Comparing Viral Vectors and Fate Mapping Approaches for Astrocyte-to-Neuron Reprogramming in the Injured Mouse Cerebral Cortex
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animals
2.2. Labelling Endogenous Neurons with EdU
2.3. Viral Vector Preparation
2.4. Cortical Stab Wound and Viral Injections
2.5. Immunohistochemistry
2.6. Imaging, Data Analysis, and Quantification
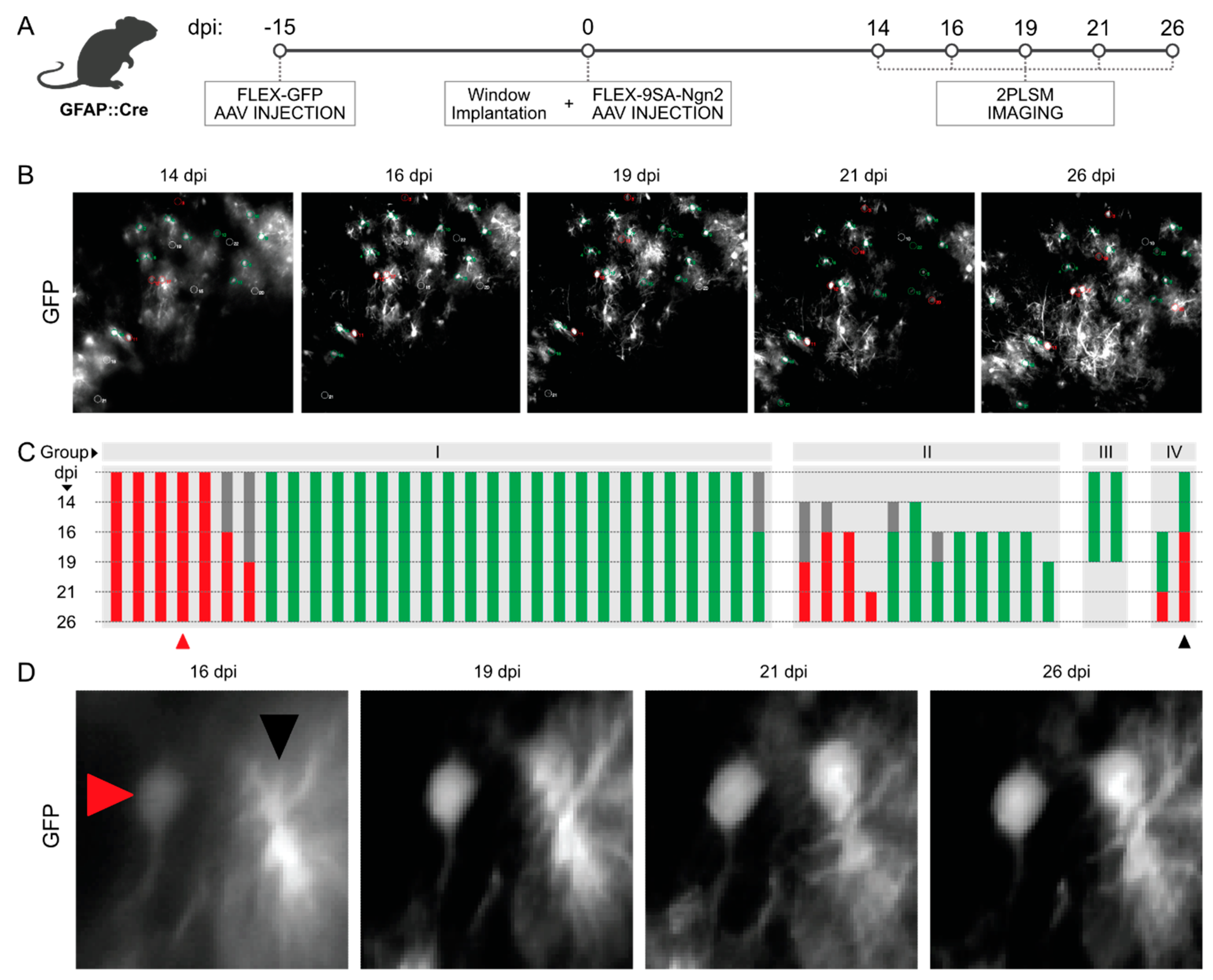
2.7. Chronic Live Imaging via 2-Photon Laser Scanning Microscopy (2PLSM)
3. Results
3.1. Fate Mapping of Cortical Astrocytes after AAV-FLEX-9SA-Ngn2 Administration
3.2. Fate Mapping of Endogenous Neurons after AAV-FLEX- 9SA-Ngn2 Administration
3.3. Longitudinal In Vivo Imaging of FLEX-AAV-Labelled Cells
3.4. Control of FLEX-AAV
3.5. Intracerebral Reprogramming with 9SA-Ngn2 Using Mo-MLVs
4. Discussion
4.1. Reprogramming via Mo-MLVs Is Reliable and 9SA-Ngn2 Is More Potent Than Its Wildtype Counterpart
4.2. Expression of Phospho-Resistant Ngn2 by AAV Labels Endogenous Neurons and Does Not Result in Direct Reprogramming
4.3. Technical Consideration of the Fate Mapping Controls
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Heins, N.; Malatesta, P.; Cecconi, F.; Nakafuku, M.; Tucker, K.L.; Hack, M.A.; Chapouton, P.; Barde, Y.-A.; Götz, M. Glial cells generate neurons: The role of the transcription factor Pax6. Nat. Neurosci. 2002, 5, 308–315. [Google Scholar] [CrossRef] [PubMed]
- Buffo, A.; Vosko, M.R.; Ertürk, D.; Hamann, G.F.; Jucker, M.; Rowitch, D.; Götz, M. Expression pattern of the transcription factor Olig2 in response to brain injuries: Implications for neuronal repair. Proc. Natl. Acad. Sci. USA 2005, 102, 18183–18188. [Google Scholar] [CrossRef] [PubMed]
- Roe, T.; Reynolds, T.; Yu, G.; Brown, P. Integration of murine leukemia virus DNA depends on mitosis. EMBO J. 1993, 12, 2099–2108. [Google Scholar] [CrossRef]
- Suzuki, Y.; Craigie, R. The road to chromatin—Nuclear entry of retroviruses. Nat. Rev. Microbiol. 2007, 5, 187–196. [Google Scholar] [CrossRef]
- Heinrich, C.; Bergami, M.; Gascón, S.; Lepier, A.; Viganò, F.; Dimou, L.; Sutor, B.; Berninger, B.; Götz, M. Sox2-Mediated Conversion of NG2 Glia into Induced Neurons in the Injured Adult Cerebral Cortex. Stem Cell Rep. 2014, 3, 1000–1014. [Google Scholar] [CrossRef] [PubMed]
- Gascón, S.; Murenu, E.; Masserdotti, G.; Ortega, F.; Russo, G.L.; Petrik, D.; Deshpande, A.; Heinrich, C.; Karow, M.; Robertson, S.P.; et al. Identification and Successful Negotiation of a Metabolic Checkpoint in Direct Neuronal Reprogramming. Cell Stem Cell 2015, 18, 396–409. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Lei, Z.; Guo, Z.; Pei, Z.; Chen, Y.; Zhang, F.; Cai, A.; Mok, G.; Lee, G.; Swaminathan, V.; et al. Development of Neuroregenerative Gene Therapy to Reverse Glial Scar Tissue Back to Neuron-Enriched Tissue. Front. Cell. Neurosci. 2020, 14. [Google Scholar] [CrossRef]
- Mattugini, N.; Bocchi, R.; Scheuss, V.; Russo, G.L.; Torper, O.; Lao, C.L.; Götz, M. Inducing Different Neuronal Subtypes from Astrocytes in the Injured Mouse Cerebral Cortex. Neuron 2019, 103, 1086–1095.e5. [Google Scholar] [CrossRef]
- Choudhury, S.R.; Hudry, E.; Maguire, C.A.; Sena-Esteves, M.; Breakefield, X.O.; Grandi, P. Viral vectors for therapy of neurologic diseases. Neuropharmacology 2017, 120, 63–80. [Google Scholar] [CrossRef]
- Merienne, N.; Delzor, A.; Viret, A.; Dufour, N.; Rey, M.; Hantraye, P.; Déglon, N. Gene transfer engineering for astrocyte-specific silencing in the CNS. Gene Ther. 2015, 22, 830–839. [Google Scholar] [CrossRef]
- Lee, Y.; Messing, A.; Su, M.; Brenner, M. GFAP promoter elements required for region-specific and astrocyte-specific expression. Glia 2008, 56, 481–493. [Google Scholar] [CrossRef]
- Wang, L.-L.; Serrano, C.; Zhong, X.; Ma, S.; Zou, Y.; Zhang, C.-L. Revisiting astrocyte to neuron conversion with lineage tracing in vivo. Cell 2021, 184, 5465–5481.e16. [Google Scholar] [CrossRef]
- Su, M.; Hu, H.; Lee, Y.; D’azzo, A.; Messing, A.; Brenner, M. Expression Specificity of GFAP Transgenes. Neurochem. Res. 2004, 29, 2075–2093. [Google Scholar] [CrossRef] [PubMed]
- Leib, D.; Chen, Y.H.; Monteys, A.M.; Davidson, B.L. Limited astrocyte-to-neuron conversion in the mouse brain using NeuroD1 overexpression. Mol. Ther. 2022, 30, 982–986. [Google Scholar] [CrossRef] [PubMed]
- Yang, G.; Yan, Z.; Wu, X.; Zhang, M.; Xu, C.; Shi, L.; Yang, H.; Fang, K. Ptbp1 knockdown failed to induce astrocytes to neurons in vivo. Gene Ther. 2023, 30, 801–806. [Google Scholar] [CrossRef] [PubMed]
- Hoang, T.; Kim, D.W.; Appel, H.; Ozawa, M.; Zheng, S.; Kim, J.; Blackshaw, S. Ptbp1 deletion does not induce astrocyte-to-neuron conversion. Nature 2023, 618, E1–E7. [Google Scholar] [CrossRef]
- Bocchi, R.; Masserdotti, G.; Götz, M. Direct neuronal reprogramming: Fast forward from new concepts toward therapeutic approaches. Neuron 2021, 110, 366–393. [Google Scholar] [CrossRef]
- Hand, R.; Bortone, D.; Mattar, P.; Nguyen, L.; Heng, J.I.-T.; Guerrier, S.; Boutt, E.; Peters, E.; Barnes, A.P.; Parras, C.; et al. Phosphorylation of Neurogenin2 Specifies the Migration Properties and the Dendritic Morphology of Pyramidal Neurons in the Neocortex. Neuron 2005, 48, 45–62. [Google Scholar] [CrossRef]
- Ali, F.; Hindley, C.; McDowell, G.; Deibler, R.; Jones, A.; Kirschner, M.; Guillemot, F.; Philpott, A. Cell cycle-regulated multi-site phosphorylation of Neurogenin 2 coordinates cell cycling with differentiation during neurogenesis. Development 2011, 138, 4267–4277. [Google Scholar] [CrossRef]
- Hindley, C.; Ali, F.; McDowell, G.; Cheng, K.; Jones, A.; Guillemot, F.; Philpott, A. Post-translational modification of Ngn2 differentially affects transcription of distinct targets to regulate the balance between progenitor maintenance and differentiation. Development 2012, 139, 1718–1723. [Google Scholar] [CrossRef]
- Ma, Y.-C.; Song, M.-R.; Park, J.P.; Ho, H.-Y.H.; Hu, L.; Kurtev, M.V.; Zieg, J.; Ma, Q.; Pfaff, S.L.; Greenberg, M.E. Regulation of Motor Neuron Specification by Phosphorylation of Neurogenin 2. Neuron 2008, 58, 65–77. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Mattar, P.; Zinyk, D.; Singh, K.; Chaturvedi, C.-P.; Kovach, C.; Dixit, R.; Kurrasch, D.M.; Ma, Y.-C.; Chan, J.A.; et al. GSK3 Temporally Regulates Neurogenin 2 Proneural Activity in the Neocortex. J. Neurosci. 2012, 32, 7791–7805. [Google Scholar] [CrossRef] [PubMed]
- Pereira, A.; Diwakar, J.; Masserdotti, G.; Beşkardeş, S.; Simon, T.; So, Y.; Martín-Loarte, L.; Bergemann, F.; Vasan, L.; Schauer, T.; et al. Direct neuronal reprogramming of mouse astrocytes is associated with multiscale epigenome remodeling and requires Yy1. Nat. Neurosci. 2024, 27, 1260–1273. [Google Scholar] [CrossRef]
- Gregorian, C.; Nakashima, J.; Le Belle, J.; Ohab, J.; Kim, R.; Liu, A.; Smith, K.B.; Groszer, M.; Garcia, A.D.; Sofroniew, M.V.; et al. Pten Deletion in Adult Neural Stem/Progenitor Cells Enhances Constitutive Neurogenesis. J. Neurosci. 2009, 29, 1874–1886. [Google Scholar] [CrossRef]
- Nakamura, T.; Colbert, M.C.; Robbins, J. Neural Crest Cells Retain Multipotential Characteristics in the Developing Valves and Label the Cardiac Conduction System. Circ. Res. 2006, 98, 1547–1554. [Google Scholar] [CrossRef]
- Saunders, A.; Johnson, C.A.; Sabatini, B.L. Novel recombinant adeno-associated viruses for Cre activated and inactivated transgene expression in neurons. Front. Neural Circuits 2012, 6, 31179. [Google Scholar] [CrossRef] [PubMed]
- Hol, E.M.; Pekny, M. Glial fibrillary acidic protein (GFAP) and the astrocyte intermediate filament system in diseases of the central nervous system. Curr. Opin. Cell Biol. 2015, 32, 121–130. [Google Scholar] [CrossRef] [PubMed]
- Su, E.; Akdemir, A.; Yu-Szu, H.; Deneen, B. Astrocytogenesis: Where, when, and how [version 1; peer review: 2 approved]. F1000Research 2020, 9. [Google Scholar] [CrossRef]
- Magrinelli, E.; Baumann, N.; Wagener, R.J.; Glangetas, C.; Bellone, C.; Jabaudon, D.; Klingler, E. Heterogeneous fates of simultaneously-born neurons in the cortical ventricular zone. Sci. Rep. 2022, 12, 6022. [Google Scholar] [CrossRef]
- Miller, M.W.; Nowakowski, R. Use of bromodeoxyuridine-immunohistochemistry to examine the proliferation, migration and time of origin of cells in the central nervous system. Brain Res. 1988, 457, 44–52. [Google Scholar] [CrossRef]
- Govindan, S.; Oberst, P.; Jabaudon, D. In vivo pulse labeling of isochronic cohorts of cells in the central nervous system using FlashTag. Nat. Protoc. 2018, 13, 2297–2311. [Google Scholar] [CrossRef] [PubMed]
- Canhos, L.L.; Chen, M.; Falk, S.; Popper, B.; Straub, T.; Götz, M.; Sirko, S. Repetitive injury and absence of monocytes promote astrocyte self-renewal and neurological recovery. Glia 2020, 69, 165–181. [Google Scholar] [CrossRef] [PubMed]
- Bardehle, S.; Krüger, M.; Buggenthin, F.; Schwausch, J.; Ninkovic, J.; Clevers, H.; Snippert, H.J.; Theis, F.J.; Meyer-Luehmann, M.; Bechmann, I.; et al. Live imaging of astrocyte responses to acute injury reveals selective juxtavascular proliferation. Nat. Neurosci. 2013, 16, 580–586. [Google Scholar] [CrossRef] [PubMed]
- Fischer, K.B.; Collins, H.K.; Callaway, E.M. Sources of off-target expression from recombinase-dependent AAV vectors and mitigation with cross-over insensitive ATG-out vectors. Proc. Natl. Acad. Sci. USA 2019, 116, 52. [Google Scholar] [CrossRef]
- Lentini, C.; D’orange, M.; Marichal, N.; Trottmann, M.-M.; Vignoles, R.; Foucault, L.; Verrier, C.; Massera, C.; Raineteau, O.; Conzelmann, K.-K.; et al. Reprogramming reactive glia into interneurons reduces chronic seizure activity in a mouse model of mesial temporal lobe epilepsy. Cell Stem Cell 2021, 28, 2104–2121.e10. [Google Scholar] [CrossRef] [PubMed]
- Sonsalla, G.; Malpartida, A.B.; Riedemann, T.; Gusic, M.; Rusha, E.; Bulli, G.; Najas, S.; Janjic, A.; Hersbach, B.A.; Smialowski, P.; et al. Direct neuronal reprogramming of NDUFS4 patient cells identifies the unfolded protein response as a novel general reprogramming hurdle. Neuron 2024, 112, 1117–1132.e9. [Google Scholar] [CrossRef]
- Xiang, Z.; He, S.; Chen, R.; Liu, S.; Liu, M.; Xu, L.; Zheng, J.; Jiang, Z.; Ma, L.; Sun, Y.; et al. Two-photon live imaging of direct glia-to-neuron conversion in the mouse cortex. Neural Regen. Res. 2023, 19, 1781–1788. [Google Scholar] [CrossRef]
- Zwirner, J.; Lier, J.; Franke, H.; Hammer, N.; Matschke, J.; Trautz, F.; Tse, R.; Ondruschka, B. GFAP positivity in neurons following traumatic brain injuries. Int. J. Leg. Med. 2021, 135, 2323–2333. [Google Scholar] [CrossRef]
- Middeldorp, J.; Berge, S.A.v.D.; Aronica, E.; Speijer, D.; Hol, E.M. Specific Human Astrocyte Subtype Revealed by Affinity Purified GFAP+1 Antibody; Unpurified Serum Cross-Reacts with Neurofilament-L in Alzheimer. PLoS ONE 2009, 4, e7663. [Google Scholar] [CrossRef]
- Hol, E.M.; Roelofs, R.F.; Moraal, E.; Sonnemans, M.A.; Sluijs, J.A.; Proper, E.A.; De Graan, P.N.; Fischer, D.F.; Van Leeuwen, F.W. Neuronal expression of GFAP in patients with Alzheimer pathology and identification of novel GFAP splice forms. Mol. Psychiatry 2003, 8, 786–796. [Google Scholar] [CrossRef]
- Gleichman, A.J.; Kawaguchi, R.; Sofroniew, M.V.; Carmichael, S.T. A toolbox of astrocyte-specific, serotype-independent adeno-associated viral vectors using microRNA targeting sequences. Nat. Commun. 2023, 14, 7426. [Google Scholar] [CrossRef] [PubMed]
- Kawabata, H.; Konno, A.; Matsuzaki, Y.; Sato, Y.; Kawachi, M.; Aoki, R.; Tsutsumi, S.; Togai, S.; Kobayashi, R.; Horii, T.; et al. Improving cell-specific recombination using AAV vectors in the murine CNS by capsid and expression cassette optimization. Mol. Ther.-Methods Clin. Dev. 2024, 32, 101185. [Google Scholar] [CrossRef]
- Taschenberger, G.; Tereshchenko, J.; Kügler, S. A MicroRNA124 Target Sequence Restores Astrocyte Specificity of gfaABC1D-Driven Transgene Expression in AAV-Mediated Gene Transfer. Mol. Ther.-Nucleic Acids 2017, 8, 13–25. [Google Scholar] [CrossRef]
- Botterill, J.J.; Khlaifia, A.; Walters, B.J.; Brimble, M.A.; Scharfman, H.E.; Arruda-Carvalho, M. Off-target expression of cre-dependent adeno-associated viruses in wild-type C57BL/6J mice. Eneuro 2021, 8, 6. [Google Scholar] [CrossRef]
- Brenner, M.; Messing, A. Regulation of GFAP Expression. ASN Neuro 2021, 13. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.; Sheng, S.; Wang, Y.; Ding, L.; Xu, X.; Xia, X.; Zheng, J.C. Astrocyte-derived extracellular vesicles: A double-edged sword in central nervous system disorders. Neurosci. Biobehav. Rev. 2021, 125, 148–159. [Google Scholar] [CrossRef]
- Zhou, C.; Huang, M.; Wang, S.; Chu, S.; Zhang, Z.; Chen, N. Tunneling nanotubes: The transport highway for astrocyte-neuron communication in the central nervous system. Brain Res. Bull. 2024, 209, 110921. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Cao, J. Astrocyte-to-neuron transportation of enhanced green fluorescent protein in cerebral cortex requires F-actin dependent tunneling nanotubes. Sci. Rep. 2021, 11, 16798. [Google Scholar] [CrossRef]
- Ridder, K.; Keller, S.; Dams, M.; Rupp, A.-K.; Schlaudraff, J.; Del Turco, D.; Starmann, J.; Macas, J.; Karpova, D.; Devraj, K.; et al. Extracellular Vesicle-Mediated Transfer of Genetic Information between the Hematopoietic System and the Brain in Response to Inflammation. PLoS Biol. 2014, 12, e1001874. [Google Scholar] [CrossRef]
- Hu, N.-Y.; Chen, Y.-T.; Wang, Q.; Jie, W.; Liu, Y.-S.; You, Q.-L.; Li, Z.-L.; Li, X.-W.; Reibel, S.; Pfrieger, F.W.; et al. Expression Patterns of Inducible Cre Recombinase Driven by Differential Astrocyte-Specific Promoters in Transgenic Mouse Lines. Neurosci. Bull. 2019, 36, 530–544. [Google Scholar] [CrossRef]
- Halley-Stott, R.P.; Jullien, J.; Pasque, V.; Gurdon, J. Mitosis Gives a Brief Window of Opportunity for a Change in Gene Transcription. PLoS Biol. 2014, 12, e1001914. [Google Scholar] [CrossRef] [PubMed]
- Sirko, S.; Behrendt, G.; Johansson, P.A.; Tripathi, P.; Costa, M.R.; Bek, S.; Heinrich, C.; Tiedt, S.; Colak, D.; Dichgans, M.; et al. Reactive Glia in the Injured Brain Acquire Stem Cell Properties in Response to Sonic Hedgehog. Cell Stem Cell 2013, 12, 426–439. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Li, W.; Wang, T.; Liao, J.-C.; Wang, X.; Wang, Q.-S.; Wan, K.-Y.; Yang, Y.-Y.; He, Q.; Zhang, J.-X. Unexpected BrdU inhibition on astrocyte-to-neuron conversion. Neural Regen. Res. 2022, 17, 1526–1534. [Google Scholar] [CrossRef]
- Ghazale, H.; Park, E.; Vasan, L.; Mester, J.; Saleh, F.; Trevisiol, A.; Zinyk, D.; Chinchalongporn, V.; Liu, M.; Fleming, T.; et al. Ascl1 phospho-site mutations enhance neuronal conversion of adult cortical astrocytes in vivo. Front. Neurosci. 2022, 16, 917071. [Google Scholar] [CrossRef] [PubMed]
- Galante, C.; Marichal, N.; Schuurmans, C.; Berninger, B.; Péron, S. Low-efficiency conversion of proliferative glia into induced neurons by Ascl1 in the postnatal mouse cerebral cortex in vivo. bioRxiv 2022. [Google Scholar] [CrossRef]
- Marichal, N.; Peron, S.; Beltran Arranz, A.; Galante, C.; Scarante, F.F.; Wiffen, R.; Schuurmans, C.; Karow, M.; Gascon, S.; Berninger, B. Reprogramming early cortical astroglia into neurons with hallmarks of fast-spiking parvalbumin-positive interneurons by phospho-site deficient Ascl1. bioRxiv 2023. [Google Scholar] [CrossRef]



| Virus Type | Mix | Virus Name | Virus Titre | Final Volume |
|---|---|---|---|---|
| AAVs | Control | CAG:FLEX-mScarlet or GFP | 3.12 × 1010 gc/mouse | 500 nL |
| Reprogramming | CAG:FLEX-mScarlet or GFP | 3.12 × 1010 gc/mouse | 500 nL | |
| CAG:FLEX-9SA-Ngn2 | 4.90 × 1011 gc/mouse | |||
| Mo-MLVs | Reprogramming | CAG-9SA-Ngn2-IRES-mScarlet | 5.8 × 106 TU/mouse | 500 nL |
| Antibody/DAPI | Species/Isotype | Source | Identifier | Dilution |
|---|---|---|---|---|
| Anti-GFP | Chicken | AvesLab | GFP-1020 | 1:1000 |
| Anti-RFP | Rabbit | VWR | ROCK600-401-379S | 1:1000 |
| Anti-NeuN | Mouse-IgG1 | Merck Millipore | MAB377 | 1:250 |
| Anti-Sox9 | Rabbit | Merck Millipore | ab5535 | 1:1500 |
| Anti-GFAP | Rabbit | Dako | Z0334 | 1:500 |
| Anti-chicken-488 | Donkey | Dianova | 703-545-155 | 1:1000 |
| Anti-rabbit-598 | Donkey | Thermo Fisher | A-21207 | 1:1000 |
| Anti-mouse IgG1-647 | Goat | Thermo Fisher | A-21240 | 1:1000 |
| DAPI | - | Sigma | 28718-90-3 | 1:1000 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Puglisi, M.; Lao, C.L.; Wani, G.; Masserdotti, G.; Bocchi, R.; Götz, M. Comparing Viral Vectors and Fate Mapping Approaches for Astrocyte-to-Neuron Reprogramming in the Injured Mouse Cerebral Cortex. Cells 2024, 13, 1408. https://doi.org/10.3390/cells13171408
Puglisi M, Lao CL, Wani G, Masserdotti G, Bocchi R, Götz M. Comparing Viral Vectors and Fate Mapping Approaches for Astrocyte-to-Neuron Reprogramming in the Injured Mouse Cerebral Cortex. Cells. 2024; 13(17):1408. https://doi.org/10.3390/cells13171408
Chicago/Turabian StylePuglisi, Matteo, Chu Lan Lao, Gulzar Wani, Giacomo Masserdotti, Riccardo Bocchi, and Magdalena Götz. 2024. "Comparing Viral Vectors and Fate Mapping Approaches for Astrocyte-to-Neuron Reprogramming in the Injured Mouse Cerebral Cortex" Cells 13, no. 17: 1408. https://doi.org/10.3390/cells13171408
APA StylePuglisi, M., Lao, C. L., Wani, G., Masserdotti, G., Bocchi, R., & Götz, M. (2024). Comparing Viral Vectors and Fate Mapping Approaches for Astrocyte-to-Neuron Reprogramming in the Injured Mouse Cerebral Cortex. Cells, 13(17), 1408. https://doi.org/10.3390/cells13171408






