A Humanized Yeast Model for Studying TRAPP Complex Mutations; Proof-of-Concept Using Variants from an Individual with a TRAPPC1-Associated Neurodevelopmental Syndrome
Abstract
:1. Introduction
2. Materials and Methods
2.1. Preparation of sgRNAs That Target the Yeast TRAPP Genes
2.2. Preparation of Repair Template DNA
2.3. Yeast Transformations
2.4. Primer Design for Confirmation via Colony PCR
2.5. Colony Screening via PCR
2.6. Yeast Liquid Growth Assays
2.7. Autophagy Assays in Yeast
2.8. Preparation of Yeast Cell Lysates
2.9. Invertase Secretion Assay
2.10. Western Blot Analysis
2.11. Genetic Studies
2.12. Cell Culture
2.13. Autophagy Assays in Mammalian Cells
2.14. Antibodies and Dyes
2.15. Immunofluorescence Microscopy
2.16. Retention Using Selective Hooks (RUSH) Assay
3. Results
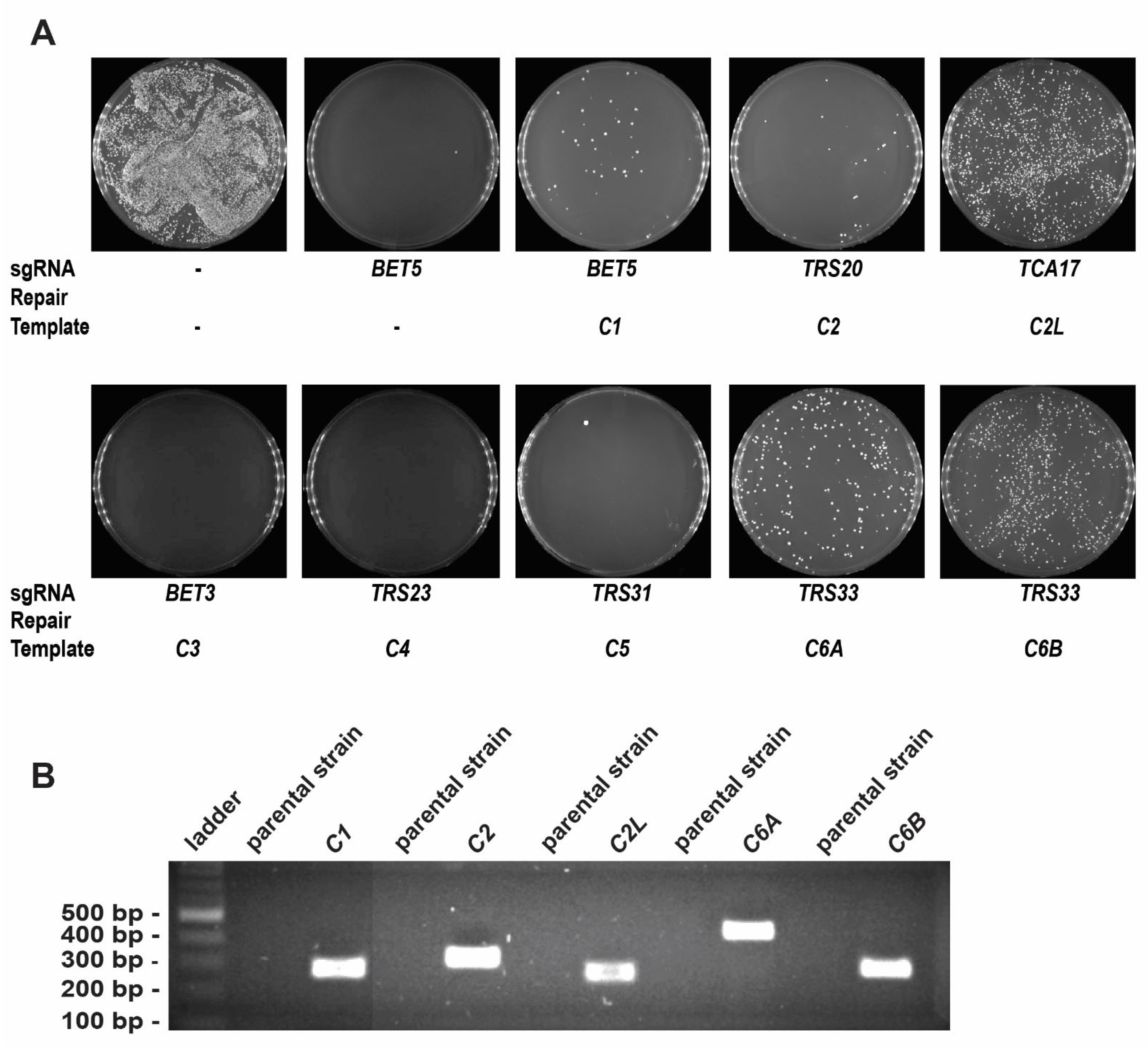
3.1. TRAPPC1, TRAPPC2, TRAPPC2L, TRAPPC6A, and TRAPPC6B Successfully Replace Their Yeast Orthologs BET5, TRS20, TCA17, and TRS33, Respectively
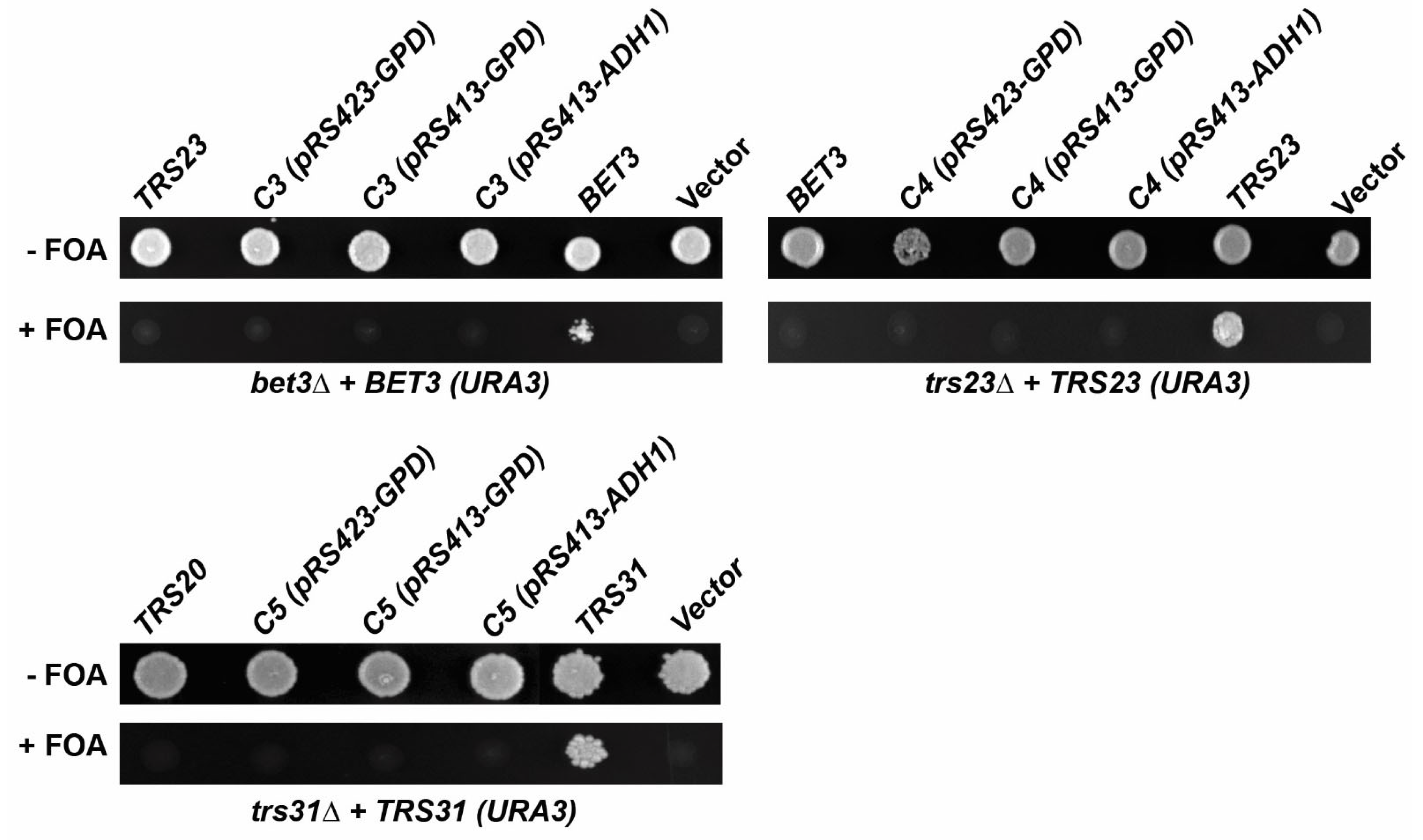
3.2. TRAPPC3, TRAPPC4, and TRAPPC5 Cannot Complement the Loss of BET3, TRS23, and TRS31, Respectively, in Yeast
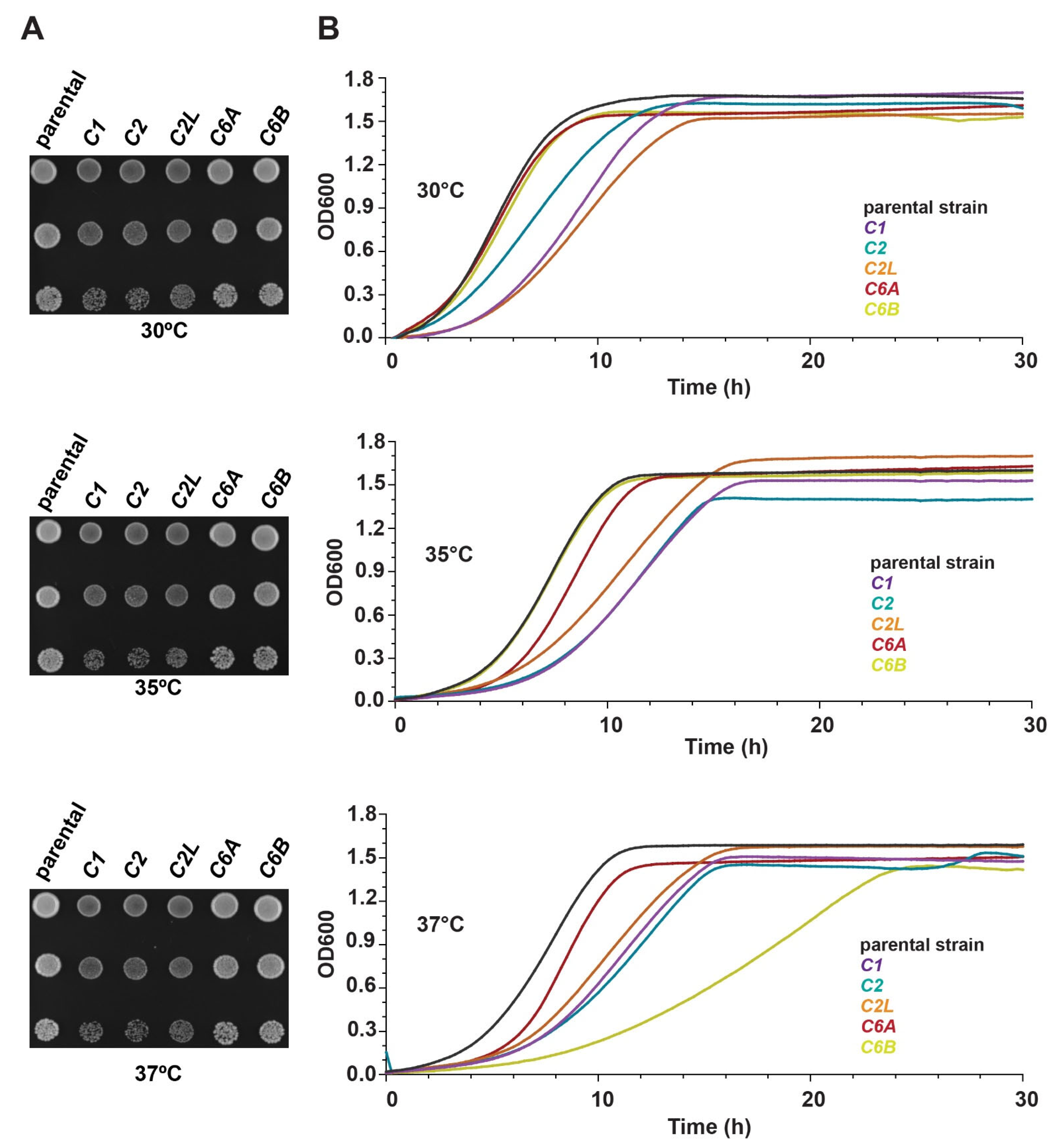
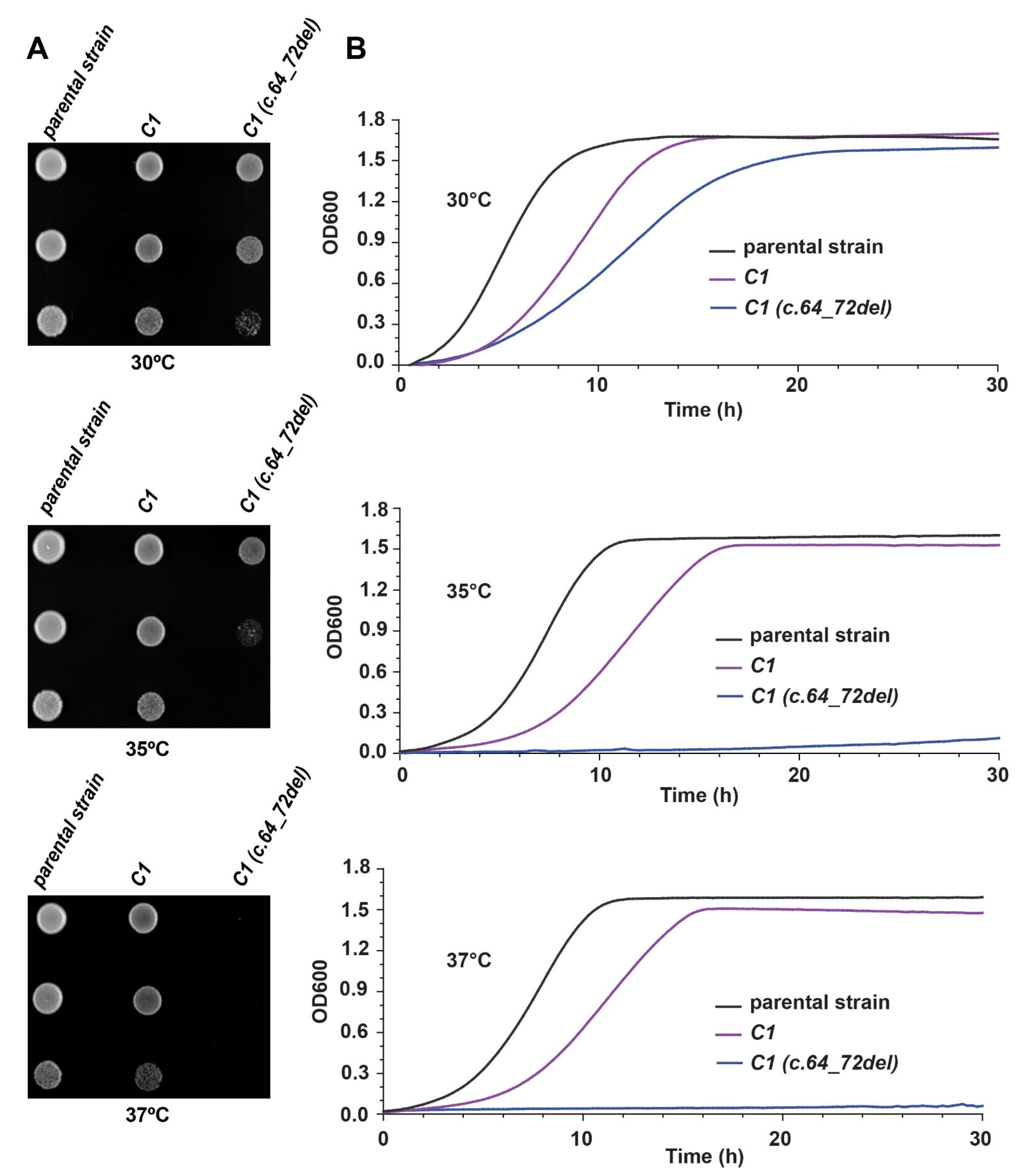
3.3. Humanized Yeast Strains Show a Slower Growth Phenotype Compared to the Parental Yeast Strains
3.4. A Novel Disorder Associated with Biallelic TRAPPC1 Variants
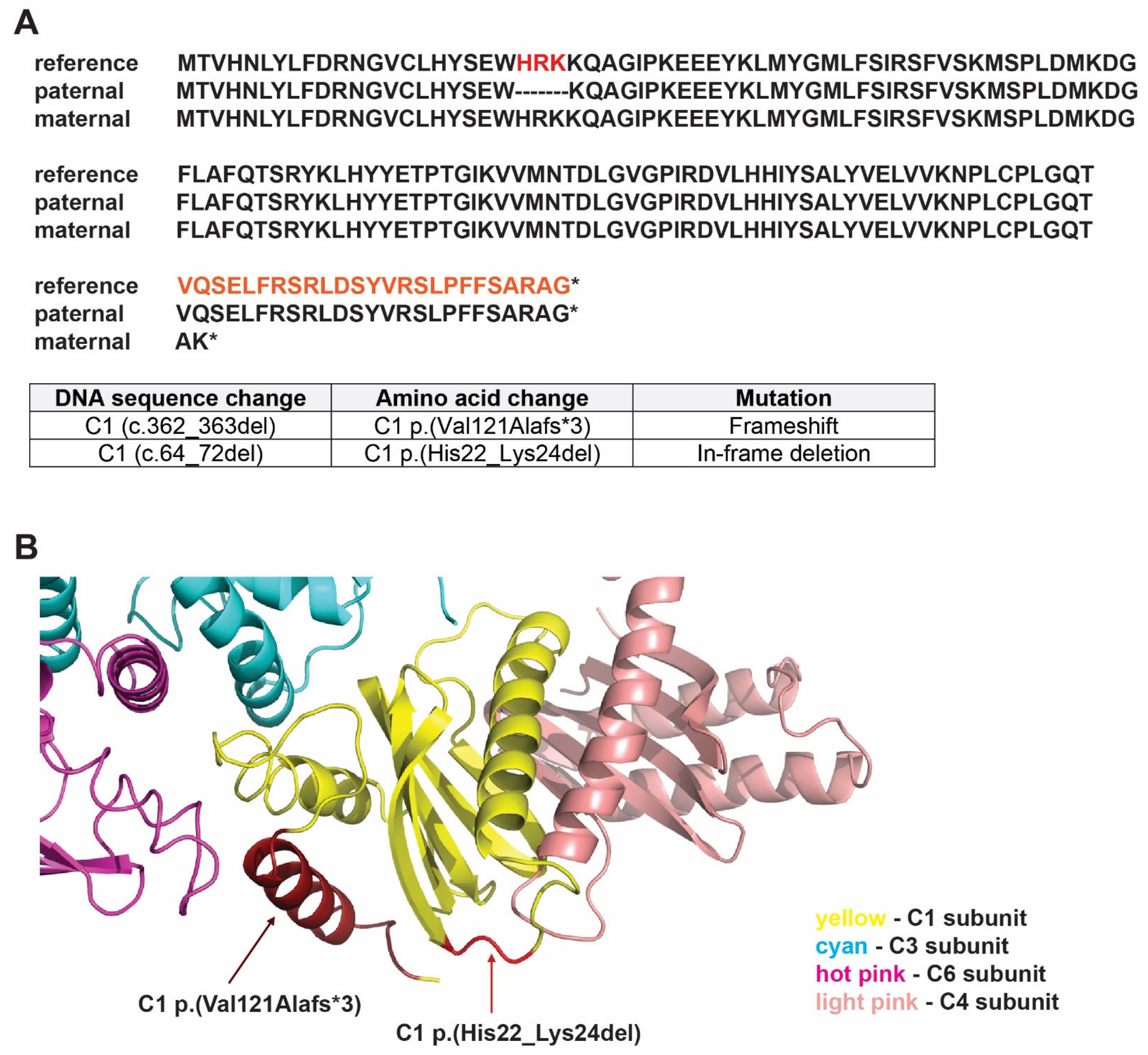
3.5. Humanized Yeast as a Platform for Characterizing the First Reported Mutation in TRAPPC1
3.6. Humanized Yeast with C1 (c.64_72del) Is Conditional-Lethal
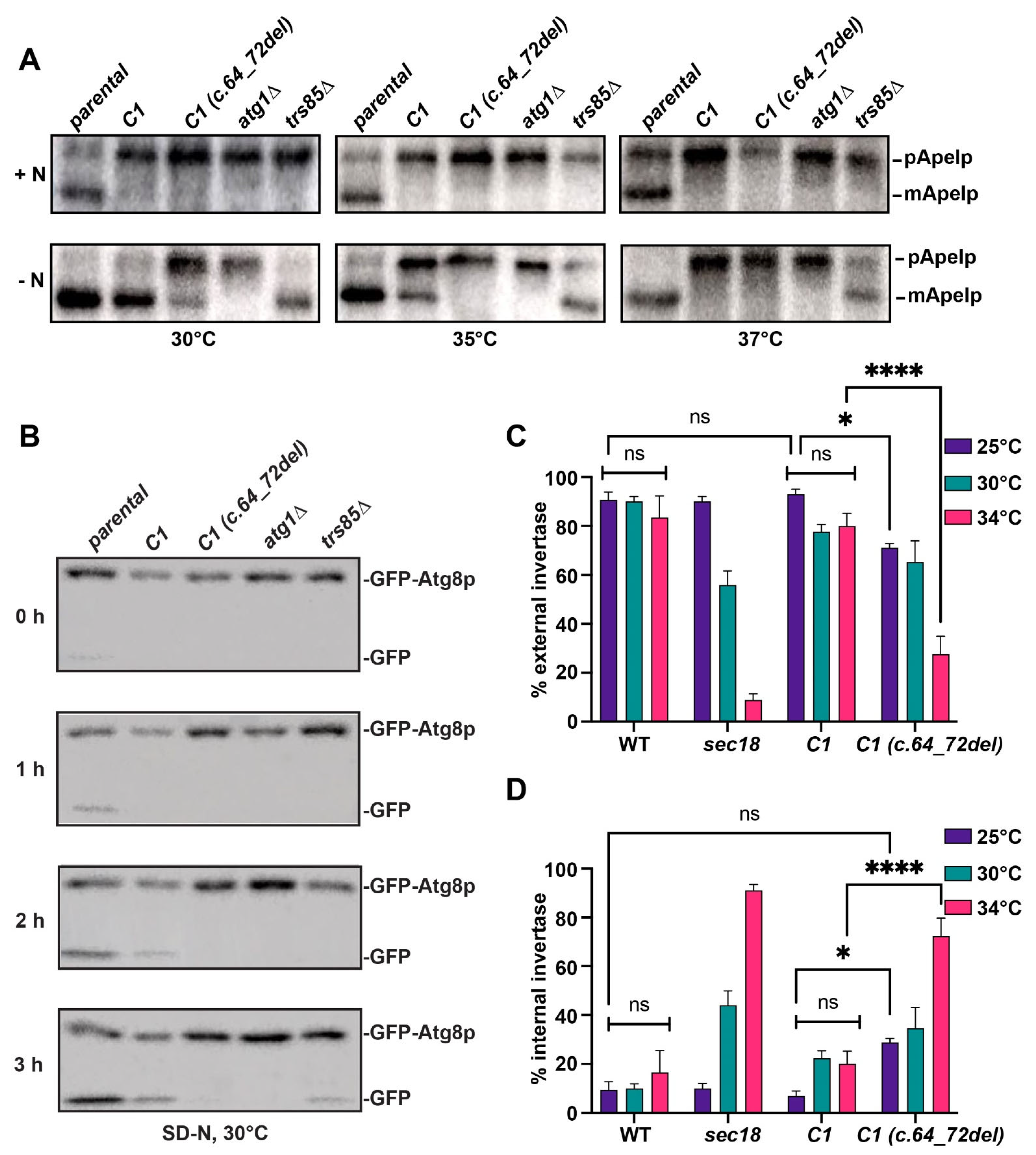
3.7. Non-Selective Autophagy Is Defective in the Humanized C1 (c.64_72del) Yeast Strain
3.8. Secretion Is Defective in the Humanized C1 (c.64_72del) Yeast Strain
3.9. Fibroblasts from the Affected Individual with TRAPPC1 Mutations Do Not Show an Accumulation of LC3-II during Starvation
3.10. Wild-Type TRAPPC1 Rescues a Membrane Trafficking Defect and Golgi Fragmentation in Patient Fibroblasts
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Khosla, N.; Valdez, R. A compilation of national plans, policies and government actions for rare diseases in 23 countries. Intractable Rare Dis. Res. 2018, 7, 213–222. [Google Scholar] [CrossRef]
- Nguengang Wakap, S.; Lambert, D.M.; Olry, A.; Rodwell, C.; Gueydan, C.; Lanneau, V.; Murphy, D.; Le Cam, Y.; Rath, A. Estimating cumulative point prevalence of rare diseases: Analysis of the Orphanet database. Eur. J. Hum. Genet. 2020, 28, 165–173. [Google Scholar] [CrossRef] [PubMed]
- Talarico, R.; Aguilera, S.; Alexander, T.; Amoura, Z.; Andersen, J.; Arnaud, L.; Avcin, T.; Marsal Barril, S.; Beretta, L.; Bom-bardieri, S.; et al. The added value of a European Reference Network on rare and complex connective tissue and musculo-skeletal diseases: Insights after the first 5 years of the ERN ReCONNET. Clin. Exp. Rheumatol. 2022, 40 (Suppl. S134), 3–11. [Google Scholar] [CrossRef] [PubMed]
- Yang, G.; Cintina, I.; Pariser, A.; Oehrlein, E.; Sullivan, J.; Kennedy, A. The national economic burden of rare disease in the United States in 2019. Orphanet J. Rare Dis. 2022, 17, 163. [Google Scholar] [CrossRef] [PubMed]
- Haendel, M.; Vasilevsky, N.; Unni, D.; Bologa, C.; Harris, N.; Rehm, H.; Hamosh, A.; Baynam, G.; Groza, T.; McMurry, J.; et al. How many rare diseases are there? Nat. Rev. Drug Discov. 2020, 19, 77–78. [Google Scholar] [CrossRef]
- Lipatova, Z.; Van Bergen, N.; Stanga, D.; Sacher, M.; Christodoulou, J.; Segev, N. TRAPPing a neurological disorder: From yeast to humans. Autophagy 2020, 16, 965–966. [Google Scholar] [CrossRef] [PubMed]
- Cardinali, P.; Migliorini, L.; Rania, N. The Caregiving Experiences of Fathers and Mothers of Children with Rare Diseases in Italy: Challenges and Social Support Perceptions. Front. Psychol. 2019, 10, 1780. [Google Scholar] [CrossRef]
- Garrino, L.; Picco, E.; Finiguerra, I.; Rossi, D.; Simone, P.; Roccatello, D. Living with and treating rare diseases: Experiences of patients and professional health care providers. Qual. Health Res. 2015, 25, 636–651. [Google Scholar] [CrossRef]
- Hoffman-Andrews, L. The known unknown: The challenges of genetic variants of uncertain significance in clinical practice. J. Law Biosci. 2018, 4, 648–657. [Google Scholar] [CrossRef]
- Palade, G. Intracellular aspects of the process of protein synthesis. Science 1975, 189, 347–358. [Google Scholar] [CrossRef]
- García-Cazorla, A.; Oyarzábal, A.; Saudubray, J.-M.; Martinelli, D.; Dionisi-Vici, C. Genetic disorders of cellular trafficking. Trends Genet. 2022, 38, 724–751. [Google Scholar] [CrossRef] [PubMed]
- Yarwood, R.; Hellicar, J.; Woodman, P.G.; Lowe, M. Membrane trafficking in health and disease. Dis. Model Mech. 2020, 13, dmm043448. [Google Scholar] [CrossRef] [PubMed]
- Condò, I. Rare Monogenic Diseases: Molecular Pathophysiology and Novel Therapies. Int. J. Mol. Sci. 2022, 23, 6525. [Google Scholar] [CrossRef] [PubMed]
- Olkkonen, V.M.; Ikonen, E. When intracellular logistics fails—Genetic defects in membrane trafficking. J. Cell Sci. 2006, 119, 5031–5045. [Google Scholar] [CrossRef]
- Sacher, M.; Shahrzad, N.; Kamel, H.; Milev, M.P. TRAPPopathies: An emerging set of disorders linked to variations in the genes encoding transport protein particle (TRAPP)-associated proteins. Traffic 2019, 20, 5–26. [Google Scholar] [CrossRef]
- Kim, J.J.; Lipatova, Z.; Segev, N. TRAPP Complexes in Secretion and Autophagy. Front. Cell Dev. Biol. 2016, 4, 20. [Google Scholar] [CrossRef]
- Choi, C.; Davey, M.; Schluter, C.; Pandher, P.; Fang, Y.; Foster, L.J.; Conibear, E. Organization and assembly of the TRAPPII complex. Traffic 2011, 12, 715–725. [Google Scholar] [CrossRef]
- Liang, Y.; Morozova, N.; Tokarev, A.A.; Mulholland, J.W.; Segev, N. The role of Trs65 in the Ypt/Rab guanine nucleotide exchange factor function of the TRAPP II complex. Mol. Biol. Cell 2007, 18, 2533–2541. [Google Scholar] [CrossRef]
- Montpetit, B.; Conibear, E. Identification of the Novel TRAPP Associated Protein Tca17. Traffic 2009, 10, 713–723. [Google Scholar] [CrossRef]
- Scrivens, P.J.; Shahrzad, N.; Moores, A.; Morin, A.; Brunet, S.; Sacher, M. TRAPPC2L is a novel, highly conserved TRAPP-interacting protein. Traffic 2009, 10, 724–736. [Google Scholar] [CrossRef]
- Lynch-Day, M.A.; Bhandari, D.; Menon, S.; Huang, J.; Cai, H.; Bartholomew, C.R.; Brumell, J.H.; Ferro-Novick, S.; Klionsky, D.J. Trs85 directs a Ypt1 GEF, TRAPPIII, to the phagophore to promote autophagy. Proc. Natl. Acad. Sci. USA 2010, 107, 7811–7816. [Google Scholar] [CrossRef]
- Galindo, A.; Munro, S. The TRAPP complexes: Oligomeric exchange factors that activate the small GTPases Rab1 and Rab11. FEBS Lett. 2023, 597, 734–749. [Google Scholar] [CrossRef]
- Brunet, S.; Sacher, M. Are all multisubunit tethering complexes bona fide tethers? Traffic 2014, 15, 1282–1287. [Google Scholar] [CrossRef] [PubMed]
- Cai, H.; Zhang, Y.; Pypaert, M.; Walker, L.; Ferro-Novick, S. Mutants in trs120 disrupt traffic from the early endosome to the late Golgi. J. Cell Biol. 2005, 171, 823–833. [Google Scholar] [CrossRef]
- Brunet, S.; Shahrzad, N.; Saint-Dic, D.; Dutczak, H.; Sacher, M. A trs20 mutation that mimics an SEDT-causing mutation blocks selective and non-selective autophagy: A model for TRAPP III organization. Traffic 2013, 14, 1091–1104. [Google Scholar] [CrossRef]
- Joiner, A.M.; Phillips, B.P.; Yugandhar, K.; Sanford, E.J.; Smolka, M.B.; Yu, H.; Miller, E.A.; Fromme, J.C. Structural basis of TRAPPIII-mediated Rab1 activation. EMBO J. 2021, 40, e107607. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.; Li, C.M.; Luo, X.M.; Siu, G.K.Y.; Gan, W.J.; Zhang, L.; Wu, W.K.K.; Chan, H.C.; Yu, S. Mammalian TRAPPIII Complex positively modulates the recruitment of Sec13/31 onto COPII vesicles. Sci. Rep. 2017, 7, 43207. [Google Scholar] [CrossRef] [PubMed]
- Allan, B.B.; Moyer, B.D.; Balch, W.E. Rab1 recruitment of p115 into a cis-SNARE complex: Programming budding COPII vesicles for fusion. Science 2000, 289, 444–448. [Google Scholar] [CrossRef]
- Wang, J.; Menon, S.; Yamasaki, A.; Chou, H.T.; Walz, T.; Jiang, Y.; Ferro-Novick, S. Ypt1 recruits the Atg1 kinase to the preautophagosomal structure. Proc. Natl. Acad. Sci. USA 2013, 110, 9800–9805. [Google Scholar] [CrossRef]
- Bassik, M.C.; Kampmann, M.; Lebbink, R.J.; Wang, S.; Hein, M.Y.; Poser, I.; Weibezahn, J.; Horlbeck, M.A.; Chen, S.; Mann, M.; et al. A Systematic Mammalian Genetic Interaction Map Reveals Pathways Underlying Ricin Susceptibility. Cell 2013, 152, 909–922. [Google Scholar] [CrossRef]
- Zappa, F.; Wilson, C.; Di Tullio, G.; Santoro, M.; Pucci, P.; Monti, M.; D’Amico, D.; Pisonero-Vaquero, S.; De Cegli, R.; Romano, A.; et al. The TRAPP complex mediates secretion arrest induced by stress granule assembly. EMBO J. 2019, 38, e101704. [Google Scholar] [CrossRef] [PubMed]
- Gedeon, Á.K.; Colley, A.; Jamieson, R.; Thompson, E.M.; Rogers, J.; Sillence, D.; Tiller, G.E.; Mulley, J.C.; Gécz, J. Identification of the gene (SEDL) causing X-linked spondyloepiphyseal dysplasia tarda. Nat. Genet. 1999, 22, 400–404. [Google Scholar] [CrossRef]
- Ghosh, S.G.; Scala, M.; Beetz, C.; Helman, G.; Stanley, V.; Yang, X.; Breuss, M.W.; Mazaheri, N.; Selim, L.; Hadipour, F.; et al. A relatively common homozygous TRAPPC4 splicing variant is associated with an early-infantile neurodegenerative syndrome. Eur. J. Hum. Genet. 2021, 29, 271–279. [Google Scholar] [CrossRef] [PubMed]
- Van Bergen, N.J.; Guo, Y.; Al-Deri, N.; Lipatova, Z.; Stanga, D.; Zhao, S.; Murtazina, R.; Gyurkovska, V.; Pehlivan, D.; Mitani, T.; et al. Deficiencies in vesicular transport mediated by TRAPPC4 are associated with severe syndromic intellectual disability. Brain 2020, 143, 112–130. [Google Scholar] [CrossRef]
- Almousa, H.; Lewis, S.A.; Bakhtiari, S.; Nordlie, S.H.; Pagnozzi, A.; Magee, H.; Efthymiou, S.; Heim, J.A.; Cornejo, P.; Zaki, M.S.; et al. TRAPPC6B biallelic variants cause a neurodevelopmental disorder with TRAPP II and trafficking disruptions. Brain 2023, 147, 311–324. [Google Scholar] [CrossRef]
- Aridon, P.; De Fusco, M.; Winkelmann, J.W.; Zucconi, M.; Arnao, V.; Ferini-Strambi, L.; Casari, G. A TRAPPC6B splicing variant associates to restless legs syndrome. Park. Relat. Disord. 2016, 31, 135–138. [Google Scholar] [CrossRef] [PubMed]
- Marin-Valencia, I.; Novarino, G.; Johansen, A.; Rosti, B.; Issa, M.Y.; Musaev, D.; Bhat, G.; Scott, E.; Silhavy, J.L.; Stanley, V.; et al. A homozygous founder mutation in TRAPPC6B associates with a neurodevelopmental disorder characterised by micro-cephaly, epilepsy and autistic features. J. Med. Genet. 2018, 55, 48–54. [Google Scholar] [CrossRef]
- Mohamoud, H.S.; Ahmed, S.; Jelani, M.; Alrayes, N.; Childs, K.; Vadgama, N.; Almramhi, M.M.; Al-Aama, J.Y.; Goodbourn, S.; Nasir, J. A missense mutation in TRAPPC6A leads to build-up of the protein, in patients with a neurodevelopmental syndrome and dysmorphic features. Sci. Rep. 2018, 8, 2053. [Google Scholar] [CrossRef]
- Bögershausen, N.; Shahrzad, N.; Chong, J.X.; Von Kleist-Retzow, J.C.; Stanga, D.; Li, Y.; Bernier, F.P.; Loucks, C.M.; Wirth, R.; Puffenberger, E.G.; et al. Recessive TRAPPC11 mutations cause a disease spectrum of limb girdle muscular dystrophy and myopathy with movement disorder and intellectual disability. Am. J. Hum. Genet. 2013, 93, 181–190. [Google Scholar] [CrossRef]
- Larson, A.A.; Baker, P.R.; Milev, M.P.; Press, C.A.; Sokol, R.J.; Cox, M.O.; Lekostaj, J.K.; Stence, A.A.; Bossler, A.D.; Mueller, J.M.; et al. TRAPPC11 and GOSR2 mutations associate with hypoglycosylation of α-dystroglycan and muscular dystrophy. Skelet. Muscle 2018, 8, 17. [Google Scholar] [CrossRef]
- Liang, W.-C.; Zhu, W.; Mitsuhashi, S.; Noguchi, S.; Sacher, M.; Ogawa, M.; Shih, H.-H.; Jong, Y.-J.; Nishino, I. Congenital muscular dystrophy with fatty liver and infantile-onset cataract caused by TRAPPC11 mutations: Broadening of the pheno-type. Skelet. Muscle 2015, 5, 29. [Google Scholar] [CrossRef] [PubMed]
- Matalonga, L.; Bravo, M.; Serra-Peinado, C.; García-Pelegrí, E.; Ugarteburu, O.; Vidal, S.; Llambrich, M.; Quintana, E.; Fuster-Jorge, P.; Gonzalez-Bravo, M.N.; et al. Mutations in TRAPPC11 are associated with a congenital disorder of glycosylation. Hum. Mutat. 2017, 38, 148–151. [Google Scholar] [CrossRef]
- Milev, M.P.; Grout, M.E.; Saint-Dic, D.; Cheng, Y.-H.H.; Glass, I.A.; Hale, C.J.; Hanna, D.S.; Dorschner, M.O.; Prematilake, K.; Shaag, A.; et al. Mutations in TRAPPC12 Manifest in Progressive Childhood Encephalopathy and Golgi Dysfunction. Am. J. Hum. Genet. 2017, 101, 291–299. [Google Scholar] [CrossRef]
- Milev, M.P.; Stanga, D.; Schänzer, A.; Nascimento, A.; Saint-Dic, D.; Ortez, C.; Natera-de Benito, D.; Barrios, D.G.; Colomer, J.; Badosa, C.; et al. Characterization of three TRAPPC11 variants suggests a critical role for the extreme carboxy terminus of the protein. Sci. Rep. 2019, 9, 14036. [Google Scholar] [CrossRef]
- Munot, P.; McCrea, N.; Torelli, S.; Manzur, A.; Sewry, C.; Chambers, D.; Feng, L.; Ala, P.; Zaharieva, I.; Ragge, N.; et al. TRAPPC11-related muscular dystrophy with hypoglycosylation of alpha-dystroglycan in skeletal muscle and brain. Neuropathol. Appl. Neurobiol. 2022, 48, e12771. [Google Scholar] [CrossRef] [PubMed]
- Fee, D.B.; Harmelink, M.; Monrad, P.; Pyzik, E. Siblings with Mutations in TRAPPC11 Presenting With Limb-Girdle Muscular Dystrophy 2S. J. Clin. Neuromuscul. Dis. 2017, 19, 27–30. [Google Scholar] [CrossRef] [PubMed]
- Hentschel, A.; Czech, A.; Münchberg, U.; Freier, E.; Schara-Schmidt, U.; Sickmann, A.; Reimann, J.; Roos, A. Protein signature of human skin fibroblasts allows the study of the molecular etiology of rare neurological diseases. Orphanet J. Rare Dis. 2021, 16, 73. [Google Scholar] [CrossRef] [PubMed]
- Auburger, G.; Klinkenberg, M.; Drost, J.; Marcus, K.; Morales-Gordo, B.; Kunz, W.S.; Brandt, U.; Broccoli, V.; Reichmann, H.; Gispert, S.; et al. Primary skin fibroblasts as a model of Parkinson’s disease. Mol. Neurobiol. 2012, 46, 20–27. [Google Scholar] [CrossRef]
- Vangipuram, M.; Ting, D.; Kim, S.; Diaz, R.; Schüle, B. Skin punch biopsy explant culture for derivation of primary human fibroblasts. J. Vis. Exp. 2013, 77, e3779. [Google Scholar] [CrossRef]
- Al-Deri, N.; Okur, V.; Ahimaz, P.; Milev, M.; Valivullah, Z.; Hagen, J.; Sheng, Y.; Chung, W.; Sacher, M.; Ganapathi, M. A novel homozygous variant in TRAPPC2L results in a neurodevelopmental disorder and disrupts TRAPP complex function. J. Med. Genet. 2021, 58, 592–601. [Google Scholar] [CrossRef]
- Mesdom, P.; Colle, R.; Lebigot, E.; Trabado, S.; Deflesselle, E.; Fève, B.; Becquemont, L.; Corruble, E.; Verstuyft, C. Human Dermal Fibroblast: A Promising Cellular Model to Study Biological Mechanisms of Major Depression and Antidepressant Drug Response. Curr. Neuropharmacol. 2020, 18, 301–318. [Google Scholar] [CrossRef]
- Kachroo, A.H.; Vandeloo, M.; Greco, B.M.; Abdullah, M. Humanized yeast to model human biology, disease and evolution. Dis. Model Mech. 2022, 15, dmm049309. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Park, J.; Kim, T.; Lee, J.-S. The functional study of human proteins using humanized yeast. J. Microbiol. 2020, 58, 343–349. [Google Scholar] [CrossRef] [PubMed]
- Foury, F. Human genetic diseases: A cross-talk between man and yeast. Gene 1997, 195, 1–10. [Google Scholar] [CrossRef]
- Laurent, J.M.; Young, J.H.; Kachroo, A.H.; Marcotte, E.M. Efforts to make and apply humanized yeast. Brief Funct. Genom. 2016, 15, 155–163. [Google Scholar] [CrossRef] [PubMed]
- Garge, R.K.; Cha, H.J.; Lee, C.; Gollihar, J.D.; Kachroo, A.H.; Wallingford, J.B.; Marcotte, E.M. Discovery of new vascular disrupting agents based on evolutionarily conserved drug action, pesticide resistance mutations, and humanized yeast. Genetics 2021, 219, iyab101. [Google Scholar] [CrossRef]
- Kachroo, A.H.; Laurent, J.M.; Yellman, C.M.; Meyer, A.G.; Wilke, C.O.; Marcotte, E.M. Systematic humanization of yeast genes reveals conserved functions and genetic modularity. Science 2015, 348, 921–925. [Google Scholar] [CrossRef]
- DiCarlo, J.E.; Norville, J.E.; Mali, P.; Rios, X.; Aach, J.; Church, G.M. Genome engineering in Saccharomyces cerevisiae using CRISPR-Cas systems. Nucleic Acids Res. 2013, 41, 4336–4343. [Google Scholar] [CrossRef]
- Akhmetov, A.; Laurent, J.M.; Gollihar, J.; Gardner, E.C.; Garge, R.K.; Ellington, A.D.; Kachroo, A.H.; Marcotte, E.M. Single-step Precision Genome Editing in Yeast Using CRISPR-Cas9. Bio. Protoc. 2018, 8, e2765. [Google Scholar] [CrossRef]
- Brzáčová, Z.; Peťková, M.; Veljačiková, K.; Zajičková, T.; Tomáška, Ľ. Reconstruction of human genome evolution in yeast: An educational primer for use with “systematic humanization of the yeast cytoskeleton discerns functionally replaceable from divergent human genes”. Genetics 2021, 219, iyab118. [Google Scholar] [CrossRef]
- Doench, J.G.; Fusi, N.; Sullender, M.; Hegde, M.; Vaimberg, E.W.; Donovan, K.F.; Smith, I.; Tothova, Z.; Wilen, C.; Orchard, R.; et al. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat. Biotechnol. 2016, 34, 184–191. [Google Scholar] [CrossRef]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef] [PubMed]
- Klionsky, D.J.; Cueva, R.; Yaver, D.S. Aminopeptidase I of Saccharomyces cerevisiae is localized to the vacuole independent of the secretory pathway. J. Cell Biol. 1992, 119, 287–299. [Google Scholar] [CrossRef]
- Munn, A.L.; Heese-Peck, A.; Stevenson, B.J.; Pichler, H.; Riezman, H. Specific sterols required for the internalization step of endocytosis in yeast. Mol. Biol. Cell 1999, 10, 3943–3957. [Google Scholar] [CrossRef] [PubMed]
- Boncompain, G.; Divoux, S.; Gareil, N.; de Forges, H.; Lescure, A.; Latreche, L.; Mercanti, V.; Jollivet, F.; Raposo, G.; Perez, F. Synchronization of secretory protein traffic in populations of cells. Nat. Methods 2012, 9, 493–498. [Google Scholar] [CrossRef] [PubMed]
- Boonekamp, F.J.; Knibbe, E.; Vieira-Lara, M.A.; Wijsman, M.; Luttik, M.A.H.; van Eunen, K.; Ridder, M.d.; Bron, R.; Almonacid Suarez, A.M.; van Rijn, P.; et al. Full humanization of the glycolytic pathway in Saccharomyces cerevisiae. Cell Rep. 2022, 39, 111010. [Google Scholar] [CrossRef] [PubMed]
- Luque, J.; Mendes, I.; Gomez, B.; Morte, B.; Lopez de Heredia, M.; Herreras, E.; Corrochano, V.; Bueren, J.; Gallano, P.; Artuch, R.; et al. CIBERER: Spanish national network for research on rare diseases: A highly productive collaborative initiative. Clin. Genet. 2022, 101, 481–493. [Google Scholar] [CrossRef]
- Filippini, F.; Rossi, V.; Galli, T.; Budillon, A.; D’Urso, M.; D’Esposito, M. Longins: A new evolutionary conserved VAMP family sharing a novel SNARE domain. Trends Biochem. Sci. 2001, 26, 407–409. [Google Scholar] [CrossRef]
- Rossi, V.; Banfield, D.K.; Vacca, M.; Dietrich, L.E.P.; Ungermann, C.; D’Esposito, M.; Galli, T.; Filippini, F. Longins and their longin domains: Regulated SNAREs and multifunctional SNARE regulators. Trends Biochem. Sci. 2004, 29, 682–688. [Google Scholar] [CrossRef]
- Schlenker, O.; Hendricks, A.; Sinning, I.; Wild, K. The structure of the mammalian signal recognition particle (SRP) receptor as prototype for the interaction of small GTPases with Longin domains. J. Biol. Chem. 2006, 281, 8898–8906. [Google Scholar] [CrossRef]
- Cheong, H.; Klionsky, D.J. Biochemical methods to monitor autophagy-related processes in yeast. Methods Enzym. 2008, 451, 1–26. [Google Scholar] [CrossRef]
- Scott, S.V.; Hefner-Gravink, A.; Morano, K.A.; Noda, T.; Ohsumi, Y.; Klionsky, D.J. Cytoplasm-to-vacuole targeting and au-tophagy employ the same machinery to deliver proteins to the yeast vacuole. Proc. Natl. Acad. Sci. USA 1996, 93, 12304–12308. [Google Scholar] [CrossRef]
- Klionsky, D.J.; Abdel-Aziz, A.K.; Abdelfatah, S.; Abdellatif, M.; Abdoli, A.; Abel, S.; Abeliovich, H.; Abildgaard, M.H.; Abudu, Y.P.; Acevedo-Arozena, A.; et al. Guidelines for the use and interpretation of assays for monitoring autophagy (4th edition)1. Autophagy 2021, 17, 1–382. [Google Scholar] [CrossRef] [PubMed]
- Garge, R.K.; Laurent, J.M.; Kachroo, A.H.; Marcotte, E.M. Systematic Humanization of the Yeast Cytoskeleton Discerns Functionally Replaceable from Divergent Human Genes. Genetics 2020, 215, 1153–1169. [Google Scholar] [CrossRef]
- DuBay, K.H.; Bowman, G.R.; Geissler, P.L. Fluctuations within folded proteins: Implications for thermodynamic and allosteric regulation. Acc. Chem. Res. 2015, 48, 1098–1105. [Google Scholar] [CrossRef]
- Guarnera, E.; Berezovsky, I.N. On the perturbation nature of allostery: Sites, mutations, and signal modulation. Curr. Opin. Struct. Biol. 2019, 56, 18–27. [Google Scholar] [CrossRef] [PubMed]
- Prabantu, V.M.; Naveenkumar, N.; Srinivasan, N. Influence of Disease-Causing Mutations on Protein Structural Networks. Front. Mol. Biosci. 2020, 7, 620554. [Google Scholar] [CrossRef]
- Bagde, S.R.; Fromme, J.C. Structure of a TRAPPII-Rab11 activation intermediate reveals GTPase substrate selection mecha-nisms. Sci. Adv. 2022, 8, eabn7446. [Google Scholar] [CrossRef]
- Cai, Y.; Chin, H.F.; Lazarova, D.; Menon, S.; Fu, C.; Cai, H.; Sclafani, A.; Rodgers, D.W.; De La Cruz, E.M.; Ferro-Novick, S.; et al. The structural basis for activation of the Rab Ypt1p by the TRAPP membrane tethering complexes. Cell 2008, 133, 1202–1213. [Google Scholar] [CrossRef]
- Galindo, A.; Planelles-Herrero, V.J.; Degliesposti, G.; Munro, S. Cryo-EM structure of metazoan TRAPPIII, the multi-subunit complex that activates the GTPase Rab1. EMBO J. 2021, 40, e107608. [Google Scholar] [CrossRef]
- Kim, Y.-G.; Raunser, S.; Munger, C.; Wagner, J.; Song, Y.-L.; Cygler, M.; Walz, T.; Oh, B.-H.; Sacher, M. The architecture of the multisubunit TRAPP I complex suggests a model for vesicle tethering. Cell 2006, 127, 817–830. [Google Scholar] [CrossRef] [PubMed]
- Fassio, A.; Falace, A.; Esposito, A.; Aprile, D.; Guerrini, R.; Benfenati, F. Emerging Role of the Autophagy/Lysosomal Deg-radative Pathway in Neurodevelopmental Disorders with Epilepsy. Front. Cell Neurosci. 2020, 14, 39. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Stanford, K.R.; Kundu, M. ER-to-Golgi Trafficking and Its Implication in Neurological Diseases. Cells 2020, 9, 408. [Google Scholar] [CrossRef] [PubMed]



| Mammalian Subunits | Yeast Subunits | TRAPP Complex | Essential in Yeast? | %Identity/Similarity of Core Subunits |
|---|---|---|---|---|
| TRAPPC1 | Bet5p | II/III | yes | 33/54 |
| TRAPPC2 | Trs20p | II/III | yes | 34/52 |
| TRAPPC2L | Tca17p | II | no | 24/43 |
| TRAPPC3 | Bet3p | II/III | yes | 56/76 |
| TRAPPC4 | Trs23p | II/III | yes | 29/44 |
| TRAPPC5 | Trs31p | II/III | yes | 31/52 |
| TRAPPC6A/C6B | Trs33p | II/III | no | 35/51 and 33/47 |
| TRAPPC8 | Trs85p | III | no | - |
| TRAPPC9 | Trs120p | II | yes | - |
| TRAPPC10 | Trs130p | II | yes | - |
| TRAPPC11 | - | III | - | - |
| TRAPPC12 | - | III | - | - |
| TRAPPC13 | Trs65 | III/II * | no | - |
| Plasmid | Construct or Backbone |
|---|---|
| MSB2 | pRS315 |
| MSB206 | pRS313 |
| MSB230 | pRS313-TRS20 |
| MSB1253 | pRS416-GFP-ATG8 |
| MSB1790 | pmRFP-N1-TRAPPC1 |
| MSB1791 | pmRFP-C1-TRAPPC1 |
| MSB1797 | pRS413-ADH1-TRAPPC1 |
| MSB1800 | pRS413-ADH1-TRAPPC1 (c.362_363del) |
| MSB1801 | pRS413-ADH1-TRAPPC1 (c.64_72del) |
| MSB1805 | pCEN6-Cas9-GFP-KanMX |
| MSB1806 | pCEN6-Cas9-GFP-KanMX BET5 sgRNA1 |
| MSB1820 | pCEN6-Cas9-GFP-KanMX TRS20 sgRNA1 |
| MSB1821 | pCEN6-Cas9-GFP-KanMX TRS23 sgRNA1 |
| MSB1823 | pCEN6-Cas9-GFP-KanMX BET3 sgRNA1 |
| MSB1824 | pCEN6-Cas9-GFP-KanMX TRS31 sgRNA1 |
| MSB1826 | pCEN6-Cas9-GFP-KanMX TCA17 sgRNA1 |
| MSB1829 | pCEN6-Cas9-GFP-KanMX TRS33 sgRNA1 |
| MSB1839 | pRS413-ADH1-TRAPPC5 |
| MSB1840 | pRS413-GPD-TRAPPC5 |
| MSB1841 | pRS423-GPD-TRAPPC5 |
| MSB1855 | pRS413-ADH1-TRAPPC3 |
| MSB1856 | pRS413-GPD-TRAPPC3 |
| MSB1857 | pRS423-GPD-TRAPPC3 |
| MSB1858 | pRS413-ADH1-TRAPPC4 |
| MSB1859 | pRS413-GPD-TRAPPC4 |
| MSB1860 | pRS423-GPD-TRAPPC4 |
| Yeast Strain | Genotype | Source |
|---|---|---|
| MSY61 | MATα his3Δ1 leu2Δ0 ura3Δ0 MET15 bet3∆::KanMX pRS315-BET3 | Sacher Lab |
| MSY62 | MATα his3Δ1 leu2Δ0 ura3Δ0 MET15 trs23∆::KanMX pRS316-TRS23 | Sacher Lab |
| MSY64 | MATα his3Δ1 leu2Δ0 ura3Δ0 MET15 bet5∆::KanMX pRS316-BET5 | Sacher Lab |
| MSY65 | MATα his3Δ1 leu2Δ0 ura3Δ0 MET15 trs31∆::KanMX pRS316-TRS31 | Sacher Lab |
| MSY135 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 | Sacher Lab |
| MSY362 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 trs85∆::KanMX | Titorenko Lab |
| MSY563 | MATα his3Δ1 leu2Δ0 ura3ΔMET15 trs23Δ::KanMX pRS315-TRS23 TRS85–3xHA::HIS3 | Sacher Lab |
| MSY706 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 atg1∆::KanMX | Brett Lab |
| MSY710 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 atg1∆::KanMX pRS416-GFP-ATG8 | Sacher Lab |
| MSY740 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 trs85∆::KanMX pRS416-GFP-ATG8 | Sacher Lab |
| MSY766 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 pRS416-GFP-ATG8 | Sacher Lab |
| MSY901 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 TRS130-3xHA::HIS3 | Sacher Lab |
| MSY950 | MATα his3Δ1 leu2Δ0 ura3Δ0 MET15 bet5∆::KanMX pRS316-BET5 pRS413-ADH1 | Zykaj E. |
| MSY951 | MATα his3Δ1 leu2Δ0 ura3Δ0 MET15 bet5∆::KanMX pRS316-BET5 pRS413-ADH1-TRAPPC1 | Zykaj E. |
| MSY952 | MATα his3Δ1 leu2Δ0 ura3Δ0 MET15 bet5∆::KanMX pRS316-BET5 pRS313-BET5 | Zykaj E. |
| MSY955 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 bet5∆ with TRAPPC1 | Zykaj E. |
| MSY956 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 bet5∆ with TRAPPC1 ∆HRK (c.64_72del) | Zykaj E. |
| MSY957 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 trs20∆ with TRAPPC2 | Zykaj E. |
| MSY958 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 TRS130-3xHA::HIS3 tca17∆ with TRAPPC2L | Zykaj E. |
| MSY959 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 TRS130-3XHA::HIS3 trs33∆ with TRAPPC6A | Zykaj E. |
| MSY960 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 TRS130-3XHA::HIS3 trs33∆ with TRAPPC6B | Zykaj E. |
| MSY963 | MATα his3Δ1 leu2Δ0 ura3Δ0 MET15 trs31∆::KanMX pRS316-TRS31 pRS413-ADH1-TRAPPC5 | Zykaj E. |
| MSY964 | MATα his3Δ1 leu2Δ0 ura3Δ0 MET15 trs31∆::KanMX pRS316-TRS31 pRS413-GPD-TRAPPC5 | Zykaj E. |
| MSY965 | MATα his3Δ1 leu2Δ0 ura3Δ0 MET15 trs31∆::KanMX pRS316-TRS31 pRS423-GPD-TRAPPC5 | Zykaj E. |
| MSY967 | MATα his3Δ1 leu2Δ0 ura3Δ0 MET15 trs31∆::KanMX pRS316-TRS31 pRS315 | Zykaj E. |
| MSY968 | MATα his3Δ1 leu2Δ0 ura3Δ0 MET15 trs31∆::KanMX pRS316-TRS31 pRS315-TRS31 | Zykaj E. |
| MSY971 | MATα his3Δ1 leu2Δ0 ura3Δ0 MET15 trs23∆::KanMX pRS316-TRS23 pRS313 | Zykaj E. |
| MSY972 | MATα his3Δ1 leu2Δ0 ura3Δ0 MET15 trs23∆::KanMX pRS316-TRS23 pRS313-TRS23 | Zykaj E. |
| MSY973 | MATα his3Δ1 leu2Δ0 ura3Δ0 MET15 trs23∆::KanMX pRS316-TRS23 pRS413-ADH1-TRAPPC4 | Zykaj E. |
| MSY974 | MATα his3Δ1 leu2Δ0 ura3Δ0 MET15 trs23∆::KanMX pRS316-TRS23 pRS413-GPD-TRAPPC4 | Zykaj E. |
| MSY975 | MATα his3Δ1 leu2Δ0 ura3Δ0 MET15 trs23∆::KanMX pRS316-TRS23 pRS423-GPD-TRAPPC4 | Zykaj E. |
| MSY976 | MATα his3Δ1 leu2Δ0 ura3Δ0 MET15 bet3∆::KanMX pRS315-BET3 pRS313 | Zykaj E. |
| MSY977 | MATα his3Δ1 leu2Δ0 ura3Δ0 MET15 bet3∆::KanMX pRS315-BET3 pRS313–BET3 | Zykaj E. |
| MSY978 | MATα his3Δ1 leu2Δ0 ura3Δ0 MET15 bet3∆::KanMX pRS315-BET3 pRS413-ADH1-TRAPPC3 | Zykaj E. |
| MSY979 | MATα his3Δ1 leu2Δ0 ura3Δ0 MET15 bet3∆::KanMX pRS315-BET3 pRS413-GPD-TRAPPC3 | Zykaj E. |
| MSY980 | MATα his3Δ1 leu2Δ0 ura3Δ0 MET15 bet3∆::KanMX pRS315-BET3 pRS423-GPD-TRAPPC3 | Zykaj E. |
| MSY981 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 bet5∆ with TRAPPC1 pRS416-GFP-ATG8 | Zykaj E. |
| MSY982 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 bet5∆ with TRAPPC1 ∆HRK (c.64_72del) pRS416-GFP-ATG8 | Zykaj E. |
| MSY983 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 TRS130-3xHA::HIS3 bet5∆ with TRAPPC1 | Zykaj E. |
| MSY984 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 TRS130-3xHA::HIS3 bet5∆ with TRAPPC1 ∆HRK (c.64_72del) | Zykaj E. |
| MSY987 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 tca17∆ with TRAPPC2L | Zykaj E. |
| MSY988 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 trs33∆ with TRAPPC6B | Zykaj E. |
| MSY993 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 trs33∆ with TRAPPC6A | Zykaj E. |
| Primer Name | Sequence |
|---|---|
| BET5 sgRNA1-Fp | GACTTTTTACACAAAAGCTCTCCAAA |
| BET5 sgRNA1-Rp | AAACTTTGGAGAGCTTTTGTGTAAAA |
| TRS20 sgRNA1-Fp | GACTTTACCAATGCAGAAAATCCACA |
| TRS20 sgRNA1-Rp | AAACTGTGGATTTTCTGCATTGGTAA |
| TCA17 sgRNA1-Fp | GACTTTGATTTACGTCCCTAATGAGG |
| TCA17 sgRNA1-Rp | AAACCCTCATTAGGGACGTAAATCAA |
| BET3 sgRNA1-Fp | GACTTTCTTCAAAGACCTCGATTGCG |
| BET3 sgRNA1-Rp | AAACCGCAATCGAGGTCTTTGAAGAA |
| TRS23 sgRNA1-Fp | GACTTTCTTGTAATAAACAAATCAGG |
| TRS23 sgRNA1-Rp | AAACCCTGATTTGTTTATTACAAGAA |
| TRS31 sgRNA1-Fp | GACTTTGCATCTGACCAACAATTCCC |
| TRS31 sgRNA1-Rp | AAACGGGAATTGTTGGTCAGATGCAA |
| TRS33 sgRNA1-Fp | GACTTTGGACCTTGTGGAGAAGATTG |
| TRS33 sgRNA1-Rp | AAACCAATCTTCTCCACAAGGTCCAA |
| Primer Name | Sequence |
|---|---|
| TRAPPC1-Fp | AATACAAAAAAAAAATACAGAACTATCGTAAGATAATCTGCTTTTGATATAAATCATAAATCAGTGGAGCATGACTGTCCACAACCTGTAC |
| TRAPPC1-Rp | TATACGATGAATGCAATTCAATTCATCGCTTCTATTTATTTTAGGTTTTTCTTCTGCTCATGGTTCGTGCTCAGCCAGCCCGGGCGGAGAAG |
| TRAPPC2-Fp | AAGAAAAAAAAGAGAAGGTAAACACTAATACGAATAGAGAATACAGAAAAAACATACAAAGCACATTGAGATGTCTGGGAGCTTCTACTTTG |
| TRAPPC2-Rp | AAAAAAAAAAAAATACATACTACATATACATATACGCCATAAAAAATCTCTGCATCTATCTTATTTCCCATCAGCTTAAAAGGTGTTTCTTC |
| TRAPPC2L-Fp | AAGTTATTGAGTTGAAAATTGAGTAAATATTCTCTCCAAAAATCAAATTGTACGTTTATATCACCAAACTATGGCGGTGTGCATCGCGGTG |
| TRAPPC2L-Rp | GCTCTCACTCCAAAGCAGTGACATTAAATTTTGCTTTCATTCGTGAAAGAATCGACTAATAAATTATCATTCAGCACACCTGTATCATCATC |
| TRAPPC3-Fp | CCATAGCTATAGAGATGGGAATGCAGAGTAAAGCTTCAAGTGTTCATTGATAACCAAAACTGGGTCAAAAATGTCGAGGCAGGCGAACCGTG |
| TRAPPC3-Rp | AAATAATGTATACCTAAATAACGACACCAATAATAACAAAAATCACGAAAAAAAAAAGCTTACATGTCCATTATTCCTCTCCAGCTGGAAG |
| TRAPPC4-Fp | AAAAGGAATCTGCCTTTGCATAAGTTCAAAAGTGCAATTTTAGTGTTGGATTTAAACGGGAAAAATTGAAATGGCGATTTTTAGTGTGTATG |
| TRAPPC4-Rp | CTTAGTTCTAGAAGTACGTAGTATTTATTTTCTTGGTGTGAATGCGTTTATCTTCCGATGGCGCGTGCGTCTATGACCCAGGTCCAAAAG |
| TRAPPC5-Fp | AGGACAAGAGACACGGAAGCGACAGAAAGACAGGGAGTACTATCACCATCCATAATACTGGAGTAACATTATGGAGGCGCGCTTCACGCGC |
| TRAPPC5-Rp | TCAACAGTAAATATACAAACTCTTTATCCTTTTCTTGGTTTTAGGTATTTACCGATGTTTTGAGATGATATCAGCGGCCCTCCAGGGCCCG |
| TRAPPC6A-Fp | AGCTGAGAGTGAAGACCTCATGCCATAAAAGAAAACATACACCGGTTATAAAGTTGGAAGATAAACAATTATGGCGGATACTGTGTTGTTTG |
| TRAPPC6A-Rp | TCTTTTATATACACACTTATATCTATCTATATGTCGATGTACATTCTTAGAACAAAAATCTGTCGGACCTTTAGGATTTCGGAATCACCACC |
| TRAPPC6B-Fp | AGCTGAGAGTGAAGACCTCATGCCATAAAAGAAAACATACACCGGTTATAAAGTTGGAAGATAAACAATTATGGCGGATGAGGCGTTGTTTTTG |
| TRAPPC6B-Rp | TCTTTTATATACACACTTATATCTATCTATATGTCGATGTACATTCTTAGAACAAAAATCTGTCGGACCTCTACAGCTTCTGTATCATCAC |
| Primer Name | Sequence |
|---|---|
| TRAPPC1-Fp | GTCCTTTACTATTGACTAGTG |
| TRAPPC1-Rp | CATCCCGTACATCAGCTTATAC |
| TRAPPC2-Fp | CCCTTTACCTCTTCAAAGCCT |
| TRAPPC2-Rp | AAGTACATGTTGTTCGATAGC |
| TRAPPC2L-Fp | AAGTTCTGCCGGAAGAGCTCATC |
| TRAPPC2L-Rp | ATTGCGGAGATCTTCTCATCC |
| TRAPPC6A-Fp | GGAATCTGGTATGCACTCAC |
| TRAPPC6A-Rp | TGTTGTCTTGCAGGACGTAG |
| TRAPPC6B-Fp | GGAATCTGGTATGCACTCAC |
| TRAPPC6B-Rp | AACTCATCCTTGAACCTTGCAG |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zykaj, E.; Abboud, C.; Asadi, P.; Warsame, S.; Almousa, H.; Milev, M.P.; Greco, B.M.; López-Sánchez, M.; Bratkovic, D.; Kachroo, A.H.; et al. A Humanized Yeast Model for Studying TRAPP Complex Mutations; Proof-of-Concept Using Variants from an Individual with a TRAPPC1-Associated Neurodevelopmental Syndrome. Cells 2024, 13, 1457. https://doi.org/10.3390/cells13171457
Zykaj E, Abboud C, Asadi P, Warsame S, Almousa H, Milev MP, Greco BM, López-Sánchez M, Bratkovic D, Kachroo AH, et al. A Humanized Yeast Model for Studying TRAPP Complex Mutations; Proof-of-Concept Using Variants from an Individual with a TRAPPC1-Associated Neurodevelopmental Syndrome. Cells. 2024; 13(17):1457. https://doi.org/10.3390/cells13171457
Chicago/Turabian StyleZykaj, Erta, Chelsea Abboud, Paria Asadi, Simane Warsame, Hashem Almousa, Miroslav P. Milev, Brittany M. Greco, Marcos López-Sánchez, Drago Bratkovic, Aashiq H. Kachroo, and et al. 2024. "A Humanized Yeast Model for Studying TRAPP Complex Mutations; Proof-of-Concept Using Variants from an Individual with a TRAPPC1-Associated Neurodevelopmental Syndrome" Cells 13, no. 17: 1457. https://doi.org/10.3390/cells13171457
APA StyleZykaj, E., Abboud, C., Asadi, P., Warsame, S., Almousa, H., Milev, M. P., Greco, B. M., López-Sánchez, M., Bratkovic, D., Kachroo, A. H., Pérez-Jurado, L. A., & Sacher, M. (2024). A Humanized Yeast Model for Studying TRAPP Complex Mutations; Proof-of-Concept Using Variants from an Individual with a TRAPPC1-Associated Neurodevelopmental Syndrome. Cells, 13(17), 1457. https://doi.org/10.3390/cells13171457







