The Nuclear NF-κB Regulator IκBζ: Updates on Its Molecular Functions and Pathophysiological Roles
Abstract
:1. Introduction
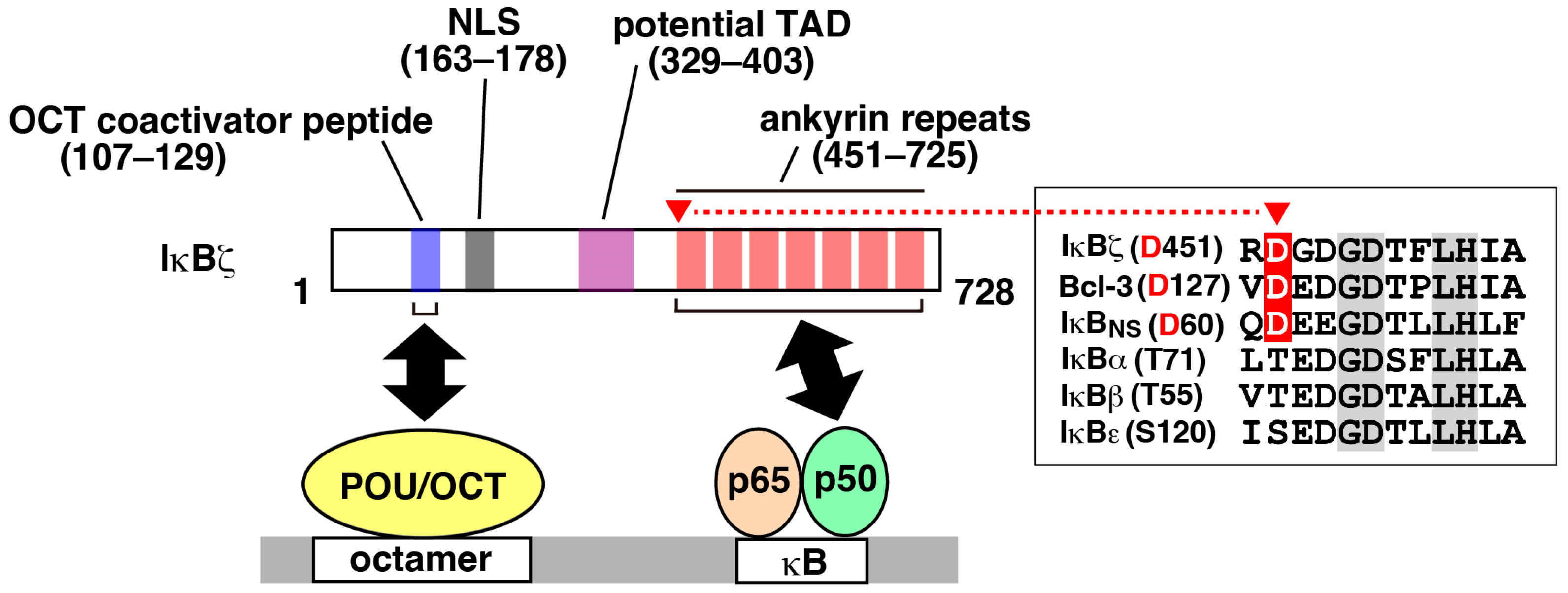
2. Identification, Structure, and Expression Properties
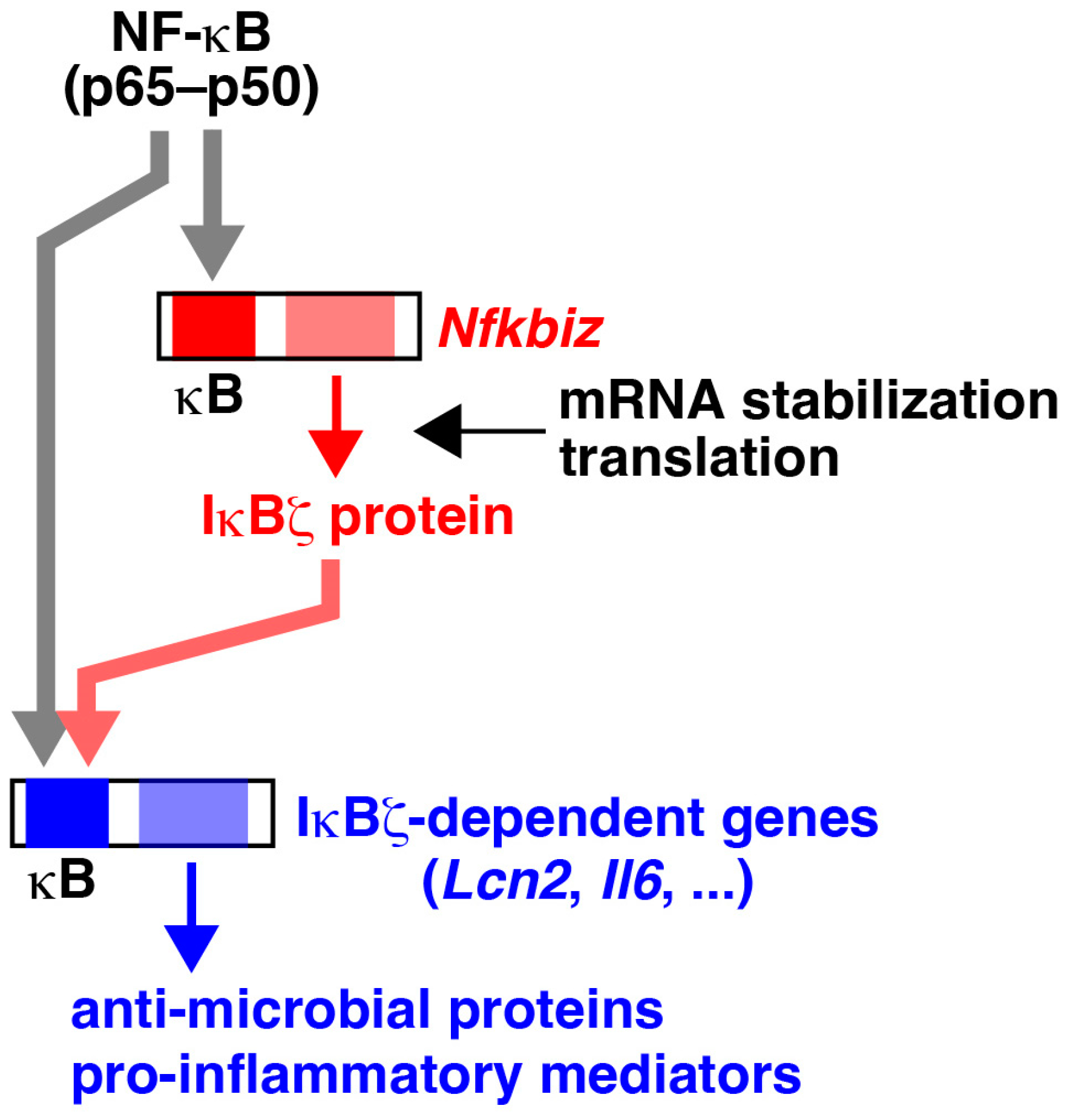
3. Molecular Functions
4. Cell Type-Specific Roles of IκBζ and Associations with Human Diseases
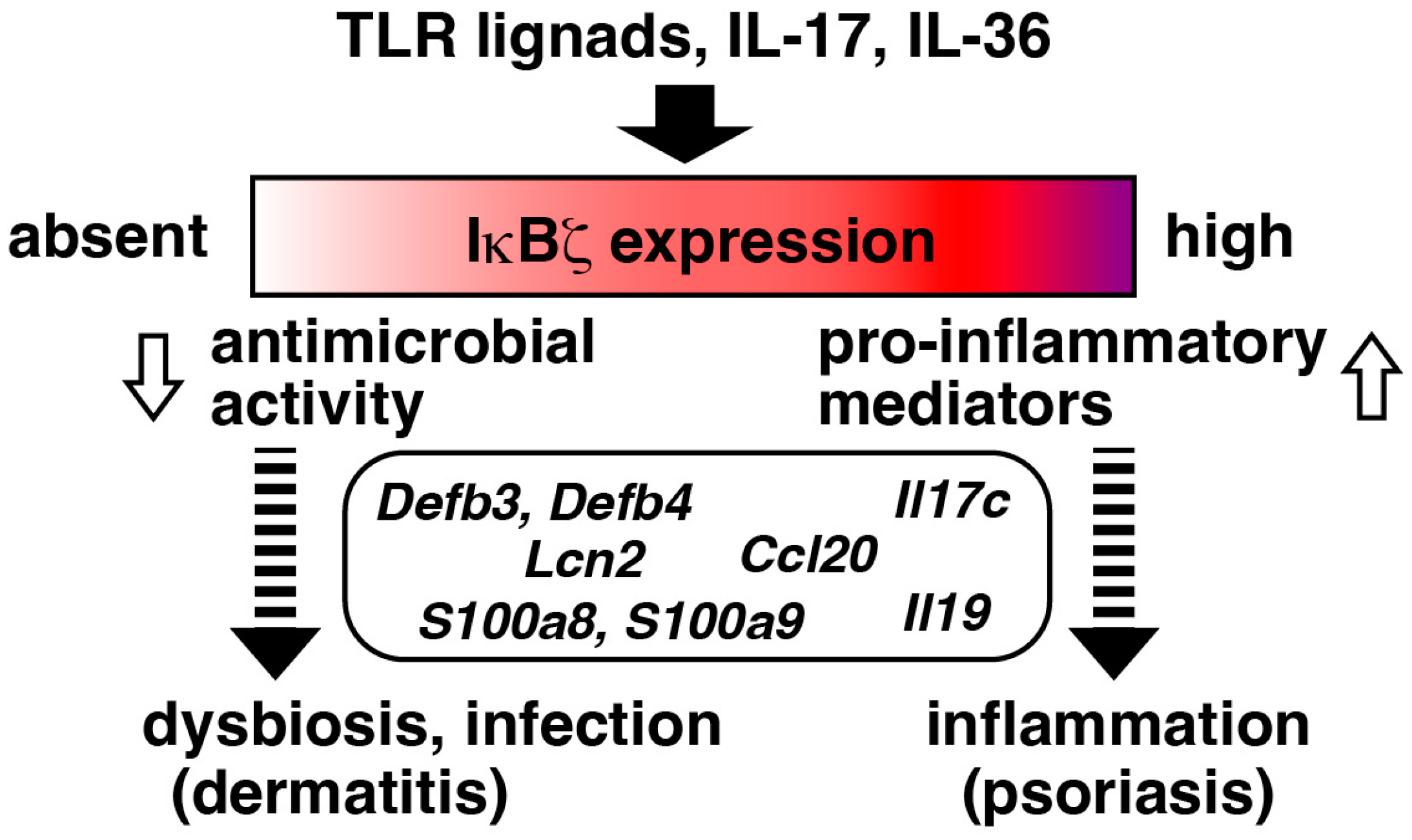
4.1. Keratinocytes
4.2. Intestinal Epithelial Cells
4.3. Oral Epithelial Cells
4.4. Natural Killer (NK) Cells
4.5. Chondrocytes
4.6. Other Types of Cells
| Tissue (Cre Driver) | Phenotypes |
|---|---|
| Epithelial cells (KRT5-Cre) | Inflammation in facial skin, dacryoadenitis similar to human Sjögren’s syndrome [23], reduced production of antimicrobial proteins, and susceptible to infection by Staphylococcus aureus [65]. |
| Keratinocytes (KRT14-Cre) | Resistance to imiquimod-induced psoriasis model [72]. |
| Intestinal epithelial cells (Vil1-Cre) | Reduced expression of anti-microbial proteins, expansion of SFB, and increase in Th17 cells [58]. |
| Oral epithelial cells (Krt13-Cre) | Increased susceptibility to oropharyngeal candidiasis [81]. |
| Chondrocytes (Col2a1-Cre) | Downregulation of matrix-degrading enzymes and alleviation of osteoarthritis model [85]. |
| Hepatocytes (Alb-Cre) | Facilitated progression of nonalcoholic fatty liver disease (NAFLD) [86]. |
| T cells (Lck-Cre) | Increased production of IFN-γ [88]. |
| B cells (Cd79a/Mb1-Cre) | Impairment of T cell-dependent type 1 antibody response and Reduction in the expression of activation-induced cytidine deaminase (AID) [89]. |
5. Discussion and Perspectives
Funding
Conflicts of Interest
References
- Hayden, M.S.; Ghosh, S. NF-κB, the first quarter-century: Remarkable progress and outstanding questions. Genes Dev. 2012, 26, 203–234. [Google Scholar] [CrossRef]
- Hinz, M.; Arslan, S.; Scheidereit, C. It takes two to tango: IκBs, the multifunctional partners of NF-κB. Immunol. Rev. 2012, 246, 59–76. [Google Scholar] [CrossRef]
- Kitamura, H.; Kanehira, K.; Okita, K.; Morimatsu, M.; Saito, M. MAIL, a novel nuclear IκB protein that potentiates LPS-induced IL-6 production. FEBS Lett. 2000, 485, 53–56. [Google Scholar] [CrossRef] [PubMed]
- Haruta, H.; Kato, A.; Todokoro, K. Isolation of a novel interleukin-1-inducible nuclear protein bearing ankyrin-repeat motifs. J. Biol. Chem. 2001, 276, 12485–12488. [Google Scholar] [CrossRef] [PubMed]
- Yamazaki, S.; Muta, T.; Takeshige, K. A novel IκB prsotein, IκB-ζ, induced by proinflammatory stimuli, negatively regulates nuclear factor-κB in the nuclei. J. Biol. Chem. 2001, 276, 27657–27662. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, S.; May, M.J.; Kopp, E.B. NF-κB and Rel proteins: Evolutionarily conserved mediators of immune responses. Annu. Rev. Immunol. 1998, 16, 225–260. [Google Scholar] [CrossRef]
- Wulczyn, F.G.; Naumann, M.; Scheidereit, C. Candidate proto-oncogene bcl-3 encodes a subunit-specific inhibitor of transcription factor NF-κB. Nature 1992, 358, 597–599. [Google Scholar] [CrossRef]
- Franzoso, G.; Bours, V.; Park, S.; Tomita-Yamaguchi, M.; Kelly, K.; Siebenlist, U. The candidate oncoprotein Bcl-3 is an antagonist of p50/NF-κB-mediated inhibition. Nature 1992, 359, 339–342. [Google Scholar] [CrossRef]
- Fiorini, E.; Schmitz, I.; Marissen, W.E.; Osborn, S.L.; Touma, M.; Sasada, T.; Reche, P.A.; Tibaldi, E.V.; Hussey, R.E.; Kruisbeek, A.M.; et al. Peptide-induced negative selection of thymocytes activates transcription of an NF-κB inhibitor. Mol. Cell 2002, 9, 637–648. [Google Scholar] [CrossRef]
- Yamamoto, M.; Yamazaki, S.; Uematsu, S.; Sato, S.; Hemmi, H.; Hoshino, K.; Kaisho, T.; Kuwata, H.; Takeuchi, O.; Takeshige, K.; et al. Regulation of Toll/IL-1-receptor-mediated gene expression by the inducible nuclear protein IκBζ. Nature 2004, 430, 218–222. [Google Scholar] [CrossRef]
- Yamamoto, M.; Takeda, K. Role of nuclear IκB proteins in the regulation of host immune responses. J. Infect. Chemother. 2008, 14, 265–269. [Google Scholar] [CrossRef]
- Kohda, A.; Yamazaki, S.; Sumimoto, H. The Nuclear Protein IκBζ Forms a Transcriptionally Active Complex with Nuclear Factor-κB (NF-κB) p50 and the Lcn2 Promoter via the N- and C-terminal Ankyrin Repeat Motifs. J. Biol. Chem. 2016, 291, 20739–20752. [Google Scholar] [CrossRef] [PubMed]
- Eto, A.; Muta, T.; Yamazaki, S.; Takeshige, K. Essential roles for NF-κB and a Toll/IL-1 receptor domain-specific signal(s) in the induction of IκB-ζ. Biochem. Biophys. Res. Commun. 2003, 301, 495–501. [Google Scholar] [CrossRef] [PubMed]
- Yamazaki, S.; Muta, T.; Matsuo, S.; Takeshige, K. Stimulus-specific induction of a novel nuclear factor-κB regulator, IκB-ζ, via Toll/Interleukin-1 receptor is mediated by mRNA stabilization. J. Biol. Chem. 2005, 280, 1678–1687. [Google Scholar] [CrossRef] [PubMed]
- Adachi, O.; Kawai, T.; Takeda, K.; Matsumoto, M.; Tsutsui, H.; Sakagami, M.; Nakanishi, K.; Akira, S. Targeted disruption of the MyD88 gene results in loss of IL-1- and IL-18-mediated function. Immunity 1998, 9, 143–150. [Google Scholar] [CrossRef] [PubMed]
- Kawai, T.; Takeuchi, O.; Fujita, T.; Inoue, J.; Mühlradt, P.F.; Sato, S.; Hoshino, K.; Akira, S. Lipopolysaccharide stimulates the MyD88-independent pathway and results in activation of IFN-regulatory factor 3 and the expression of a subset of lipopolysaccharide-inducible genes. J. Immunol. 2001, 167, 5887–5894. [Google Scholar] [CrossRef]
- MaruYama, T.; Sayama, A.; Ishii, K.J.; Muta, T. Screening of posttranscriptional regulatory molecules of IκB-ζ. Biochem. Biophys. Res. Commun. 2016, 469, 711–715. [Google Scholar] [CrossRef]
- Shen, F.; Ruddy, M.J.; Plamondon, P.; Gaffen, S.L. Cytokines link osteoblasts and inflammation: Microarray analysis of interleukin-17- and TNF-alpha-induced genes in bone cells. J. Leukoc. Biol. 2005, 77, 388–399. [Google Scholar] [CrossRef]
- Li, X.; Bechara, R.; Zhao, J.; McGeachy, M.J.; Gaffen, S.L. IL-17 receptor-based signaling and implications for disease. Nat. Immunol. 2019, 20, 1594–1602. [Google Scholar] [CrossRef]
- Novatchkova, M.; Leibbrandt, A.; Werzowa, J.; Neubüser, A.; Eisenhaber, F. The STIR-domain superfamily in signal transduction, development and immunity. Trends Biochem. Sci. 2003, 28, 226–229. [Google Scholar] [CrossRef]
- Schwandner, R.; Yamaguchi, K.; Cao, Z. Requirement of tumor necrosis factor receptor-associated factor (TRAF)6 in interleukin 17 signal transduction. J. Exp. Med. 2000, 191, 1233–1240. [Google Scholar] [CrossRef] [PubMed]
- Ohba, T.; Ariga, Y.; Maruyama, T.; Truong, N.K.; Inoue, J.; Muta, T. Identification of interleukin-1 receptor-associated kinase 1 as a critical component that induces post-transcriptional activation of IκB-ζ. FEBS J. 2012, 279, 211–222. [Google Scholar] [CrossRef]
- Okuma, A.; Hoshino, K.; Ohba, T.; Fukushi, S.; Aiba, S.; Akira, S.; Ono, M.; Kaisho, T.; Muta, T. Enhanced apoptosis by disruption of the STAT3-IκB-ζ signaling pathway in epithelial cells induces Sjögren’s syndrome-like autoimmune disease. Immunity 2013, 38, 450–460. [Google Scholar] [CrossRef]
- Muromoto, R.; Tawa, K.; Ohgakiuchi, Y.; Sato, A.; Saino, Y.; Hirashima, K.; Minoguchi, H.; Kitai, Y.; Kashiwakura, J.I.; Shimoda, K.; et al. IκB-ζ Expression Requires Both TYK2/STAT3 Activity and IL-17-Regulated mRNA Stabilization. Immunohorizons 2019, 3, 172–185. [Google Scholar] [CrossRef]
- Muromoto, R.; Sato, A.; Komori, Y.; Nariya, K.; Kitai, Y.; Kashiwakura, J.I.; Matsuda, T. Regulation of NFKBIZ gene promoter activity by STAT3, C/EBPβ, and STAT1. Biochem. Biophys. Res. Commun. 2022, 613, 61–66. [Google Scholar] [CrossRef]
- Dhamija, S.; Doerrie, A.; Winzen, R.; Dittrich-Breiholz, O.; Taghipour, A.; Kuehne, N.; Kracht, M.; Holtmann, H. IL-1-induced post-transcriptional mechanisms target overlapping translational silencing and destabilizing elements in IκBζ mRNA. J. Biol. Chem. 2010, 285, 29165–29178. [Google Scholar] [CrossRef] [PubMed]
- Amatya, N.; Childs, E.E.; Cruz, J.A.; Aggor, F.E.Y.; Garg, A.V.; Berman, A.J.; Gudjonsson, J.E.; Atasoy, U.; Gaffen, S.L. IL-17 integrates multiple self-reinforcing, feed-forward mechanisms through the RNA binding protein Arid5a. Sci. Signal 2018, 11, eaat4617. [Google Scholar] [CrossRef] [PubMed]
- Jeltsch, K.M.; Hu, D.; Brenner, S.; Zöller, J.; Heinz, G.A.; Nagel, D.; Vogel, K.U.; Rehage, N.; Warth, S.C.; Edelmann, S.L.; et al. Cleavage of roquin and regnase-1 by the paracaspase MALT1 releases their cooperatively repressed targets to promote T(H)17 differentiation. Nat. Immunol. 2014, 15, 1079–1089. [Google Scholar] [CrossRef]
- Garg, A.V.; Amatya, N.; Chen, K.; Cruz, J.A.; Grover, P.; Whibley, N.; Conti, H.R.; Hernandez Mir, G.; Sirakova, T.; Childs, E.C.; et al. MCPIP1 Endoribonuclease Activity Negatively Regulates Interleukin-17-Mediated Signaling and Inflammation. Immunity 2015, 43, 475–487. [Google Scholar] [CrossRef]
- Mino, T.; Murakawa, Y.; Fukao, A.; Vandenbon, A.; Wessels, H.H.; Ori, D.; Uehata, T.; Tartey, S.; Akira, S.; Suzuki, Y.; et al. Regnase-1 and Roquin Regulate a Common Element in Inflammatory mRNAs by Spatiotemporally Distinct Mechanisms. Cell 2015, 161, 1058–1073. [Google Scholar] [CrossRef]
- Behrens, G.; Winzen, R.; Rehage, N.; Dörrie, A.; Barsch, M.; Hoffmann, A.; Hackermüller, J.; Tiedje, C.; Heissmeyer, V.; Holtmann, H. A translational silencing function of MCPIP1/Regnase-1 specified by the target site context. Nucleic Acids Res. 2018, 46, 4256–4270. [Google Scholar] [CrossRef]
- Uehata, T.; Yamada, S.; Ori, D.; Vandenbon, A.; Giladi, A.; Jelinski, A.; Murakawa, Y.; Watanabe, H.; Takeuchi, K.; Toratani, K.; et al. Regulation of lymphoid-myeloid lineage bias through regnase-1/3-mediated control of Nfkbiz. Blood 2024, 143, 243–257. [Google Scholar] [CrossRef] [PubMed]
- Sønder, S.U.; Saret, S.; Tang, W.; Sturdevant, D.E.; Porcella, S.F.; Siebenlist, U. IL-17-induced NF-κB activation via CIKS/Act1: Physiologic significance and signaling mechanisms. J. Biol. Chem. 2011, 286, 12881–12890. [Google Scholar] [CrossRef] [PubMed]
- Dhamija, S.; Winzen, R.; Doerrie, A.; Behrens, G.; Kuehne, N.; Schauerte, C.; Neumann, E.; Dittrich-Breiholz, O.; Kracht, M.; Holtmann, H. Interleukin-17 (IL-17) and IL-1 activate translation of overlapping sets of mRNAs, including that of the negative regulator of inflammation, MCPIP1. J. Biol. Chem. 2013, 288, 19250–19259. [Google Scholar] [CrossRef] [PubMed]
- Lindenblatt, C.; Schulze-Osthoff, K.; Totzke, G. IκBζ expression is regulated by miR-124a. Cell Cycle 2009, 8, 2019–2023. [Google Scholar] [CrossRef] [PubMed]
- Rossato, M.; Curtale, G.; Tamassia, N.; Castellucci, M.; Mori, L.; Gasperini, S.; Mariotti, B.; De Luca, M.; Mirolo, M.; Cassatella, M.A.; et al. IL-10-induced microRNA-187 negatively regulates TNF-α, IL-6, and IL-12p40 production in TLR4-stimulated monocytes. Proc. Natl. Acad. Sci. USA 2012, 109, E3101–E3110. [Google Scholar] [CrossRef]
- Liu, Z.; Tang, C.; He, L.; Yang, D.; Cai, J.; Zhu, J.; Shu, S.; Liu, Y.; Yin, L.; Chen, G.; et al. The negative feedback loop of NF-κB/miR-376b/NFKBIZ in septic acute kidney injury. JCI Insight 2020, 5, e142272. [Google Scholar] [CrossRef]
- Michael, D.; Feldmesser, E.; Gonen, C.; Furth, N.; Maman, A.; Heyman, O.; Argoetti, A.; Tofield, A.; Baichman-Kass, A.; Ben-Dov, A.; et al. miR-4734 conditionally suppresses ER stress-associated proinflammatory responses. FEBS Lett. 2023, 597, 1233–1245. [Google Scholar] [CrossRef]
- Feng, Y.; Chen, Z.; Xu, Y.; Han, Y.; Jia, X.; Wang, Z.; Zhang, N.; Lv, W. The central inflammatory regulator IκBζ: Induction, regulation and physiological functions. Front. Immunol. 2023, 14, 1188253. [Google Scholar] [CrossRef]
- Bambouskova, M.; Gorvel, L.; Lampropoulou, V.; Sergushichev, A.; Loginicheva, E.; Johnson, K.; Korenfeld, D.; Mathyer, M.E.; Kim, H.; Huang, L.H.; et al. Electrophilic properties of itaconate and derivatives regulate the IκBζ-ATF3 inflammatory axis. Nature 2018, 556, 501–504. [Google Scholar] [CrossRef]
- Swain, A.; Bambouskova, M.; Kim, H.; Andhey, P.S.; Duncan, D.; Auclair, K.; Chubukov, V.; Simons, D.M.; Roddy, T.P.; Stewart, K.M.; et al. Comparative evaluation of itaconate and its derivatives reveals divergent inflammasome and type I interferon regulation in macrophages. Nat. Metab. 2020, 2, 594–602. [Google Scholar] [CrossRef] [PubMed]
- Makhezer, N.; Ben Khemis, M.; Liu, D.; Khichane, Y.; Marzaioli, V.; Tlili, A.; Mojallali, M.; Pintard, C.; Letteron, P.; Hurtado-Nedelec, M.; et al. NOX1-derived ROS drive the expression of Lipocalin-2 in colonic epithelial cells in inflammatory conditions. Mucosal. Immunol. 2019, 12, 117–131. [Google Scholar] [CrossRef]
- Arra, M.; Swarnkar, G.; Ke, K.; Otero, J.E.; Ying, J.; Duan, X.; Maruyama, T.; Rai, M.F.; O’Keefe, R.J.; Mbalaviele, G.; et al. LDHA-mediated ROS generation in chondrocytes is a potential therapeutic target for osteoarthritis. Nat. Commun. 2020, 11, 3427. [Google Scholar] [CrossRef] [PubMed]
- Kimura, A.; Kitajima, M.; Nishida, K.; Serada, S.; Fujimoto, M.; Naka, T.; Fujii-Kuriyama, Y.; Sakamato, S.; Ito, T.; Handa, H.; et al. NQO1 inhibits the TLR-dependent production of selective cytokines by promoting IκB-ζ degradation. J. Exp. Med. 2018, 215, 2197–2209. [Google Scholar] [CrossRef]
- He, Y.; Feng, D.; Hwang, S.; Mackowiak, B.; Wang, X.; Xiang, X.; Rodrigues, R.M.; Fu, Y.; Ma, J.; Ren, T.; et al. Interleukin-20 exacerbates acute hepatitis and bacterial infection by downregulating IκBζ target genes in hepatocytes. J. Hepatol. 2021, 75, 163–176. [Google Scholar] [CrossRef]
- Motoyama, M.; Yamazaki, S.; Eto-Kimura, A.; Takeshige, K.; Muta, T. Positive and negative regulation of nuclear factor-κB-mediated transcription by IκB-ζ, an inducible nuclear protein. J. Biol. Chem. 2005, 280, 7444–7451. [Google Scholar] [CrossRef] [PubMed]
- Totzke, G.; Essmann, F.; Pohlmann, S.; Lindenblatt, C.; Jänicke, R.U.; Schulze-Osthoff, K. A novel member of the IκB family, human IκB-ζ, inhibits transactivation of p65 and its DNA binding. J. Biol. Chem. 2006, 281, 12645–12654. [Google Scholar] [CrossRef] [PubMed]
- Cowland, J.B.; Muta, T.; Borregaard, N. IL-1β-specific up-regulation of neutrophil gelatinase-associated lipocalin is controlled by IκB-ζ. J. Immunol. 2006, 176, 5559–5566. [Google Scholar] [CrossRef]
- Matsuo, S.; Yamazaki, S.; Takeshige, K.; Muta, T. Crucial roles of binding sites for NF-κB and C/EBPs in IκB-ζ-mediated transcriptional activation. Biochem. J. 2007, 405, 605–615. [Google Scholar] [CrossRef]
- Yamazaki, S.; Akira, S.; Sumimoto, H. Glucocorticoid augments lipopolysaccharide-induced activation of the IκBζ-dependent genes encoding the anti-microbial glycoproteins lipocalin 2 and pentraxin 3. J. Biochem. 2015, 157, 399–410. [Google Scholar] [CrossRef]
- Kohda, A.; Yamazaki, S.; Sumimoto, H. DNA element downstream of the κB site in the Lcn2 promoter is required for transcriptional activation by IκBζ and NF-κB p50. Genes Cells 2014, 19, 620–628. [Google Scholar] [CrossRef] [PubMed]
- Kayama, H.; Ramirez-Carrozzi, V.R.; Yamamoto, M.; Mizutani, T.; Kuwata, H.; Iba, H.; Matsumoto, M.; Honda, K.; Smale, S.T.; Takeda, K. Class-specific regulation of pro-inflammatory genes by MyD88 pathways and IκBζ. J. Biol. Chem. 2008, 283, 12468–12477. [Google Scholar] [CrossRef] [PubMed]
- Yamazaki, S.; Matsuo, S.; Muta, T.; Yamamoto, M.; Akira, S.; Takeshige, K. Gene-specific requirement of a nuclear protein, IκB-ζ, for promoter association of inflammatory transcription regulators. J. Biol. Chem. 2008, 283, 32404–32411. [Google Scholar] [CrossRef]
- Tartey, S.; Matsushita, K.; Vandenbon, A.; Ori, D.; Imamura, T.; Mino, T.; Standley, D.M.; Hoffmann, J.A.; Reichhart, J.M.; Akira, S.; et al. Akirin2 is critical for inducing inflammatory genes by bridging IκB-ζ and the SWI/SNF complex. Embo. J. 2014, 33, 2332–2348. [Google Scholar] [CrossRef] [PubMed]
- Saccani, S.; Pantano, S.; Natoli, G. Two waves of nuclear factor κB recruitment to target promoters. J. Exp. Med. 2001, 193, 1351–1359. [Google Scholar] [CrossRef] [PubMed]
- Ramirez-Carrozzi, V.R.; Nazarian, A.A.; Li, C.C.; Gore, S.L.; Sridharan, R.; Imbalzano, A.N.; Smale, S.T. Selective and antagonistic functions of SWI/SNF and Mi-2β nucleosome remodeling complexes during an inflammatory response. Genes. Dev. 2006, 20, 282–296. [Google Scholar] [CrossRef]
- Yamazaki, S.; Takeshige, K. Protein synthesis inhibitors enhance the expression of mRNAs for early inducible inflammatory genes via mRNA stabilization. Biochim. Biophys. Acta 2008, 1779, 108–114. [Google Scholar] [CrossRef]
- Yamazaki, S.; Inohara, N.; Ohmuraya, M.; Tsuneoka, Y.; Yagita, H.; Katagiri, T.; Nishina, T.; Mikami, T.; Funato, H.; Araki, K.; et al. IκBζ controls IL-17-triggered gene expression program in intestinal epithelial cells that restricts colonization of SFB and prevents Th17-associated pathologies. Mucosal. Immunol. 2022, 15, 1321–1337. [Google Scholar] [CrossRef]
- Daly, A.E.; Yeh, G.; Soltero, S.; Smale, S.T. Selective regulation of a defined subset of inflammatory and immunoregulatory genes by an NF-κB p50-IκBζ pathway. Genes Dev. 2024. [CrossRef]
- Zhu, N.; Rogers, W.E.; Heidary, D.K.; Huxford, T. Structural and biochemical analyses of the nuclear IκBζ protein in complex with the NF-κB p50 homodimer. Genes Dev. 2024. [CrossRef]
- Alpsoy, A.; Wu, X.S.; Pal, S.; Klingbeil, O.; Kumar, P.; El Demerdash, O.; Nalbant, B.; Vakoc, C.R. IκBζ is a dual-use coactivator of NF-κB and POU transcription factors. Mol. Cell 2024, 84, 1149–1157.e7. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Nagahama, Y.; Singh, S.K.; Kozakai, Y.; Nabeshima, H.; Fukushima, K.; Tanaka, H.; Motooka, D.; Fukui, E.; Vivier, E.; et al. Deletion of the mRNA endonuclease Regnase-1 promotes NK cell anti-tumor activity via OCT2-dependent transcription of Ifng. Immunity 2024, 57, 1360–1377.e13. [Google Scholar] [CrossRef] [PubMed]
- Shiina, T.; Konno, A.; Oonuma, T.; Kitamura, H.; Imaoka, K.; Takeda, N.; Todokoro, K.; Morimatsu, M. Targeted disruption of MAIL, a nuclear IκB protein, leads to severe atopic dermatitis-like disease. J. Biol. Chem. 2004, 279, 55493–55498. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.; Lee, Y.S.; Yang, J.Y.; Lee, S.H.; Park, Y.Y.; Kweon, M.N. The resident pathobiont Staphylococcus xylosus in Nfkbiz-deficient skin accelerates spontaneous skin inflammation. Sci. Rep. 2017, 7, 6348. [Google Scholar] [CrossRef]
- Terui, H.; Yamasaki, K.; Wada-Irimada, M.; Onodera-Amagai, M.; Hatchome, N.; Mizuashi, M.; Yamashita, R.; Kawabe, T.; Ishii, N.; Abe, T.; et al. Staphylococcus aureus skin colonization promotes SLE-like autoimmune inflammation via neutrophil activation and the IL-23/IL-17 axis. Sci. Immunol. 2022, 7, eabm9811. [Google Scholar] [CrossRef] [PubMed]
- Tsoi, L.C.; Spain, S.L.; Ellinghaus, E.; Stuart, P.E.; Capon, F.; Knight, J.; Tejasvi, T.; Kang, H.M.; Allen, M.H.; Lambert, S.; et al. Enhanced meta-analysis and replication studies identify five new psoriasis susceptibility loci. Nat. Commun. 2015, 6, 7001. [Google Scholar] [CrossRef]
- Chiricozzi, A.; Guttman-Yassky, E.; Suárez-Fariñas, M.; Nograles, K.E.; Tian, S.; Cardinale, I.; Chimenti, S.; Krueger, J.G. Integrative responses to IL-17 and TNF-α in human keratinocytes account for key inflammatory pathogenic circuits in psoriasis. J. Investig. Dermatol. 2011, 131, 677–687. [Google Scholar] [CrossRef]
- Sanford, M.; McKeage, K. Secukinumab: First global approval. Drugs 2015, 75, 329–338. [Google Scholar] [CrossRef]
- Mease, P.J. Inhibition of interleukin-17, interleukin-23 and the TH17 cell pathway in the treatment of psoriatic arthritis and psoriasis. Curr. Opin. Rheumatol. 2015, 27, 127–133. [Google Scholar] [CrossRef] [PubMed]
- Bertelsen, T.; Ljungberg, C.; Litman, T.; Huppertz, C.; Hennze, R.; Rønholt, K.; Iversen, L.; Johansen, C. IκBζ is a key player in the antipsoriatic effects of secukinumab. J. Allergy Clin. Immunol. 2020, 145, 379–390. [Google Scholar] [CrossRef]
- Johansen, C.; Mose, M.; Ommen, P.; Bertelsen, T.; Vinter, H.; Hailfinger, S.; Lorscheid, S.; Schulze-Osthoff, K.; Iversen, L. IκBζ is a key driver in the development of psoriasis. Proc. Natl. Acad. Sci. USA 2015, 112, E5825–E5833. [Google Scholar] [CrossRef] [PubMed]
- Lorscheid, S.; Müller, A.; Löffler, J.; Resch, C.; Bucher, P.; Kurschus, F.C.; Waisman, A.; Schäkel, K.; Hailfinger, S.; Schulze-Osthoff, K.; et al. Keratinocyte-derived IκBζ drives psoriasis and associated systemic inflammation. JCI Insight 2019, 4, e130835. [Google Scholar] [CrossRef] [PubMed]
- Müller, A.; Hennig, A.; Lorscheid, S.; Grondona, P.; Schulze-Osthoff, K.; Hailfinger, S.; Kramer, D. IκBζ is a key transcriptional regulator of IL-36-driven psoriasis-related gene expression in keratinocytes. Proc. Natl. Acad. Sci. USA 2018, 115, 10088–10093. [Google Scholar] [CrossRef]
- Mandal, A.; Kumbhojkar, N.; Reilly, C.; Dharamdasani, V.; Ukidve, A.; Ingber, D.E.; Mitragotri, S. Treatment of psoriasis with NFKBIZ siRNA using topical ionic liquid formulations. Sci. Adv. 2020, 6, eabb6049. [Google Scholar] [CrossRef] [PubMed]
- Nanki, K.; Fujii, M.; Shimokawa, M.; Matano, M.; Nishikori, S.; Date, S.; Takano, A.; Toshimitsu, K.; Ohta, Y.; Takahashi, S.; et al. Somatic inflammatory gene mutations in human ulcerative colitis epithelium. Nature 2020, 577, 254–259. [Google Scholar] [CrossRef] [PubMed]
- Kakiuchi, N.; Yoshida, K.; Uchino, M.; Kihara, T.; Akaki, K.; Inoue, Y.; Kawada, K.; Nagayama, S.; Yokoyama, A.; Yamamoto, S.; et al. Frequent mutations that converge on the NFKBIZ pathway in ulcerative colitis. Nature 2020, 577, 260–265. [Google Scholar] [CrossRef]
- Ivanov, I.I.; Atarashi, K.; Manel, N.; Brodie, E.L.; Shima, T.; Karaoz, U.; Wei, D.; Goldfarb, K.C.; Santee, C.A.; Lynch, S.V.; et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell 2009, 139, 485–498. [Google Scholar] [CrossRef]
- Kumar, P.; Monin, L.; Castillo, P.; Elsegeiny, W.; Horne, W.; Eddens, T.; Vikram, A.; Good, M.; Schoenborn, A.A.; Bibby, K.; et al. Intestinal Interleukin-17 Receptor Signaling Mediates Reciprocal Control of the Gut Microbiota and Autoimmune Inflammation. Immunity 2016, 44, 659–671. [Google Scholar] [CrossRef]
- Hueber, W.; Sands, B.E.; Lewitzky, S.; Vandemeulebroecke, M.; Reinisch, W.; Higgins, P.D.; Wehkamp, J.; Feagan, B.G.; Yao, M.D.; Karczewski, M.; et al. Secukinumab, a human anti-IL-17A monoclonal antibody, for moderate to severe Crohn’s disease: Unexpected results of a randomised, double-blind placebo-controlled trial. Gut 2012, 61, 1693–1700. [Google Scholar] [CrossRef]
- Targan, S.R.; Feagan, B.; Vermeire, S.; Panaccione, R.; Melmed, G.Y.; Landers, C.; Li, D.; Russell, C.; Newmark, R.; Zhang, N.; et al. A Randomized, Double-Blind, Placebo-Controlled Phase 2 Study of Brodalumab in Patients with Moderate-to-Severe Crohn’s Disease. Am. J. Gastroenterol. 2016, 111, 1599–1607. [Google Scholar] [CrossRef]
- Taylor, T.C.; Coleman, B.M.; Arunkumar, S.P.; Dey, I.; Dillon, J.T.; Ponde, N.O.; Poholek, A.C.; Schwartz, D.M.; McGeachy, M.J.; Conti, H.R.; et al. IκBζ is an essential mediator of immunity to oropharyngeal candidiasis. Cell Host Microbe 2023, 31, 1700–1713.e4. [Google Scholar] [CrossRef]
- Miyake, T.; Satoh, T.; Kato, H.; Matsushita, K.; Kumagai, Y.; Vandenbon, A.; Tani, T.; Muta, T.; Akira, S.; Takeuchi, O. IκBζ is essential for natural killer cell activation in response to IL-12 and IL-18. Proc. Natl. Acad. Sci. USA 2010, 107, 17680–17685. [Google Scholar] [CrossRef]
- Kannan, Y.; Yu, J.; Raices, R.M.; Seshadri, S.; Wei, M.; Caligiuri, M.A.; Wewers, M.D. IκBζ augments IL-12- and IL-18-mediated IFN-γ production in human NK cells. Blood 2011, 117, 2855–2863. [Google Scholar] [CrossRef] [PubMed]
- Raices, R.M.; Kannan, Y.; Bellamkonda-Athmaram, V.; Seshadri, S.; Wang, H.; Guttridge, D.C.; Wewers, M.D. A novel role for IκBζ in the regulation of IFNγ production. PLoS ONE 2009, 4, e6776. [Google Scholar] [CrossRef]
- Choi, M.C.; MaruYama, T.; Chun, C.H.; Park, Y. Alleviation of Murine Osteoarthritis by Cartilage-Specific Deletion of IκBζ. Arthritis Rheumatol. 2018, 70, 1440–1449. [Google Scholar] [CrossRef] [PubMed]
- Ishikawa, H.; Hayakawa, M.; Baatartsogt, N.; Kakizawa, N.; Ohto-Ozaki, H.; Maruyama, T.; Miura, K.; Suzuki, K.; Rikiyama, T.; Ohmori, T. IκBζ regulates the development of nonalcoholic fatty liver disease through the attenuation of hepatic steatosis in mice. Sci. Rep. 2022, 12, 11634. [Google Scholar] [CrossRef] [PubMed]
- Okamoto, K.; Iwai, Y.; Oh-Hora, M.; Yamamoto, M.; Morio, T.; Aoki, K.; Ohya, K.; Jetten, A.M.; Akira, S.; Muta, T.; et al. IκBζ regulates T(H)17 development by cooperating with ROR nuclear receptors. Nature 2010, 464, 1381–1385. [Google Scholar] [CrossRef]
- MaruYama, T.; Kobayashi, S.; Ogasawara, K.; Yoshimura, A.; Chen, W.; Muta, T. Control of IFN-γ production and regulatory function by the inducible nuclear protein IκB-ζ in T cells. J. Leukoc. Biol. 2015, 98, 385–393. [Google Scholar] [CrossRef]
- Hanihara, F.; Takahashi, Y.; Okuma, A.; Ohba, T.; Muta, T. Transcriptional and post-transcriptional regulation of IκB-ζ upon engagement of the BCR, TLRs and FcγR. Int. Immunol. 2013, 25, 531–544. [Google Scholar] [CrossRef]
- Ohto-Ozaki, H.; Hayakawa, M.; Kamoshita, N.; Maruyama, T.; Tominaga, S.I.; Ohmori, T. Induction of IκBζ Augments Cytokine and Chemokine Production by IL-33 in Mast Cells. J. Immunol. 2020, 204, 2033–2042. [Google Scholar] [CrossRef]
- Martin-Oliva, D.; Aguilar-Quesada, R.; O’Valle, F.; Muñoz-Gámez, J.A.; Martínez-Romero, R.; García Del Moral, R.; Ruiz de Almodóvar, J.M.; Villuendas, R.; Piris, M.A.; Oliver, F.J. Inhibition of poly(ADP-ribose) polymerase modulates tumor-related gene expression, including hypoxia-inducible factor-1 activation, during skin carcinogenesis. Cancer Res. 2006, 66, 5744–5756. [Google Scholar] [CrossRef] [PubMed]
- Göransson, M.; Andersson, M.K.; Forni, C.; Ståhlberg, A.; Andersson, C.; Olofsson, A.; Mantovani, R.; Aman, P. The myxoid liposarcoma FUS-DDIT3 fusion oncoprotein deregulates NF-kappaB target genes by interaction with NFKBIZ. Oncogene 2009, 28, 270–278. [Google Scholar] [CrossRef] [PubMed]
- Nogai, H.; Wenzel, S.S.; Hailfinger, S.; Grau, M.; Kaergel, E.; Seitz, V.; Wollert-Wulf, B.; Pfeifer, M.; Wolf, A.; Frick, M.; et al. IκB-ζ controls the constitutive NF-κB target gene network and survival of ABC DLBCL. Blood 2013, 122, 2242–2250. [Google Scholar] [CrossRef] [PubMed]
- Willems, M.; Dubois, N.; Musumeci, L.; Bours, V.; Robe, P.A. IκBζ: An emerging player in cancer. Oncotarget 2016, 7, 66310–66322. [Google Scholar] [CrossRef]
- Cataisson, C.; Salcedo, R.; Michalowski, A.M.; Klosterman, M.; Naik, S.; Li, L.; Pan, M.J.; Sweet, A.; Chen, J.Q.; Kostecka, L.G.; et al. T-Cell Deletion of MyD88 Connects IL17 and IκBζ to RAS Oncogenesis. Mol. Cancer Res. 2019, 17, 1759–1773. [Google Scholar] [CrossRef]
- Xu, T.; Rao, T.; Yu, W.M.; Ning, J.Z.; Yu, X.; Zhu, S.M.; Yang, K.; Bai, T.; Cheng, F. Upregulation of NFKBIZ affects bladder cancer progression via the PTEN/PI3K/Akt signaling pathway. Int. J. Mol. Med. 2021, 47, 109. [Google Scholar] [CrossRef]
- Gautam, P.; Maenner, S.; Cailotto, F.; Reboul, P.; Labialle, S.; Jouzeau, J.Y.; Bourgaud, F.; Moulin, D. Emerging role of IκBζ in inflammation: Emphasis on psoriasis. Clin. Transl. Med. 2022, 12, e1032. [Google Scholar] [CrossRef]
- Johansen, C. IκBζ: A key protein in the pathogenesis of psoriasis. Cytokine 2016, 78, 20–21. [Google Scholar] [CrossRef]
- Johansen, C.; Bertelsen, T.; Ljungberg, C.; Mose, M.; Iversen, L. Characterization of TNF-α- and IL-17A-Mediated Synergistic Induction of DEFB4 Gene Expression in Human Keratinocytes through IκBζ. J. Investig. Dermatol. 2016, 136, 1608–1616. [Google Scholar] [CrossRef]
- Bertelsen, T.; Ljungberg, C.; Boye Kjellerup, R.; Iversen, L.; Johansen, C. IL-17F regulates psoriasis-associated genes through IκBζ. Exp. Dermatol. 2017, 26, 234–241. [Google Scholar] [CrossRef]
- Coto-Segura, P.; Gonzalez-Lara, L.; Gómez, J.; Eiris, N.; Batalla, A.; Gómez, C.; Requena, S.; Queiro, R.; Alonso, B.; Iglesias, S.; et al. NFKBIZ in Psoriasis: Assessing the association with gene polymorphisms and report of a new transcript variant. Hum. Immunol. 2017, 78, 435–440. [Google Scholar] [CrossRef] [PubMed]
- Bertelsen, T.; Iversen, L.; Johansen, C. The human IL-17A/F heterodimer regulates psoriasis-associated genes through IκBζ. Exp. Dermatol. 2018, 27, 1048–1052. [Google Scholar] [CrossRef] [PubMed]
- Coto-Segura, P.; González-Lara, L.; Batalla, A.; Eiris, N.; Queiro, R.; Coto, E. NFKBIZ and CW6 in Adalimumab Response among Psoriasis Patients: Genetic Association and Alternative Transcript Analysis. Mol. Diagn. Ther. 2019, 23, 627–633. [Google Scholar] [CrossRef] [PubMed]

| IκB Subfamily | Nuclear IκB | Cytoplasmic IκB |
|---|---|---|
| Members | IκBζ, Bcl-3, IκBNS | IκBα, IκBβ, IκBε |
| Intracellular distribution | Nucleus | Cytoplasm |
| NF-κB regulation | Modulation of NF-κB- mediated transcription | Inhibition of NF-κB nuclear translocation |
| Preference of binding to NF-κB subunit | p50 | p65, c-Rel |
| Basal expression | Very low | Consistent levels |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yamazaki, S. The Nuclear NF-κB Regulator IκBζ: Updates on Its Molecular Functions and Pathophysiological Roles. Cells 2024, 13, 1467. https://doi.org/10.3390/cells13171467
Yamazaki S. The Nuclear NF-κB Regulator IκBζ: Updates on Its Molecular Functions and Pathophysiological Roles. Cells. 2024; 13(17):1467. https://doi.org/10.3390/cells13171467
Chicago/Turabian StyleYamazaki, Soh. 2024. "The Nuclear NF-κB Regulator IκBζ: Updates on Its Molecular Functions and Pathophysiological Roles" Cells 13, no. 17: 1467. https://doi.org/10.3390/cells13171467





