Breast Cancer Stem Cells Upregulate IRF6 in Stromal Fibroblasts to Induce Stromagenesis
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. BCSC/Fibroblast Co-Culture
2.3. Lentiviral Production and Transduction
2.4. Flow Cytometry
2.5. Rapid MACE-Seq
2.6. RNA Preparation and qPCR
2.7. Immunofluorescence Staining
2.8. Immunohistochemistry
2.9. Protein Isolation and Western Blotting
2.10. Orthotopic Breast Cancer Xenografts
2.11. Statistical Analysis
3. Results
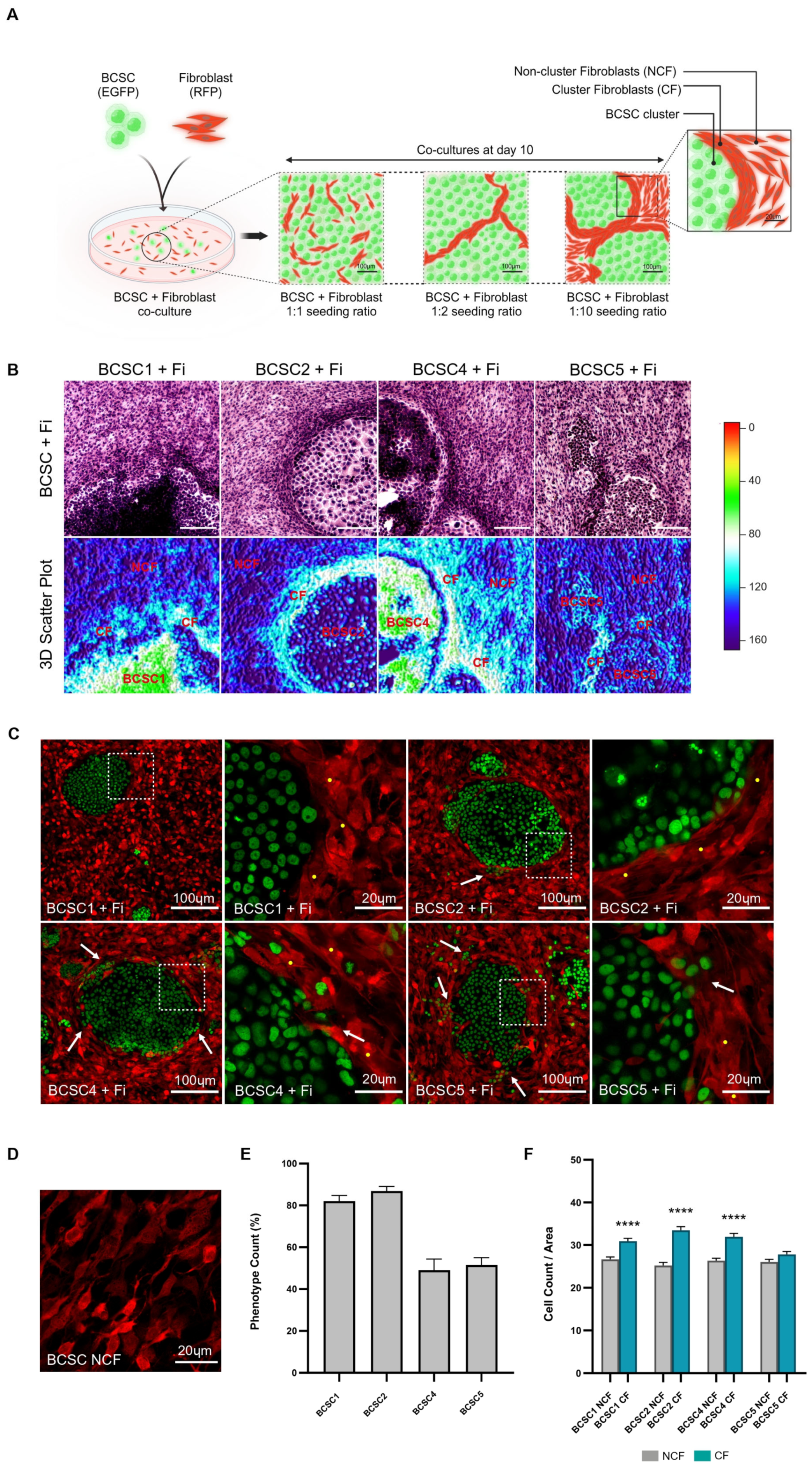
3.1. Fibroblasts Densely Aggregate along the Periphery of BCSC Clusters
3.2. CF Population Creates an ECM-Rich Region around BCSC Clusters
3.3. Gene Expression Profiling of CF Population Reveals Upregulation of CAF Characteristics and Genes Regulating Fibrosis
3.4. IRF6 Knockdown Fibroblasts Fail to Aggregate at the BCSC Cluster Periphery
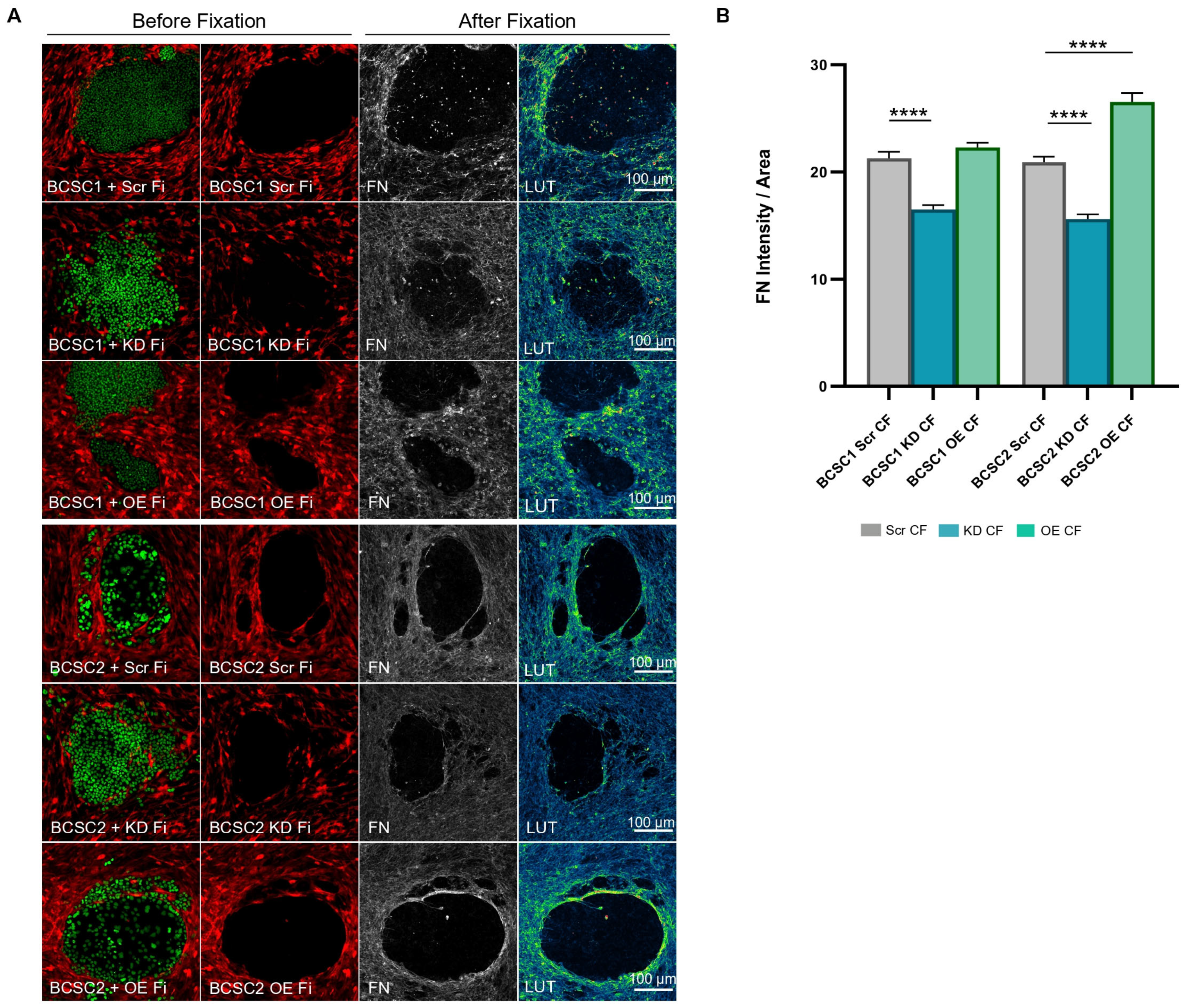
3.5. BCSC KD CF Show Reduced Fibronectin Deposition
3.6. IRF6 in Stromal Fibroblasts of Xenograft Models Regulate Stromal Development and Necrosis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| BCSC | Breast cancer stem cells |
| Fi | Fibroblast |
| BCSC + Fi | BCSC/fibroblast co-culture |
| CAF | Cancer-associated fibroblasts |
| CSC | Cancer stem cells |
| COL1A1 | Collagen type I alpha 1 chain |
| ECM | Extracellular matrix |
| FN | Fibronectin |
| DEG | Differentially expressed gene |
| GO | Gene ontology |
| IRF6 | Interferon regulatory factor 6 |
| CF | Cluster fibroblasts |
| NCF | Non-cluster fibroblasts |
| Co Fi | Control fibroblasts |
| KD Fi | Knockdown fibroblasts |
| OE Fi | Overexpression fibroblasts |
| Scr CF | Scrambled cluster fibroblasts |
| KD CF | Knockdown cluster fibroblasts |
| OE CF | Overexpression cluster fibroblasts |
| SPE | Stromal phenotype of encapsulation |
| TACS | Tumor-associated collagen signatures |
| TME | Tumor microenvironment |
| TNBC | Triple-negative breast cancer |
References
- Aleskandarany, M.A.; Vandenberghe, M.E.; Marchiò, C.; Ellis, I.O.; Sapino, A.; Rakha, E.A. Tumour Heterogeneity of Breast Cancer: From Morphology to Personalised Medicine. Pathobiology 2018, 85, 23–34. [Google Scholar] [CrossRef] [PubMed]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Abd El-Rehim, D.M.; Pinder, S.E.; Paish, C.E.; Bell, J.; Blamey, R.; Robertson, J.F.; Nicholson, R.I.; Ellis, I.O. Expression of luminal and basal cytokeratins in human breast carcinoma. J. Pathol. 2004, 203, 661–671. [Google Scholar] [CrossRef] [PubMed]
- Rakha, E.A.; Reis-Filho, J.S.; Ellis, I.O. Basal-like Breast Cancer: A Critical Review. J. Clin. Oncol. 2008, 26, 2568–2581. [Google Scholar] [CrossRef]
- Fulford, L.G.; Easton, D.F.; Reis-Filho, J.S.; Sofronis, A.; Gillett, C.E.; Lakhani, S.R.; Hanby, A. Specific morphological features predictive for the basal phenotype in grade 3 invasive ductal carcinoma of breast. Histopathology 2006, 49, 22–34. [Google Scholar] [CrossRef]
- Tsuda, H.; Takarabe, T.; Hasegawa, T.; Murata, T.; Hirohashi, S. Myoepithelial differentiation in high-grade invasive ductal carcinomas with large central acellular zones. Hum. Pathol. 1999, 30, 1134–1139. [Google Scholar] [CrossRef]
- Tsuda, H.; Takarabe, T.; Hasegawa, F.; Fukutomi, T.; Hirohashi, S. Large, Central Acellular Zones Indicating Myoepithelial Tumor Differentiation in High-Grade Invasive Ductal Carcinomas as Markers of Predisposition to Lung and Brain Metastases. Am. J. Surg. Pathol. 2000, 24, 197–202. [Google Scholar] [CrossRef]
- Takai, K.; Le, A.; Weaver, V.M.; Werb, Z. Targeting the cancer-associated fibroblasts as a treatment in triple-negative breast cancer. Oncotarget 2016, 7, 82889–82901. [Google Scholar] [CrossRef]
- Ding, J.-H.; Xiao, Y.; Zhao, S.; Xu, Y.; Xiao, Y.-L.; Shao, Z.-M.; Jiang, Y.-Z.; Di, G.-H. Integrated analysis reveals the molecular features of fibrosis in triple-negative breast cancer. Mol. Ther.—Oncolytics 2022, 24, 624–635. [Google Scholar] [CrossRef]
- O’Brien, C.A.; Kreso, A.; Jamieson CH, M. Cancer Stem Cells and Self-renewal. Clin. Cancer Res. 2010, 16, 3113–3120. [Google Scholar] [CrossRef]
- Honeth, G.; Bendahl, P.-O.; Ringnér, M.; Saal, L.H.; Gruvberger-Saal, S.K.; Lövgren, K.; Grabau, D.; Fernö, M.; Borg, Å.; Hegardt, C. The CD44+/CD24−phenotype is enriched in basal-like breast tumors. Breast Cancer Res. 2008, 10, R53. [Google Scholar] [CrossRef]
- Lawson, D.A.; Bhakta, N.R.; Kessenbrock, K.; Prummel, K.D.; Yu, Y.; Takai, K.; Zhou, A.; Eyob, H.; Balakrishnan, S.; Wang, C.-Y.; et al. Single-cell analysis reveals a stem-cell program in human metastatic breast cancer cells. Nature 2015, 526, 131–135. [Google Scholar] [CrossRef] [PubMed]
- Al-Hajj, M.; Wicha, M.S.; Benito-Hernandez, A.; Morrison, S.J.; Clarke, M.F. Prospective identification of tumorigenic breast cancer cells. Proc. Natl. Acad. Sci. USA 2003, 100, 3983–3988. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Cong, Y.; Wang, D.; Sun, Y.; Deng, L.; Liu, Y.; Martin-Trevino, R.; Shang, L.; McDermott, S.P.; Landis, M.D.; et al. Breast Cancer Stem Cells Transition between Epithelial and Mesenchymal States Reflective of their Normal Counterparts. Stem Cell Rep. 2014, 2, 78–91. [Google Scholar] [CrossRef] [PubMed]
- Malla, R.; Puvalachetty, K.; Vempati, R.K.; Marni, R.; Merchant, N.; Nagaraju, G.P. Cancer Stem Cells and Circulatory Tumor Cells Promote Breast Cancer Metastasis. Clin. Breast Cancer 2022, 22, 507–514. [Google Scholar] [CrossRef]
- Kai, K.; Arima, Y.; Kamiya, T.; Saya, H. Breast cancer stem cells. Breast Cancer 2010, 17, 80–85. [Google Scholar] [CrossRef]
- Velasco-Velázquez, M.A.; Popov, V.M.; Lisanti, M.P.; Pestell, R.G. The Role of Breast Cancer Stem Cells in Metastasis and Therapeutic Implications. Am. J. Pathol. 2011, 179, 2–11. [Google Scholar] [CrossRef]
- Vermeulen, L.; De Sousa EMelo, F.; van der Heijden, M.; Cameron, K.; de Jong, J.H.; Borovski, T.; Tuynman, J.B.; Todaro, M.; Merz, C.; Rodermond, H.; et al. Wnt activity defines colon cancer stem cells and is regulated by the microenvironment. Nat. Cell Biol. 2010, 12, 468–476. [Google Scholar] [CrossRef]
- Su, S.; Chen, J.; Yao, H.; Liu, J.; Yu, S.; Lao, L.; Wang, M.; Luo, M.; Xing, Y.; Chen, F.; et al. CD10+GPR77+ Cancer-Associated Fibroblasts Promote Cancer Formation and Chemoresistance by Sustaining Cancer Stemness. Cell 2018, 172, 841–856.e16. [Google Scholar] [CrossRef]
- Huang, M.; Li, Y.; Zhang, H.; Nan, F. Breast cancer stromal fibroblasts promote the generation of CD44+CD24− cells through SDF-1/CXCR4 interaction. J. Exp. Clin. Cancer Res. 2010, 29, 80. [Google Scholar] [CrossRef]
- Nallanthighal, S.; Heiserman, J.P.; Cheon, D.-J. The Role of the Extracellular Matrix in Cancer Stemness. Front. Cell Dev. Biol. 2019, 7, 86. [Google Scholar] [CrossRef]
- Ronnov-Jessen, L.; Petersen, O.W.; Bissell, M.J. Cellular changes involved in conversion of normal to malignant breast: Importance of the stromal reaction. Physiol. Rev. 1996, 76, 69–125. [Google Scholar] [CrossRef]
- De Vincenzo, A.; Belli, S.; Franco, P.; Telesca, M.; Iaccarino, I.; Botti, G.; Carriero, M.V.; Ranson, M.; Stoppelli, M.P. Paracrine recruitment and activation of fibroblasts by c-Myc expressing breast epithelial cells through the IGFs/IGF-1R axis. Int. J. Cancer 2019, 145, 2827–2839. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Balagam, R.; He, T.-F.; Lee, P.P.; Igoshin, O.A.; Levine, H. On the mechanism of long-range orientational order of fibroblasts. Proc. Natl. Acad. Sci. USA 2017, 114, 8974–8979. [Google Scholar] [CrossRef]
- Sappino, A.-P.; Skalli, O.; Jackson, B.; Schürch, W.; Gabbiani, G. Smooth-muscle differentiation in stromal cells of malignant and non-malignant breast tissues. Int. J. Cancer 1988, 41, 707–712. [Google Scholar] [CrossRef]
- Brown, L.F.; Guidi, A.J.; Schnitt, S.J.; Van De Water, L.; Iruela-Arispe, M.L.; Yeo, T.K.; Tognazzi, K.; Dvorak, H.F. Vascular stroma formation in carcinoma in situ, invasive carcinoma, and metastatic carcinoma of the breast. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 1999, 5, 1041–1056. [Google Scholar]
- Kalluri, R. The biology and function of fibroblasts in cancer. Nat. Rev. Cancer 2016, 16, 582–598. [Google Scholar] [CrossRef] [PubMed]
- Provenzano, P.P.; Inman, D.R.; Eliceiri, K.W.; Knittel, J.G.; Yan, L.; Rueden, C.T.; White, J.G.; Keely, P.J. Collagen density promotes mammary tumor initiation and progression. BMC Med. 2008, 6, 11. [Google Scholar] [CrossRef]
- Ahirwar, D.K.; Nasser, M.W.; Ouseph, M.M.; Elbaz, M.; Cuitiño, M.C.; Kladney, R.D.; Varikuti, S.; Kaul, K.; Satoskar, A.R.; Ramaswamy, B.; et al. Fibroblast-derived CXCL12 promotes breast cancer metastasis by facilitating tumor cell intravasation. Oncogene 2018, 37, 4428–4442. [Google Scholar] [CrossRef]
- Shani, O.; Vorobyov, T.; Monteran, L.; Lavie, D.; Cohen, N.; Raz, Y.; Tsarfaty, G.; Avivi, C.; Barshack, I.; Erez, N. Fibroblast-Derived IL33 Facilitates Breast Cancer Metastasis by Modifying the Immune Microenvironment and Driving Type 2 Immunity. Cancer Res. 2020, 80, 5317–5329. [Google Scholar] [CrossRef]
- Levental, K.R.; Yu, H.; Kass, L.; Lakins, J.N.; Egeblad, M.; Erler, J.T.; Fong, S.F.T.; Csiszar, K.; Giaccia, A.; Weninger, W.; et al. Matrix Crosslinking Forces Tumor Progression by Enhancing Integrin Signaling. Cell 2009, 139, 891–906. [Google Scholar] [CrossRef] [PubMed]
- Metzger, E.; Stepputtis, S.S.; Strietz, J.; Preca, B.-T.; Urban, S.; Willmann, D.; Allen, A.; Zenk, F.; Iovino, N.; Bronsert, P.; et al. KDM4 Inhibition Targets Breast Cancer Stem–like Cells. Cancer Res. 2017, 77, 5900–5912. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Strietz, J.; Bleilevens, A.; Stickeler, E.; Maurer, J. Chemotherapeutic Stress Influences Epithelial–Mesenchymal Transition and Stemness in Cancer Stem Cells of Triple-Negative Breast Cancer. Int. J. Mol. Sci. 2020, 21, 404. [Google Scholar] [CrossRef]
- Strietz, J.; Stepputtis, S.S.; Follo, M.; Bronsert, P.; Stickeler, E.; Maurer, J. Human Primary Breast Cancer Stem Cells Are Characterized by Epithelial-Mesenchymal Plasticity. Int. J. Mol. Sci. 2021, 22, 1808. [Google Scholar] [CrossRef]
- Ghassibe-Sabbagh, M.; El Hajj, J.; Al Saneh, M.; El Baba, N.; Abou Issa, J.; Al Haddad, M.; El Atat, O.; Sabbagh, J.; Abou Chebel, N.; El-Sibai, M. Altered regulation of cell migration in IRF6-mutated orofacial cleft patients-derived primary cells reveals a novel role of Rho GTPases in cleft/lip palate development. Cells Dev. 2021, 166, 203674. [Google Scholar] [CrossRef]
- Rhea, L.; Canady, F.J.; Le, M.; Reeb, T.; Canady, J.W.; Kacmarynski, D.S.F.; Avvari, R.; Biggs, L.C.; Dunnwald, M. Interferon regulatory factor 6 is required for proper wound healing in vivo. Dev. Dyn. 2020, 249, 509–522. [Google Scholar] [CrossRef] [PubMed]
- Jiang, W.; Hua, R.; Wei, M.; Li, C.; Qiu, Z.; Yang, X.; Zhang, C. An optimized method for high-titer lentivirus preparations without ultracentrifugation. Sci. Rep. 2015, 5, 13875. [Google Scholar] [CrossRef]
- Babraham Bioinformatics, FASTQC. Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 25 April 2024).
- Patro, R.; Duggal, G.; Love, M.I.; Irizarry, R.A.; Kingsford, C. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 2017, 14, 417–419. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. Feature Counts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- DICE Tools. Available online: https://tools.dice-database.org/GOnet/ (accessed on 26 April 2024).
- Harmonizome 3.0, Fibrosis Gene Set. Available online: https://maayanlab.cloud/Harmonizome/gene_set/Fibrosis/CTD+Gene-Disease+Associations (accessed on 26 April 2024).
- Wang, G.; Zhang, H.; Shen, X.; Jin, W.; Wang, X.; Zhou, Z. Characterization of cancer-associated fibroblasts (CAFs) and development of a CAF-based risk model for triple-negative breast cancer. Cancer Cell Int. 2023, 23, 294. [Google Scholar] [CrossRef] [PubMed]
- Nurmik, M.; Ullmann, P.; Rodriguez, F.; Haan, S.; Letellier, E. In search of definitions: Cancer-associated fibroblasts and their markers. Int. J. Cancer 2020, 146, 895–905. [Google Scholar] [CrossRef]
- Yeung, T.-L.; Leung, C.S.; Yip, K.-P.; Sheng, J.; Vien, L.; Bover, L.C.; Birrer, M.J.; Wong ST, C.; Mok, S.C. Anticancer Immunotherapy by MFAP5 Blockade Inhibits Fibrosis and Enhances Chemosensitivity in Ovarian and Pancreatic Cancer. Clin. Cancer Res. 2019, 25, 6417–6428. [Google Scholar] [CrossRef] [PubMed]
- Principe, S.; Mejia-Guerrero, S.; Ignatchenko, V.; Sinha, A.; Ignatchenko, A.; Shi, W.; Pereira, K.; Su, S.; Huang, S.H.; O’Sullivan, B.; et al. Proteomic Analysis of Cancer-Associated Fibroblasts Reveals a Paracrine Role for MFAP5 in Human Oral Tongue Squamous Cell Carcinoma. J. Proteome Res. 2018, 17, 2045–2059. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.; Piotrowska, Z.; Hare, P.J.; Chen, H.; Mulvey, H.E.; Mayfield, A.; Noeen, S.; Kattermann, K.; Greenberg, M.; Williams, A.; et al. Three subtypes of lung cancer fibroblasts define distinct therapeutic paradigms. Cancer Cell 2021, 39, 1531–1547.e10. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Sun, J.; Yang, Z.-F.; Zhou, C.; Zhou, P.-Y.; Guan, R.-Y.; Sun, B.-Y.; Wang, Z.-T.; Zhou, J.; Fan, J.; et al. Cancer-associated fibroblast-derived CXCL11 modulates hepatocellular carcinoma cell migration and tumor metastasis through the circUBAP2/miR-4756/IFIT1/3 axis. Cell Death Dis. 2021, 12, 260. [Google Scholar] [CrossRef]
- Morgan, A.; Griffin, M.; Kameni, L.; Wan, D.C.; Longaker, M.T.; Norton, J.A. Medical Biology of Cancer-Associated Fibroblasts in Pancreatic Cancer. Biology 2023, 12, 1044. [Google Scholar] [CrossRef]
- Sun, X.; Chen, Z. Cancer associated fibroblast derived CCL5 contributes to cisplatin resistance in A549 NSCLC cells partially through upregulation of lncRNA HOTAIR expression. Oncol. Lett. 2021, 22, 696. [Google Scholar] [CrossRef]
- Xu, H.; Zhao, J.; Li, J.; Zhu, Z.; Cui, Z.; Liu, R.; Lu, R.; Yao, Z.; Xu, Q. Cancer associated fibroblast–derived CCL5 promotes hepatocellular carcinoma metastasis through activating HIF1α/ZEB1 axis. Cell Death Dis. 2022, 13, 478. [Google Scholar] [CrossRef]
- Zhang, H.; Yue, J.; Jiang, Z.; Zhou, R.; Xie, R.; Xu, Y.; Wu, S. CAF-secreted CXCL1 conferred radioresistance by regulating DNA damage response in a ROS-dependent manner in esophageal squamous cell carcinoma. Cell Death Dis. 2017, 8, e2790. [Google Scholar] [CrossRef]
- Han, L.; Guo, X.; Du, R.; Guo, K.; Qi, P.; Bian, H. Identification of key genes and pathways related to cancer-associated fibroblasts in chemoresistance of ovarian cancer cells based on GEO and TCGA databases. J. Ovarian Res. 2022, 15, 75. [Google Scholar] [CrossRef] [PubMed]
- Sun, K.; Tang, S.; Hou, Y.; Xi, L.; Chen, Y.; Yin, J.; Peng, M.; Zhao, M.; Cui, X.; Liu, M. Oxidized ATM-mediated glycolysis enhancement in breast cancer-associated fibroblasts contributes to tumor invasion through lactate as metabolic coupling. EBioMedicine 2019, 41, 370–383. [Google Scholar] [CrossRef] [PubMed]
- Cords, L.; Tietscher, S.; Anzeneder, T.; Langwieder, C.; Rees, M.; de Souza, N.; Bodenmiller, B. Cancer-associated fibroblast classification in single-cell and spatial proteomics data. Nat. Commun. 2023, 14, 4294. [Google Scholar] [CrossRef] [PubMed]
- Bertero, T.; Oldham, W.M.; Grasset, E.M.; Bourget, I.; Boulter, E.; Pisano, S.; Hofman, P.; Bellvert, F.; Meneguzzi, G.; Bulavin, D.V.; et al. Tumor-Stroma Mechanics Coordinate Amino Acid Availability to Sustain Tumor Growth and Malignancy. Cell Metab. 2019, 29, 124–140.e10. [Google Scholar] [CrossRef]
- Botti, E.; Spallone, G.; Moretti, F.; Marinari, B.; Pinetti, V.; Galanti, S.; De Meo, P.D.; De Nicola, F.; Ganci, F.; Castrignanò, T.; et al. Developmental factor IRF6 exhibits tumor suppressor activity in squamous cell carcinomas. Proc. Natl. Acad. Sci. USA 2011, 108, 13710–13715. [Google Scholar] [CrossRef]
- Biggs, L.C.; Rhea, L.; Schutte, B.C.; Dunnwald, M. Interferon Regulatory Factor 6 Is Necessary, but Not Sufficient, for Keratinocyte Differentiation. J. Investig. Dermatol. 2012, 132, 50–58. [Google Scholar] [CrossRef]
- Richardson, R.J.; Dixon, J.; Malhotra, S.; Hardman, M.J.; Knowles, L.; Boot-Handford, R.P.; Shore, P.; Whitmarsh, A.; Dixon, M.J. Irf6 is a key determinant of the keratinocyte proliferation-differentiation switch. Nat. Genet. 2006, 38, 1329–1334. [Google Scholar] [CrossRef]
- Elwakeel, E.; Weigert, A. Breast Cancer CAFs: Spectrum of Phenotypes and Promising Targeting Avenues. Int. J. Mol. Sci. 2021, 22, 11636. [Google Scholar] [CrossRef]
- Tremblay, G. Stromal aspects of breast carcinoma. Exp. Mol. Pathol. 1979, 31, 248–260. [Google Scholar] [CrossRef]
- Huang, C.; Miyazaki, K.; Akaishi, S.; Watanabe, A.; Hyakusoku, H.; Ogawa, R. Biological effects of cellular stretch on human dermal fibroblasts. J. Plast. Reconstr. Aesthetic Surg. 2013, 66, e351–e361. [Google Scholar] [CrossRef]
- Lu, P.; Weaver, V.M.; Werb, Z. The extracellular matrix: A dynamic niche in cancer progression. J. Cell Biol. 2012, 196, 395–406. [Google Scholar] [CrossRef] [PubMed]
- Lagace, R.; Grimaud, J.-A.; Schürch, W.; Seemayer, T.A. Myofibroblastic stromal reaction in carcinoma of the breast: Variations of collagenous matrix and structural glycoproteins. Virchows Arch. A Pathol. Anat. Histopathol. 1985, 408, 49–59. [Google Scholar] [CrossRef] [PubMed]
- Evans, A.; Whelehan, P.; Thomson, K.; McLean, D.; Brauer, K.; Purdie, C.; Baker, L.; Jordan, L.; Rauchhaus, P.; Thompson, A. Invasive Breast Cancer: Relationship between Shear-wave Elastographic Findings and Histologic Prognostic Factors. Radiology 2012, 263, 673–677. [Google Scholar] [CrossRef]
- Provenzano, P.P.; Eliceiri, K.W.; Campbell, J.M.; Inman, D.R.; White, J.G.; Keely, P.J. Collagen reorganization at the tumor-stromal interface facilitates local invasion. BMC Med. 2006, 4, 38. [Google Scholar] [CrossRef] [PubMed]
- Reunanen, N.; Kähäri, V.M. Matrix Metalloproteinases in Cancer Cell Invasion. In Madame Curie Bioscience Database [Internet]. Austin (TX): Landes Bioscience; 2000–2013. Available online: https://www.ncbi.nlm.nih.gov/books/NBK6598/ (accessed on 23 May 2024).
- Yan, W.; Wu, X.; Zhou, W.; Fong, M.Y.; Cao, M.; Liu, J.; Liu, X.; Chen, C.-H.; Fadare, O.; Pizzo, D.P.; et al. Cancer-cell-secreted exosomal miR-105 promotes tumour growth through the MYC-dependent metabolic reprogramming of stromal cells. Nat. Cell Biol. 2018, 20, 597–609. [Google Scholar] [CrossRef]
- Mercier, I.; Casimiro, M.C.; Wang, C.; Rosenberg, A.L.; Quong, J.; Minkeu, A.; Allen, K.G.; Danilo, C.; Sotgia, F.; Bonuccelli, G.; et al. Human breast cancer-associated fibroblasts (CAFs) show caveolin-1 down-regulation and RB tumor suppressor functional inactivation: Implications for the response to hormonal therapy. Cancer Biol. Ther. 2008, 7, 1212–1225. [Google Scholar] [CrossRef]
- Barnes, B.J.; Kellum, M.J.; Field, A.E.; Pitha, P.M. Multiple Regulatory Domains of IRF-5 Control Activation, Cellular Localization, and Induction of Chemokines That Mediate Recruitment of T Lymphocytes. Mol. Cell. Biol. 2002, 22, 5721–5740. [Google Scholar] [CrossRef]
- Jing, T.; Zhao, B.; Xu, P.; Gao, X.; Chi, L.; Han, H.; Sankaran, B.; Li, P. The Structural Basis of IRF-3 Activation upon Phosphorylation. J. Immunol. 2020, 205, 1886–1896. [Google Scholar] [CrossRef]
- Bailey, C.M.; Abbott, D.E.; Margaryan, N.V.; Khalkhali-Ellis, Z.; Hendrix MJ, C. Interferon Regulatory Factor 6 Promotes Cell Cycle Arrest and Is Regulated by the Proteasome in a Cell Cycle-Dependent Manner. Mol. Cell. Biol. 2008, 28, 2235–2243. [Google Scholar] [CrossRef]
- Bailey, C.M.; Hendrix, M.J.C. IRF6 in development and disease: A mediator of quiescence and differentiation. Cell Cycle 2008, 7, 1925–1930. [Google Scholar] [CrossRef]
- Girousi, E.; Muerner, L.; Parisi, L.; Rihs, S.; von Gunten, S.; Katsaros, C.; Degen, M. Lack of IRF6 Disrupts Human Epithelial Homeostasis by Altering Colony Morphology, Migration Pattern, and Differentiation Potential of Keratinocytes. Front. Cell Dev. Biol. 2021, 9, 718066. [Google Scholar] [CrossRef] [PubMed]
- Varani, J.; Dame, M.K.; Rittie, L.; Fligiel SE, G.; Kang, S.; Fisher, G.J.; Voorhees, J.J. Decreased Collagen Production in Chronologically Aged Skin. Am. J. Pathol. 2006, 168, 1861–1868. [Google Scholar] [CrossRef] [PubMed]
- Quan, T.; Wang, F.; Shao, Y.; Rittié, L.; Xia, W.; Orringer, J.S.; Voorhees, J.J.; Fisher, G.J. Enhancing Structural Support of the Dermal Microenvironment Activates Fibroblasts, Endothelial Cells, and Keratinocytes in Aged Human Skin In Vivo. J. Investig. Dermatol. 2013, 133, 658–667. [Google Scholar] [CrossRef] [PubMed]
- Ao, M.; Brewer, B.M.; Yang, L.; Franco Coronel, O.E.; Hayward, S.W.; Webb, D.J.; Li, D. Stretching Fibroblasts Remodels Fibronectin and Alters Cancer Cell Migration. Scientific Reports 2015, 5, 8334. [Google Scholar] [CrossRef]
- Sahai, E.; Astsaturov, I.; Cukierman, E.; DeNardo, D.G.; Egeblad, M.; Evans, R.M.; Fearon, D.; Greten, F.R.; Hingorani, S.R.; Hunter, T.; et al. A framework for advancing our understanding of cancer-associated fibroblasts. Nat. Rev. Cancer 2020, 20, 174–186. [Google Scholar] [CrossRef] [PubMed]
- Kraman, M.; Bambrough, P.J.; Arnold, J.N.; Roberts, E.W.; Magiera, L.; Jones, J.O.; Gopinathan, A.; Tuveson, D.A.; Fearon, D.T. Suppression of Antitumor Immunity by Stromal Cells Expressing Fibroblast Activation Protein–α. Science 2010, 330, 827–830. [Google Scholar] [CrossRef] [PubMed]
- Barbazan, J.; Pérez-González, C.; Gómez-González, M.; Dedenon, M.; Richon, S.; Latorre, E.; Serra, M.; Mariani, P.; Descroix, S.; Sens, P.; et al. Cancer-associated fibroblasts actively compress cancer cells and modulate mechanotransduction. Nat. Commun. 2023, 14, 6966. [Google Scholar] [CrossRef]




| No. | Gene | Description | BCSC1 CF (FC) | BCSC1 CF (p-Value) | BCSC2 CF (FC) | BCSC2 CF (p-Value) |
|---|---|---|---|---|---|---|
| 1. | AP1M2 | Adaptor Related Protein Complex 1 Subunit Mu 2 | 6.4 | 2.91 × 10−5 | 10.7 | 2.08 × 10−14 |
| 2. | BST2 | Bone Marrow Stromal Cell Antigen 2 | 8.4 | 3.85 × 10−11 | 10.2 | 3.17 × 10−16 |
| 3. | C6orf223 | Long Intergenic Non-Protein Coding RNA 3040 | 5.9 | 4.90 × 10−8 | 8.5 | 2.06 × 10−16 |
| 4. | CA2 | Carbonic Anhydrase 2 | 7.7 | 1.73 × 10−7 | 9.8 | 3.47 × 10−12 |
| 5. | CBLC | CBL Proto-Oncogene C | 7.4 | 6.48 × 10−6 | 7.7 | 9.73 × 10−7 |
| 6. | CCL5 | C-C Motif Chemokine Ligand 5 | 5.9 | 4.17 × 10−5 | 7.4 | 6.59 × 10−8 |
| 7. | CCL20 | C-C Motif Chemokine Ligand 20 | 6.2 | 1.36 × 10−5 | 6.1 | 1.28 × 10−5 |
| 8. | CD70 | Cluster of Differentiation 70 | 6.7 | 1.35 × 10−5 | 7.9 | 1.24 × 10−7 |
| 9. | COBL | Cordon-Bleu WH2 Repeat Protein | 6.3 | 1.29 × 10−4 | 9.0 | 4.00 × 10−9 |
| 10. | CPVL | Carboxypeptidase Vitellogenic Like | 5.4 | 8.33 × 10−4 | 9.1 | 6.53 × 10−10 |
| 11. | CRB3 | Crumbs Cell Polarity Complex Component | 7.3 | 1.92 × 10−6 | 8.9 | 1.77 × 10−9 |
| 12. | CXCL11 | C-X-C Motif Chemokine Ligand 11 | 6.7 | 2.97 × 10−6 | 6.2 | 1.21 × 10−5 |
| 13. | DEPP1 | DEPP Autophagy Regulator 1 | 6.7 | 3.05 × 10−5 | 10.5 | 2.97 × 10−12 |
| 14. | DSC3 | Desmocollin 3 | 6.4 | 3.05 × 10−5 | 9.3 | 1.35 × 10−10 |
| 15. | ELF3 | E74 Like ETS Transcription Factor 3 | 8.5 | 1.05 × 10−13 | 11.4 | 1.33 × 10−24 |
| 16. | EPPK1 | Epiplakin | 7.2 | 1.11 × 10−5 | 7.5 | 2.39 × 10−6 |
| 17. | FGFBP1 | Fibroblast Growth Factor Binding Protein 1 | 8.1 | 2.36 × 10−6 | 9.9 | 1.89 × 10−9 |
| 18. | FOXA1 | Forkhead box A1 | 6.1 | 1.35 × 10−4 | 9.4 | 1.47 × 10−10 |
| 19. | FXYD3 | FXYD Domain Containing Ion Transport Regulator 3 | 9.4 | 9.70 × 10−10 | 9.5 | 9.85 × 10−10 |
| 20. | GJB2 | Gap Junction Protein Beta 2 | 6.7 | 1.35 × 10−5 | 9.5 | 7.78 × 10−11 |
| 21. | GRHL2 | Grainyhead Like Transcription Factor 2 | 6.8 | 2.21 × 10−5 | 9.3 | 7.91 × 10−10 |
| 22. | IGSF3 | Immunoglobulin Superfamily Member 3 | 6.7 | 7.13 × 10−5 | 8.9 | 2.57 × 10−8 |
| 23. | IQANK1 | IQ Motif and Ankyrin Repeat Containing 1 | 6.1 | 8.73 × 10−5 | 8.7 | 2.41 × 10−9 |
| 24. | IRF6 | Interferon Regulatory Factor 6 | 7.0 | 3.40 × 10−12 | 6.2 | 1.32 × 10−9 |
| 25. | KDF1 | Keratinocyte Differentiation Factor 1 | 6.8 | 1.04 × 10−5 | 9.7 | 2.44 × 10−11 |
| 26. | KRT7 | Keratin 7 | 5.6 | 1.00 × 10−12 | 7.9 | 3.37 × 10−25 |
| 27. | LAD1 | Ladinin 1 | 8.8 | 2.14 × 10−8 | 10.2 | 3.78 × 10−11 |
| 28. | LCN2 | Lipocalin 2 | 8.8 | 8.42 × 10−9 | 13.2 | 4.43 × 10−19 |
| 29. | LTB | Lymphotoxin Beta | 6.1 | 9.54 × 10−5 | 9.8 | 1.35 × 10−11 |
| 30. | MIR205HG | MIR205 host gene | 9.9 | 1.39 × 10−10 | 6.8 | 2.35 × 10−5 |
| 31. | MISP | Mitotic Spindle Positioning | 6.7 | 5.26 × 10−5 | 8.0 | 2.91 × 10−7 |
| 32. | MPZL2 | Myelin Protein Zero Like 2 | 6.9 | 1.55 × 10−5 | 9.5 | 2.39 × 10−10 |
| 33. | MST1R | Macrophage Stimulating 1 Receptor | 5.9 | 1.71 × 10−4 | 8.5 | 6.05 × 10−9 |
| 34. | NECTIN4 | Nectin Cell Adhesion Molecule 4 | 5.7 | 3.82 × 10−5 | 7.5 | 9.49 × 10−9 |
| 35. | NMU | Neuromedin U | 7.5 | 2.97 × 10−6 | 7.5 | 2.46 × 10−6 |
| 36. | OAS1 | 2′-5′-Oligoadenylate Synthase 1 | 9.8 | 2.04 × 10−16 | 7.9 | 1.29 × 10−10 |
| 37. | OASL | 2′-5′-Oligoadenylate Synthase Like | 6.8 | 9.78 × 10−9 | 6.9 | 4.08 × 10−9 |
| 38. | PI3 | Peptidase Inhibitor 3 | 6.0 | 2.48 × 10−11 | 5.1 | 2.49 × 10−8 |
| 39. | PROSER2 | Proline And Serine Rich 2 | 6.5 | 5.60 × 10−5 | 7.4 | 2.49 × 10−6 |
| 40. | PRSS8 | Serine Protease 8 | 6.2 | 2.51 × 10−4 | 9.6 | 7.05 × 10−10 |
| 41. | RAB25 | Member RAS Oncogene Family | 7.6 | 4.76 × 10−5 | 10.5 | 2.52 × 10−9 |
| 42. | S100A14 | S100 Calcium Binding Protein A14 | 7.7 | 2.76 × 10−11 | 10.0 | 1.12 × 10−18 |
| 43. | S100A9 | S100 Calcium Binding Protein A9 | 7.2 | 1.15 × 10−6 | 10.0 | 9.77 × 10−13 |
| 44. | SAA2 | Serum Amyloid A2 | 6.7 | 2.09 × 10−5 | 8.45 | 1.55 × 10−8 |
| 45. | SCNN1A | Sodium Channel Epithelial 1 Subunit Alpha | 7.0 | 1.64 × 10−7 | 7.3 | 3.46 × 10−8 |
| 46. | SFN | Stratifin | 8.4 | 1.15 × 10−9 | 9.5 | 3.75 × 10−12 |
| 47. | SLPI | Secretory Leucocyte Peptidase Inhibitor | 7.7 | 7.67 × 10−7 | 11.1 | 1.17 × 10−13 |
| 48. | SMOC1 | SPARC Related Modular Calcium Binding 1 | 7.6 | 5.85 × 10−7 | 9.0 | 1.07 × 10−9 |
| 49. | SNX10 | Sorting Nexin 10 | 6.8 | 1.96 × 10−5 | 6.7 | 1.53 × 10−5 |
| 50. | SPINT2 | Serine Peptidase Inhibitor, Kunitz Type 2 | 5.4 | 1.06 × 10−11 | 8.5 | 2.45 × 10−28 |
| 51. | TACSTD2 | Tumor Associated Calcium Signal Transducer 2 | 6.2 | 4.29 × 10−12 | 8.5 | 2.06 × 10−22 |
| 52. | TGFA | Transforming Growth Factor Alpha | 7.0 | 4.50 × 10−6 | 9.6 | 2.93 × 10−11 |
| 53. | TINAGL1 | Tubulointerstitial Nephritis Antigen Like 1 | 9.8 | 4.63 × 10−11 | 10.8 | 2.77 × 10−13 |
| 54. | WFDC2 | WAP Four-Disulphide Core Domain 2 | 6.1 | 5.80 × 10−5 | 6.3 | 1.81 × 10−5 |
| 55. | XIST | X Inactive Specific Transcript | 6.2 | 1.36 × 10−4 | 8.3 | 4.68 × 10−8 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Muralidharan, H.; Hansen, T.; Steinle, A.; Schumacher, D.; Stickeler, E.; Maurer, J. Breast Cancer Stem Cells Upregulate IRF6 in Stromal Fibroblasts to Induce Stromagenesis. Cells 2024, 13, 1466. https://doi.org/10.3390/cells13171466
Muralidharan H, Hansen T, Steinle A, Schumacher D, Stickeler E, Maurer J. Breast Cancer Stem Cells Upregulate IRF6 in Stromal Fibroblasts to Induce Stromagenesis. Cells. 2024; 13(17):1466. https://doi.org/10.3390/cells13171466
Chicago/Turabian StyleMuralidharan, Harshini, Thomas Hansen, Anja Steinle, David Schumacher, Elmar Stickeler, and Jochen Maurer. 2024. "Breast Cancer Stem Cells Upregulate IRF6 in Stromal Fibroblasts to Induce Stromagenesis" Cells 13, no. 17: 1466. https://doi.org/10.3390/cells13171466
APA StyleMuralidharan, H., Hansen, T., Steinle, A., Schumacher, D., Stickeler, E., & Maurer, J. (2024). Breast Cancer Stem Cells Upregulate IRF6 in Stromal Fibroblasts to Induce Stromagenesis. Cells, 13(17), 1466. https://doi.org/10.3390/cells13171466









