carcinoma through the autophagy inhibition by miR-375/sorafenib in lipid-coated calcium carbonate nanoparticles. Acta Biomater. 2018, 72, 248–255. [Google Scholar] [CrossRef]
- Ding, Q.J.; Remy, M.T.; Upara, C.; Hu, J.; Mora Mata, A.; Haes, A.J.; Lanzel, E.; Sun, H.; Buchakjian, M.R.; Hong, L. CaCO3 Nanoparticles Delivering MicroRNA-200c Suppress Oral Squamous Cell Carcinoma. J. Dent. Res. 2024, 103, 147–155. [Google Scholar] [CrossRef]
- Yoo, S.S.; Razzak, R.; Bédard, E.; Guo, L.; Shaw, A.R.; Moore, R.B.; Roa, W.H. Layered gadolinium-based nanoparticle as a novel delivery platform for microRNA therapeutics. Nanotechnology 2014, 25, 425102. [Google Scholar] [CrossRef] [PubMed]
- Yin, P.T.; Shah, B.P.; Lee, K.-B. Combined magnetic nanoparticle-based microRNA and hyperthermia therapy to enhance apoptosis in brain cancer cells. Small (Weinh. Der Bergstr. Ger.) 2014, 10, 4106. [Google Scholar] [CrossRef] [PubMed]
- Kara, G.; Ozpolat, B. SPIONs: Superparamagnetic iron oxide-based nanoparticles for the delivery of microRNAi-therapeutics in cancer. Biomed. Microdevices 2024, 26, 16. [Google Scholar] [CrossRef]
- Reimondez-Troitiño, S.; González-Aramundiz, J.V.; Ruiz-Bañobre, J.; López-López, R.; Alonso, M.J.; Csaba, N.; de la Fuente, M. Versatile protamine nanocapsules to restore miR-145 levels and interfere tumor growth in colorectal cancer cells. Eur. J. Pharm. Biopharm. 2019, 142, 449–459. [Google Scholar] [CrossRef]
- Mohamed, A.; Kunda, N.K.; Ross, K.; Hutcheon, G.A.; Saleem, I.Y. Polymeric nanoparticles for the delivery of miRNA to treat Chronic Obstructive Pulmonary Disease (COPD). Eur. J. Pharm. Biopharm. 2019, 136, 1–8. [Google Scholar] [CrossRef]
- Zheng, B.; Chen, L.; Pan, C.-C.; Wang, J.-Z.; Lu, G.-R.; Yang, S.-X.; Xue, Z.-X.; Wang, F.-Y.; Xu, C.-L. Targeted delivery of miRNA-204-5p by PEGylated polymer nanoparticles for colon cancer therapy. Nanomedicine 2018, 13, 769–785. [Google Scholar] [CrossRef]
- Dhuri, K.; Vyas, R.N.; Blumenfeld, L.; Verma, R.; Bahal, R. Nanoparticle delivered anti-miR-141-3p for stroke therapy. Cells 2021, 10, 1011. [Google Scholar] [CrossRef]
- Lopez-Bertoni, H.; Kozielski, K.L.; Rui, Y.; Lal, B.; Vaughan, H.; Wilson, D.R.; Mihelson, N.; Eberhart, C.G.; Laterra, J.; Green, J.J. Bioreducible polymeric nanoparticles containing multiplexed cancer stem cell regulating miRNAs inhibit glioblastoma growth and prolong survival. Nano Lett. 2018, 18, 4086–4094. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, M.-A.; Wyatt, H.; Susser, L.; Geoffrion, M.; Rasheed, A.; Duchez, A.-C.; Cottee, M.L.; Afolayan, E.; Farah, E.; Kahiel, Z. Delivery of microRNAs by chitosan nanoparticles to functionally alter macrophage cholesterol efflux in vitro and in vivo. ACS Nano 2019, 13, 6491–6505. [Google Scholar] [CrossRef]
- Plaza-Oliver, M.; Santander-Ortega, M.J.; Lozano, M.V. Current approaches in lipid-based nanocarriers for oral drug delivery. Drug Deliv. Transl. Res. 2021, 11, 471–497. [Google Scholar] [CrossRef]
- Campani, V.; De Rosa, G.; Misso, G.; Zarone, M.R.; Grimaldi, A. Lipid nanoparticles to deliver miRNA in cancer. Curr. Pharm. Biotechnol. 2016, 17, 741–749. [Google Scholar] [CrossRef]
- Lujan, H.; Griffin, W.C.; Taube, J.H.; Sayes, C.M. Synthesis and characterization of nanometer-sized liposomes for encapsulation and microRNA transfer to breast cancer cells. Int. J. Nanomed. 2019, 14, 5159–5173. [Google Scholar] [CrossRef]
- Hsu, S.-h.; Yu, B.; Wang, X.; Lu, Y.; Schmidt, C.R.; Lee, R.J.; Lee, L.J.; Jacob, S.T.; Ghoshal, K. Cationic lipid nanoparticles for therapeutic delivery of siRNA and miRNA to murine liver tumor. Nanomed. Nanotechnol. Biol. Med. 2013, 9, 1169–1180. [Google Scholar] [CrossRef]
- Taghavi, S.; Abnous, K.; Taghdisi, S.M.; Ramezani, M.; Alibolandi, M. Hybrid carbon-based materials for gene delivery in cancer therapy. J. Control. Release 2020, 318, 158–175. [Google Scholar] [CrossRef]
- Mohajeri, M.; Behnam, B.; Sahebkar, A. Biomedical applications of carbon nanomaterials: Drug and gene delivery potentials. J. Cell. Physiol. 2019, 234, 298–319. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.-R.; Xie, Y.; Li, J.-W.; Liu, R.; Chen, M.; Ren, Y.-X.; Luo, Q.; Duan, J.-L.; Bao, C.-J.; Liu, Y.-X. Development of fullerene nanospherical miRNA and application in overcoming resistant breast cancer. Mater. Today Chem. 2022, 26, 101019. [Google Scholar] [CrossRef]
- Ju, E.; Li, T.; Liu, Z.; da Silva, S.R.; Wei, S.; Zhang, X.; Wang, X.; Gao, S.-J. Specific inhibition of viral MicroRNAs by carbon dots-mediated delivery of locked nucleic acids for therapy of virus-induced cancer. ACS Nano 2020, 14, 476–487. [Google Scholar] [CrossRef] [PubMed]
- Gao, G.; Guo, Q.; Zhi, J. Nanodiamond-based theranostic platform for drug delivery and bioimaging. Small 2019, 15, 1902238. [Google Scholar] [CrossRef] [PubMed]
- Perevedentseva, E.; Lin, Y.-C.; Jani, M.; Cheng, C.-L. Biomedical applications of nanodiamonds in imaging and therapy. Nanomedicine 2013, 8, 2041–2060. [Google Scholar] [CrossRef]
- Abate, M.; Lombardi, A.; Luce, A.; Porru, M.; Leonetti, C.; Bocchetti, M.; Campani, V.; De Rosa, G.; Graziano, S.F.; Nele, V. Fluorescent nanodiamonds as innovative delivery systems for MiR-34a replacement in breast cancer. Mol. Ther.-Nucleic Acids 2023, 33, 127–141. [Google Scholar] [CrossRef]
- Masotti, A.; Miller, M.R.; Celluzzi, A.; Rose, L.; Micciulla, F.; Hadoke, P.W.; Bellucci, S.; Caporali, A. Regulation of angiogenesis through the efficient delivery of microRNAs into endothelial cells using polyamine-coated carbon nanotubes. Nanomed. Nanotechnol. Biol. Med. 2016, 12, 1511–1522. [Google Scholar] [CrossRef] [PubMed]
- Nekoueian, K.; Amiri, M.; Sillanpää, M.; Marken, F.; Boukherroub, R.; Szunerits, S. Carbon-based quantum particles: An electroanalytical and biomedical perspective. Chem. Soc. Rev. 2019, 48, 4281–4316. [Google Scholar] [CrossRef] [PubMed]
- Dong, H.; Dai, W.; Ju, H.; Lu, H.; Wang, S.; Xu, L.; Zhou, S.-F.; Zhang, Y.; Zhang, X. Multifunctional poly (l-lactide)–polyethylene glycol-grafted graphene quantum dots for intracellular microRNA imaging and combined specific-gene-targeting agents delivery for improved therapeutics. ACS Appl. Mater. Interfaces 2015, 7, 11015–11023. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Ding, N.; Huo, D.; Yang, G.; Wei, K.; Guan, G.; Li, Y.; Yang, J.; Wang, T.; Wang, Y. Surface-Engineered Monocyte Inhibits Atherosclerotic Plaque Destabilization via Graphene Quantum Dot-Mediated MicroRNA Delivery. Adv. Healthc. Mater. 2019, 8, 1900386. [Google Scholar] [CrossRef] [PubMed]
- Rupaimoole, R.; Slack, F.J. MicroRNA therapeutics: Towards a new era for the management of cancer and other diseases. Nat. Rev. Drug Discov. 2017, 16, 203–222. [Google Scholar] [CrossRef] [PubMed]
- Segal, M.; Slack, F.J. Challenges identifying efficacious miRNA therapeutics for cancer. Expert Opin. Drug Discov. 2020, 15, 987–991. [Google Scholar] [CrossRef] [PubMed]
- Fu, Z.; Wang, L.; Li, S.; Chen, F.; Au-Yeung, K.K.-W.; Shi, C. MicroRNA as an important target for anticancer drug development. Front. Pharmacol. 2021, 12, 736323. [Google Scholar] [CrossRef] [PubMed]
- Esquela-Kerscher, A.; Trang, P.; Wiggins, J.F.; Patrawala, L.; Cheng, A.; Ford, L.; Weidhaas, J.B.; Brown, D.; Bader, A.G.; Slack, F.J. The let-7 microRNA reduces tumor growth in mouse models of lung cancer. Cell Cycle 2008, 7, 759–764. [Google Scholar] [CrossRef] [PubMed]
- Lucas, T.; Schäfer, F.; Müller, P.; Eming, S.A.; Heckel, A.; Dimmeler, S. Light-inducible antimiR-92a as a therapeutic strategy to promote skin repair in healing-impaired diabetic mice. Nat. Commun. 2017, 8, 15162. [Google Scholar] [CrossRef]
- Fu, Y.; Chen, J.; Huang, Z. Recent progress in microRNA-based delivery systems for the treatment of human disease. ExRNA 2019, 1, 1–14. [Google Scholar] [CrossRef]
- Zhang, L.; Hou, D.; Chen, X.; Li, D.; Zhu, L.; Zhang, Y.; Li, J.; Bian, Z.; Liang, X.; Cai, X. Exogenous plant MIR168a specifically targets mammalian LDLRAP1: Evidence of cross-kingdom regulation by microRNA. Cell Res. 2012, 22, 107–126. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Li, X.; Liu, J.; Dong, L.; Chen, Q.; Liu, J.; Kong, H.; Zhang, Q.; Qi, X.; Hou, D.; et al. Honeysuckle-encoded atypical microRNA2911 directly targets influenza A viruses. Cell Res. 2015, 25, 39–49. [Google Scholar] [CrossRef] [PubMed]
- Xu, T.; Zhu, Y.; Lin, Z.; Lei, J.; Li, L.; Zhu, W.; Wu, D. Evidence of Cross-Kingdom Gene Regulation by Plant MicroRNAs and Possible Reasons for Inconsistencies. J. Agric. Food Chem. 2024, 72, 4564–4573. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Lei, Z.X.; Cai, Y.L.; Cheng, D.B.; Sun, T.L. MicroRNA therapeutics and nucleic acid nano-delivery systems in bacterial infection: A review. J. Mater. Chem. B 2023, 11, 7804–7833. [Google Scholar] [CrossRef] [PubMed]
- Feliu, N.; Docter, D.; Heine, M.; Del Pino, P.; Ashraf, S.; Kolosnjaj-Tabi, J.; Macchiarini, P.; Nielsen, P.; Alloyeau, D.; Gazeau, F. In vivo degeneration and the fate of inorganic nanoparticles. Chem. Soc. Rev. 2016, 45, 2440–2457. [Google Scholar] [CrossRef] [PubMed]
- Scala-Benuzzi, M.L.; Piguillem Palacios, S.V.; Takara, E.A.; Fernández-Baldo, M.A. Biomaterials and biopolymers for the development of biosensors. In Biomaterials-Based Sensors: Recent Advances and Applications; Springer: Berlin/Heidelberg, Germany, 2023; pp. 3–24. [Google Scholar]
- Wang, H. Beneficial medicinal effects and material applications of rose. Heliyon 2024, 10, e23530. [Google Scholar] [CrossRef] [PubMed]
- Wang, H. Advantages of animal leather over alternatives and its medical applications. Eur. Polym. J. 2024, 214, 113153. [Google Scholar] [CrossRef]
- Jiang, Z.; Song, Z.; Cao, C.; Yan, M.; Liu, Z.; Cheng, X.; Wang, H.; Wang, Q.; Liu, H.; Chen, S. Multiple natural polymers in drug and gene delivery systems. Curr. Med. Chem. 2024, 31, 1691–1715. [Google Scholar] [CrossRef]
- Kučuk, N.; Primožič, M.; Knez, Ž.; Leitgeb, M. Sustainable biodegradable biopolymer-based nanoparticles for healthcare applications. Int. J. Mol. Sci. 2023, 24, 3188. [Google Scholar] [CrossRef]
- Nazari-Vanani, R.; Azarpira, N.; Heli, H. Development of self-nanoemulsifying drug delivery systems for oil extracts of Citrus aurantium L. blossoms and Rose damascena and evaluation of anticancer properties. J. Drug Deliv. Sci. Technol. 2018, 47, 330–336. [Google Scholar] [CrossRef]

| Nanotechnology | Advantage or Disadvantage |
|---|---|
| Inorganic Nanoparticles |
|
| Carbon-Based Nanoparticles |
|
| Nanopore Technology |
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, H. A Review of Nanotechnology in microRNA Detection and Drug Delivery. Cells 2024, 13, 1277. https://doi.org/10.3390/cells13151277
Wang H. A Review of Nanotechnology in microRNA Detection and Drug Delivery. Cells. 2024; 13(15):1277. https://doi.org/10.3390/cells13151277
Chicago/Turabian StyleWang, Hsiuying. 2024. "A Review of Nanotechnology in microRNA Detection and Drug Delivery" Cells 13, no. 15: 1277. https://doi.org/10.3390/cells13151277
APA StyleWang, H. (2024). A Review of Nanotechnology in microRNA Detection and Drug Delivery. Cells, 13(15), 1277. https://doi.org/10.3390/cells13151277







