The Oncoprotein Fra-2 Drives the Activation of Human Endogenous Retrovirus Env Expression in Adult T-Cell Leukemia/Lymphoma (ATLL) Patients
Abstract
:1. Introduction
2. Materials and Methods
2.1. HTLV-1 Patients
2.2. Materials
2.3. Cell Culture and Transfection
2.4. RNA Analysis
2.5. TF Binding Site Prediction
2.6. Luciferase Assay
2.7. Western Blotting
2.8. ChIP-qPCR
2.9. ELISA Assay
2.10. Statistical Analyses
3. Results
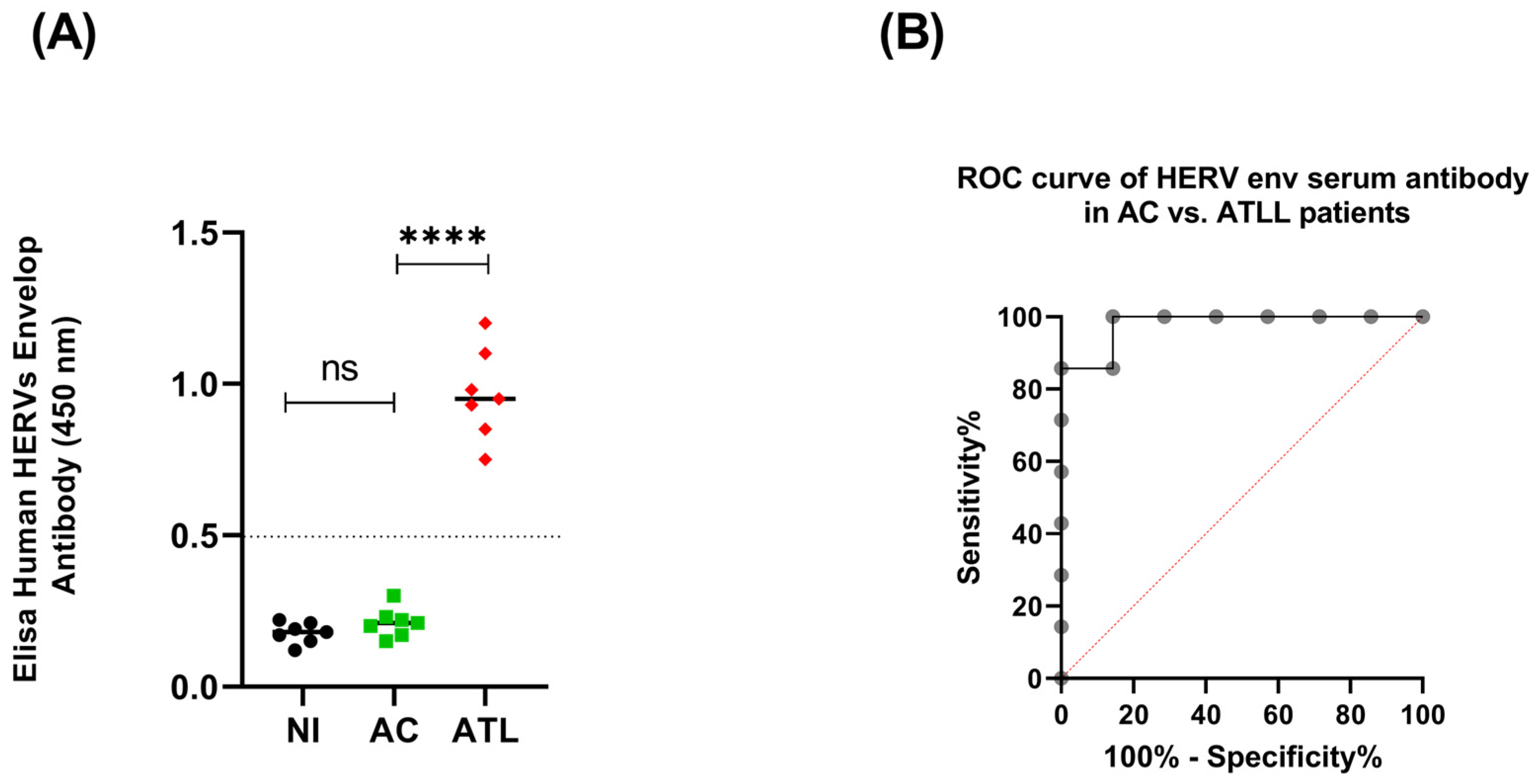
3.1. HERV Env Proteins Induce a Humoral Response in Acute ATLL Patients
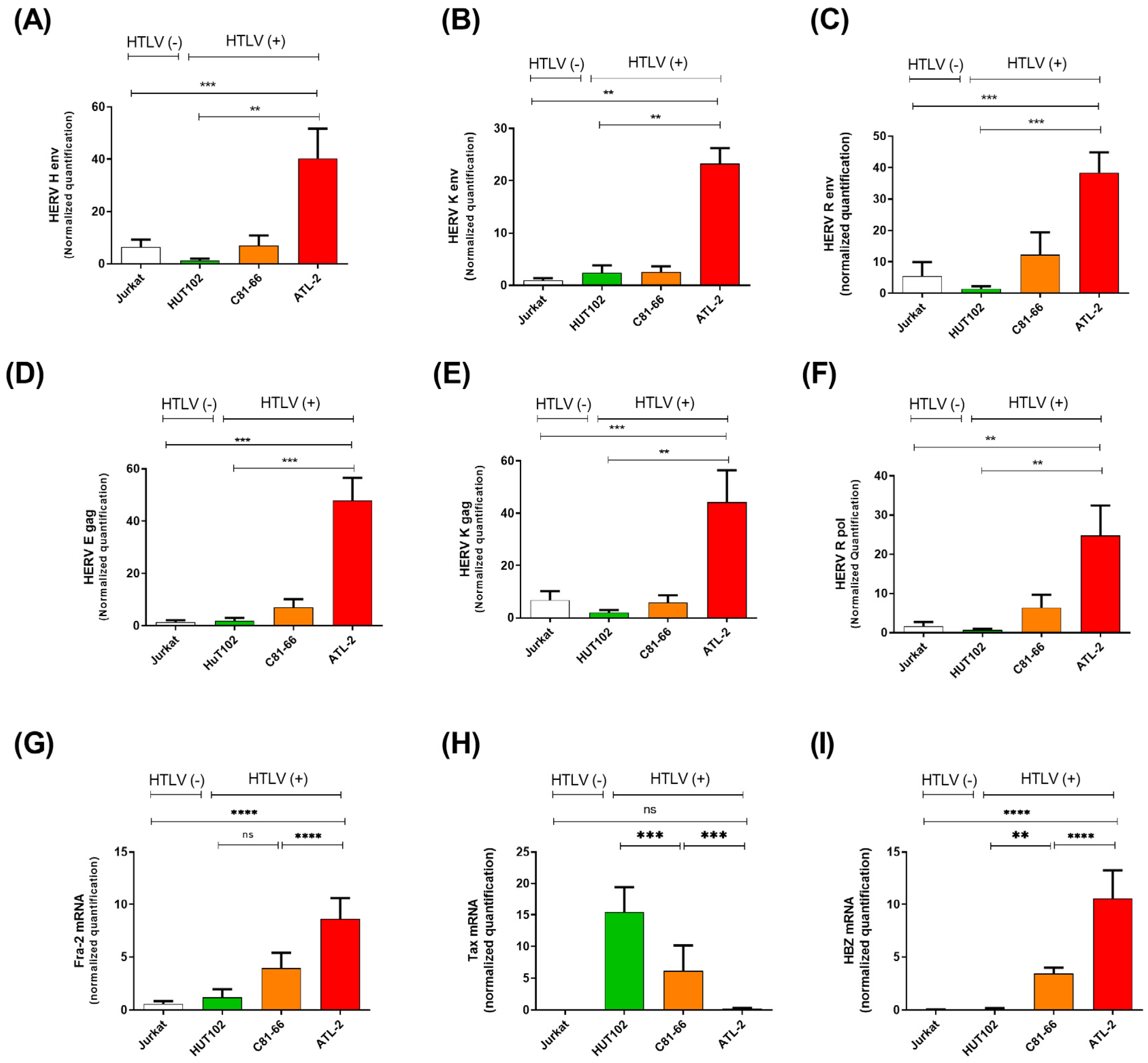
3.2. HERV Gene Expression Levels Are Increased in ATLL Patients
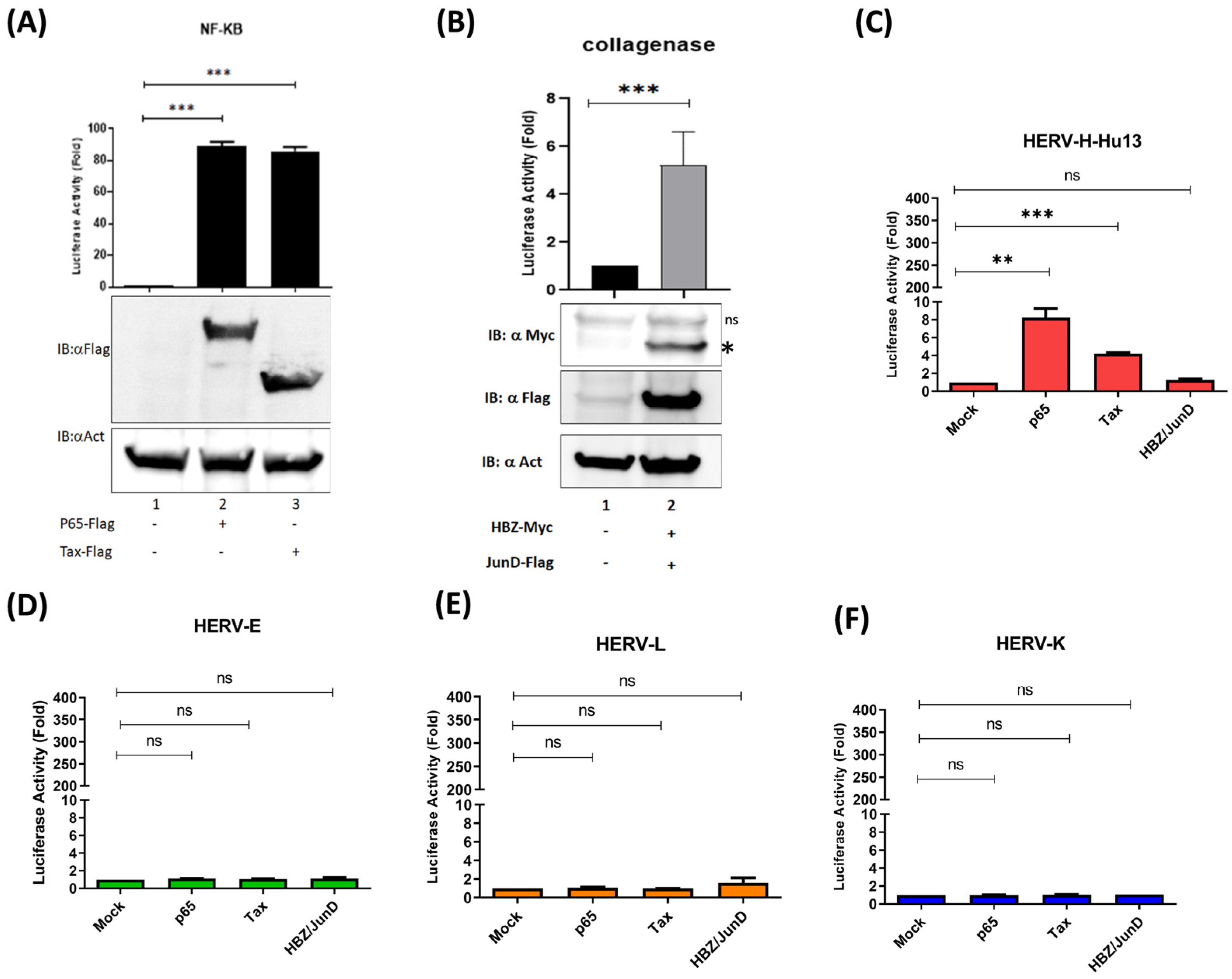
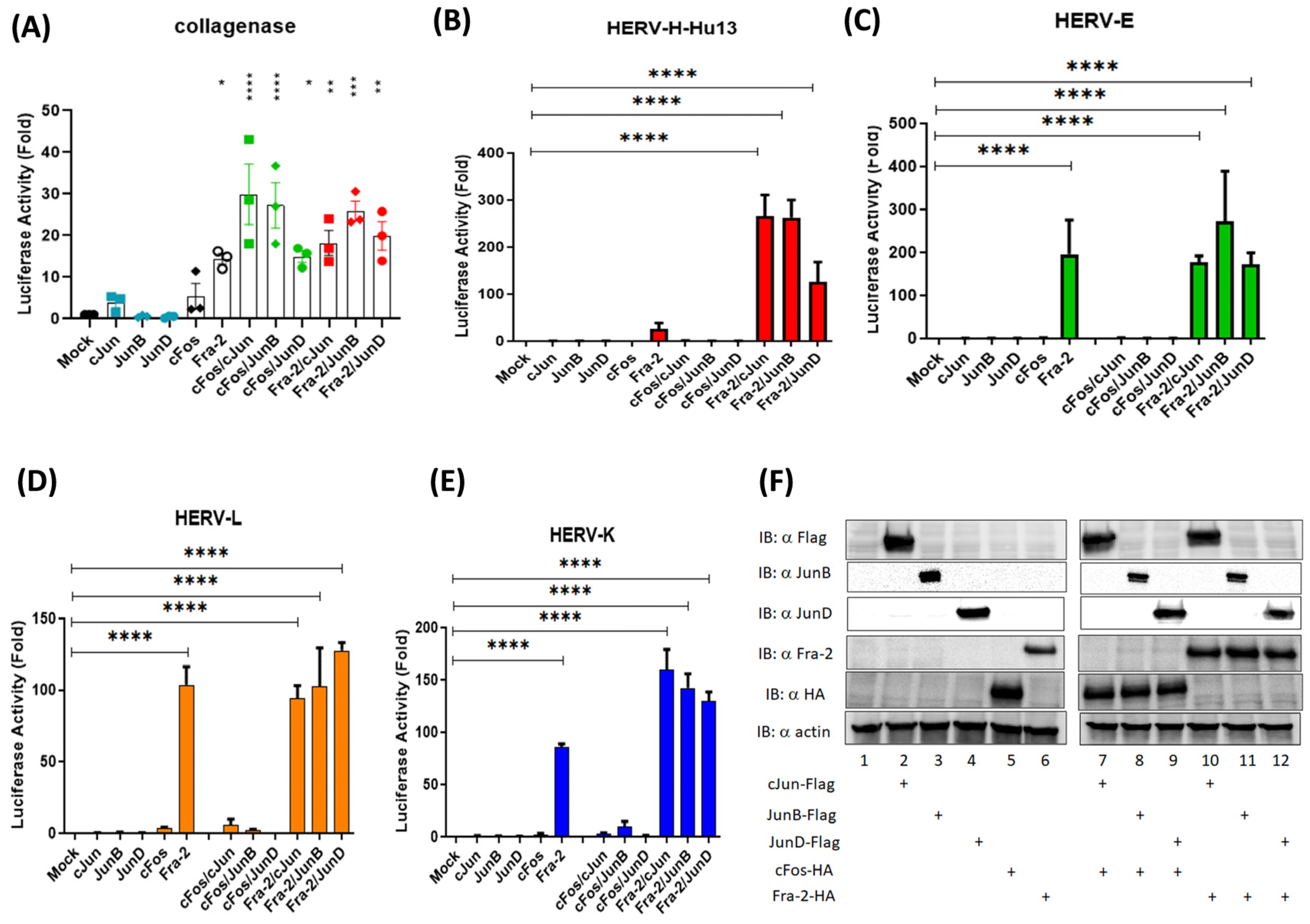
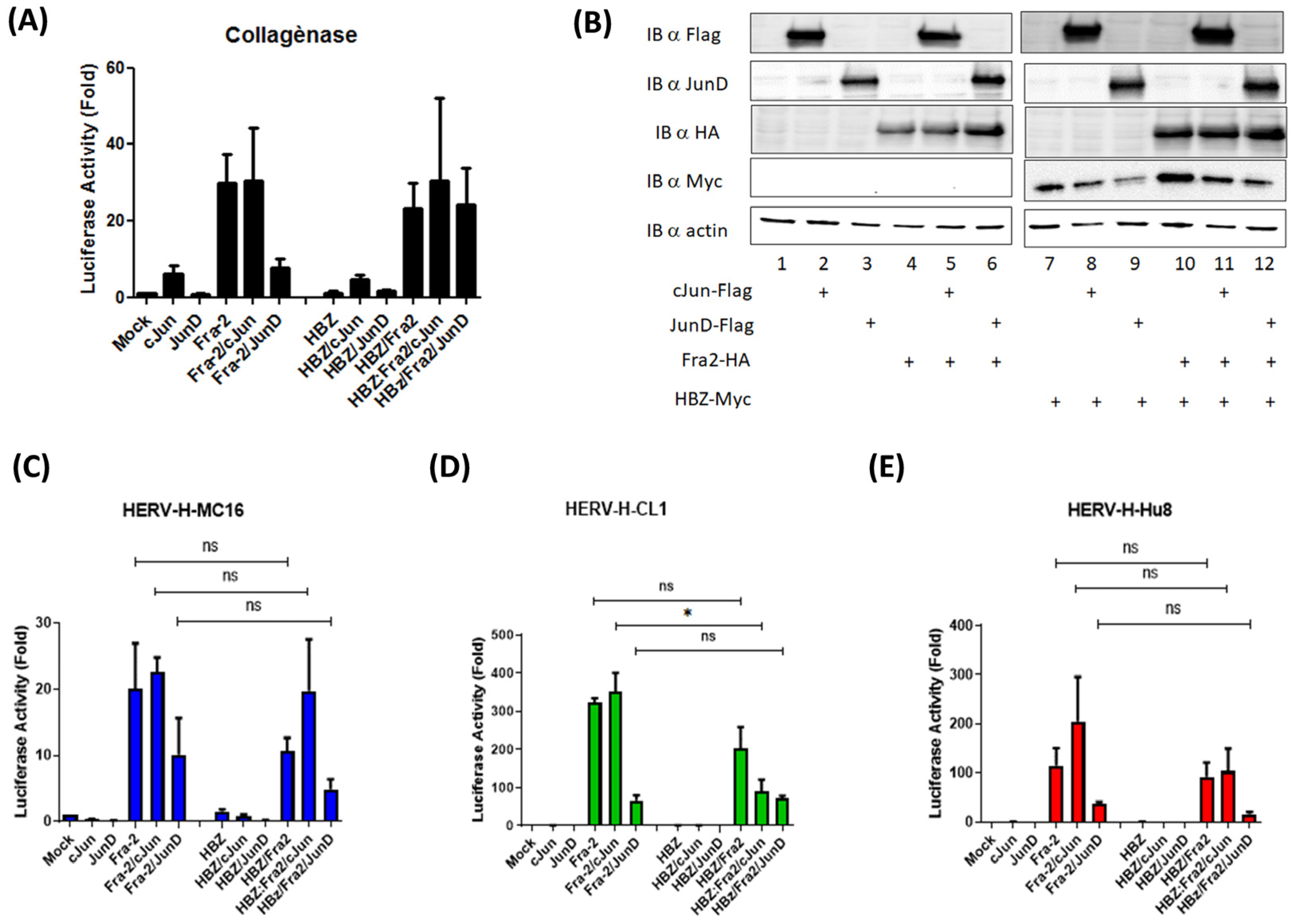
3.3. The Oncoprotein Fra-2 Activates the LTR of HERV-H, -E, -L, and -K
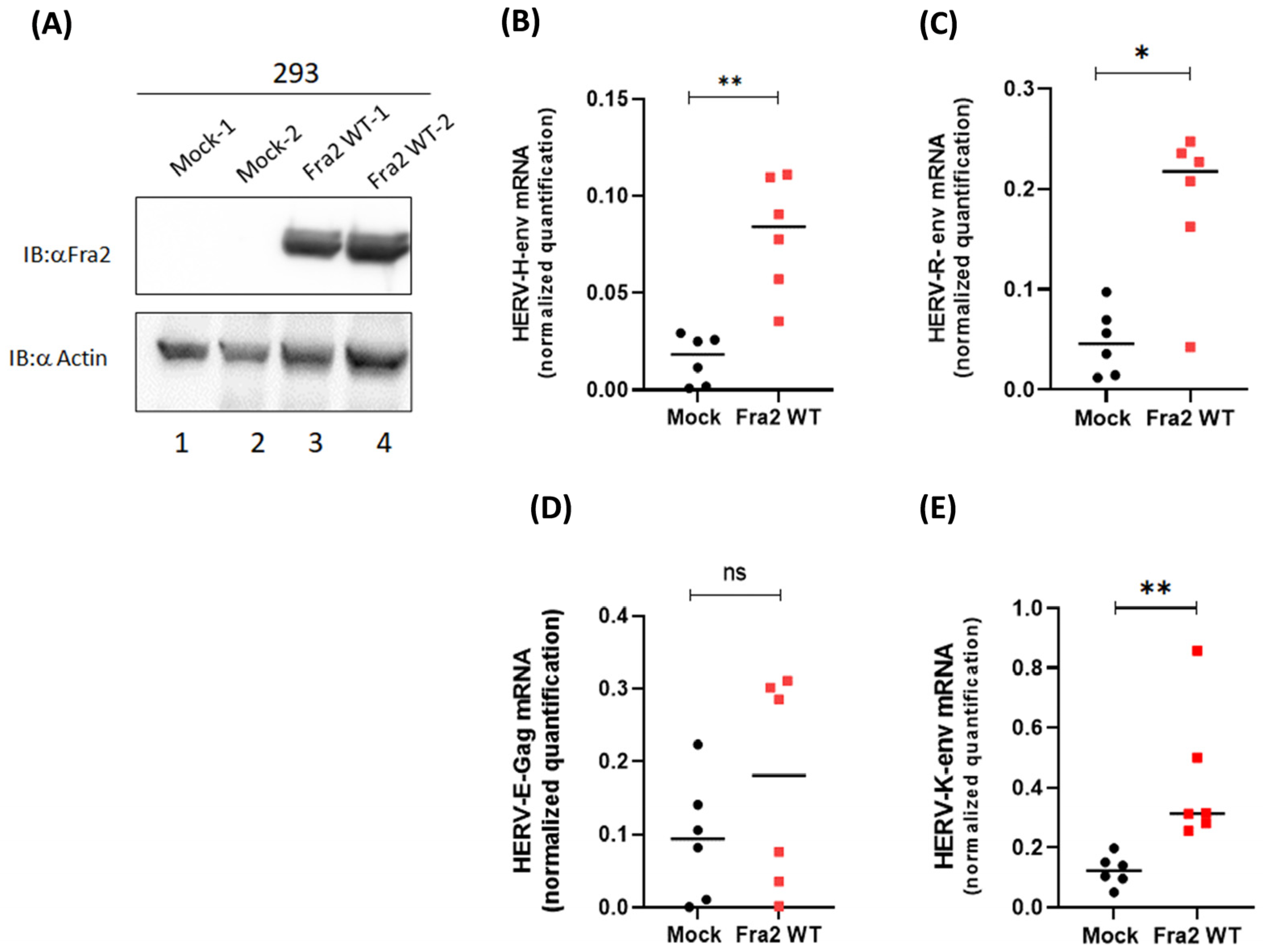
3.4. Fra-2 Upregulates HERV-H Env and HERV-K Env Genes in Primary Cells Isolated from ATLL Patients
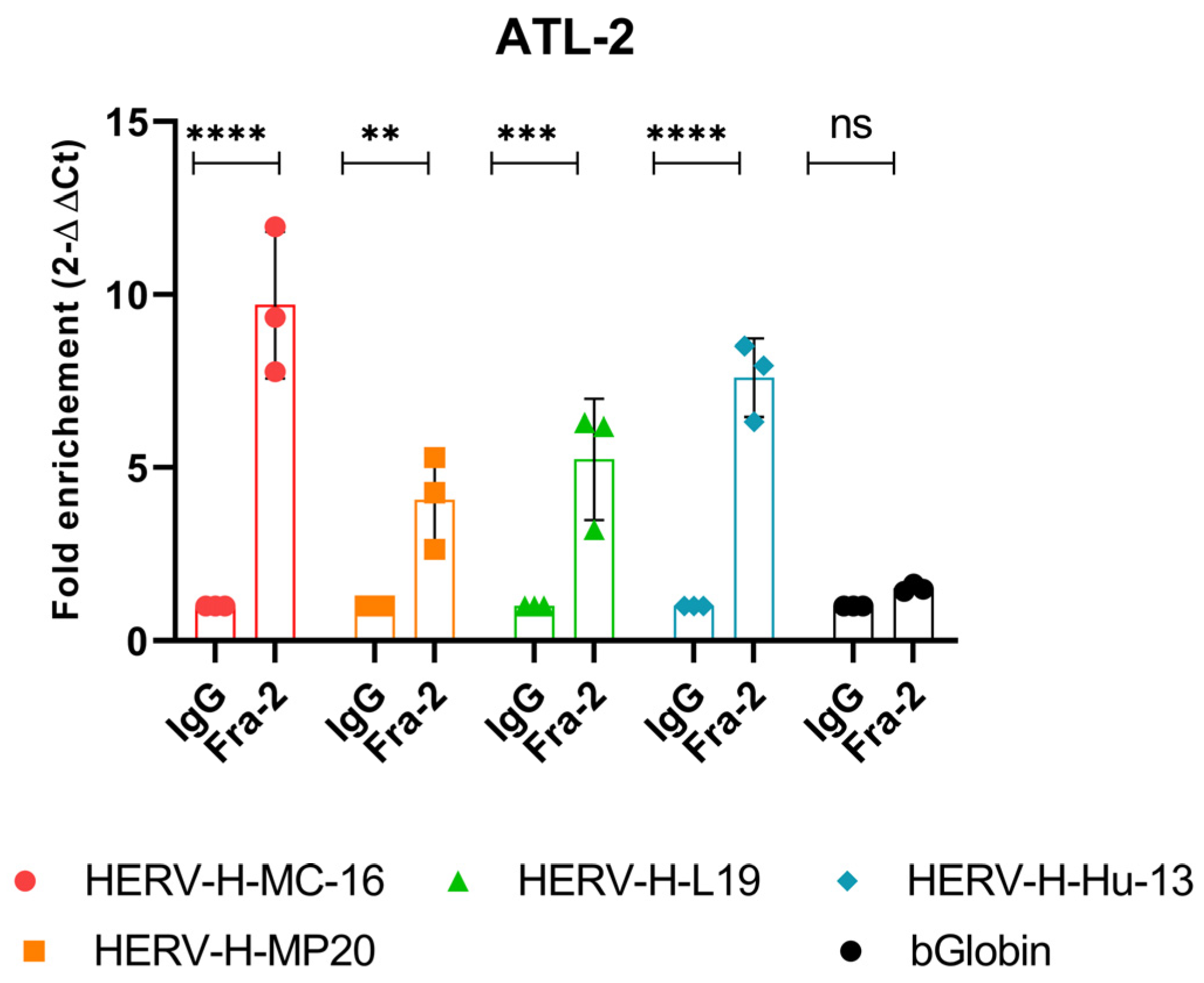
3.5. In Vivo Recruitment of Fra-2 to the HERV-H LTR
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Takasaki, Y.; Iwanaga, M.; Imaizumi, Y.; Tawara, M.; Joh, T.; Kohno, T.; Yamada, Y.; Kamihira, S.; Ikeda, S.; Tsukasaki, K.; et al. Long-term study of indolent adult T-cell leukemia-lymphoma. Blood 2010, 115, 4337–4343. [Google Scholar] [CrossRef] [PubMed]
- Tobinai, K. Current management of adult T-cell leukemia/lymphoma. Oncology 2009, 23, 1250–1256. [Google Scholar] [PubMed]
- Yasunaga, J.; Matsuoka, M. Human T-cell leukemia virus type I induces adult T-cell leukemia: From clinical aspects to molecular mechanisms. Cancer Control 2007, 14, 133–140. [Google Scholar] [CrossRef] [PubMed]
- Shimoyama, M. Diagnostic criteria and classification of clinical subtypes of adult T-cell leukaemia-lymphoma. A report from the Lymphoma Study Group (1984–87). Br. J. Haematol. 1991, 79, 428–437. [Google Scholar] [CrossRef]
- Ikeda, K.; Oka, M.; Yamada, Y.; Soda, H.; Fukuda, M.; Kinoshita, A.; Tsukamoto, K.; Noguchi, Y.; Isomoto, H.; Kohno, S.; et al. Adult T-cell leukemia cells over-express the multidrug-resistance-protein (MRP) and lung-resistance-protein (LRP) genes. Int. J. Cancer 1999, 82, 599–604. [Google Scholar] [CrossRef]
- Ishitsuka, K. Diagnosis and management of adult T-cell leukemia/lymphoma. Semin. Hematol. 2021, 58, 114–122. [Google Scholar] [CrossRef]
- Taylor, G.P.; Matsuoka, M. Natural history of adult T-cell leukemia/lymphoma and approaches to therapy. Oncogene 2005, 24, 6047–6057. [Google Scholar] [CrossRef] [PubMed]
- Bazarbachi, A.; Suarez, F.; Fields, P.; Hermine, O. How I treat adult T-cell leukemia/lymphoma. Blood 2011, 118, 1736–1745. [Google Scholar] [CrossRef]
- Grandi, N.; Tramontano, E. HERV Envelope Proteins: Physiological Role and Pathogenic Potential in Cancer and Autoimmunity. Front. Microbiol. 2018, 9, 462. [Google Scholar] [CrossRef]
- Stricker, E.; Peckham-Gregory, E.C.; Scheurer, M.E. HERVs and Cancer—A Comprehensive Review of the Relationship of Human Endogenous Retroviruses and Human Cancers. Biomedicines 2023, 11, 936. [Google Scholar] [CrossRef]
- Rivas, S.R.; Valdez, M.J.M.; Govindarajan, V.; Seetharam, D.; Doucet-O’Hare, T.T.; Heiss, J.D.; Shah, A.H. The Role of HERV-K in Cancer Stemness. Viruses 2022, 14, 2019. [Google Scholar] [CrossRef] [PubMed]
- Ko, E.-J.; Song, K.S.; Ock, M.S.; Choi, Y.H.; Kim, S.; Kim, H.-S.; Cha, H.-J. Expression profiles of human endogenous retrovirus (HERV)-K and HERV-R Env proteins in various cancers. BMB Rep. 2021, 54, 368–373. [Google Scholar] [CrossRef] [PubMed]
- Schön, U.; Seifarth, W.; Baust, C.; Hohenadl, C.; Erfle, V.; Leib-Mösch, C. Cell type-specific expression and promoter activity of human endogenous retroviral long terminal repeats. Virology 2001, 279, 280–291. [Google Scholar] [CrossRef]
- Peloponese, J.M.; Yasunaga, J.; Kinjo, T.; Watashi, K.; Jeang, K.T. Peptidylproline cis-trans-isomerase Pin1 interacts with human T-cell leukemia virus type 1 tax and modulates its activation of NF-kappaB. J. Virol. 2009, 83, 3238–3248. [Google Scholar] [CrossRef]
- Terol, M.; Gazon, H.; Lemasson, I.; Duc-Dodon, M.; Barbeau, B.; Cesaire, R.; Mesnard, J.M.; Peloponese, J.M. HBZ-mediated shift of JunD from growth suppressor to tumor promoter in leukemic cells by inhibition of ribosomal protein S25 expression. Leukemia 2017, 31, 2235–2243. [Google Scholar] [CrossRef] [PubMed]
- Belrose, G.; Gross, A.; Olindo, S.; Lézin, A.; Dueymes, M.; Komla-Soukha, I.; Smadja, D.; Tanaka, Y.; Willems, L.; Césaire, R. Effects of valproate on Tax and HBZ expression in HTLV-1 and HAM/TSP T lymphocytes. Blood 2011, 118, 2483–2491. [Google Scholar] [CrossRef]
- Maren, N.A.; Duduit, J.R.; Huang, D.; Zhao, F.; Ranney, T.G.; Liu, W. Stepwise Optimization of Real-Time RT-PCR Analysis. Methods Mol. Biol. 2023, 2653, 317–332. [Google Scholar]
- Riedel, G.; Rüdrich, U.; Fekete-Drimusz, N.; Manns, M.P.; Vondran, F.W.R.; Bock, M. An extended ΔCT-method facilitating normalisation with multiple reference genes suited for quantitative RT-PCR analyses of human hepatocyte-like cells. PLoS ONE 2014, 9, e93031. [Google Scholar] [CrossRef]
- Farré, D.; Roset, R.; Huerta, M.; Adsuara, J.E.; Roselló, L.; Albà, M.M.; Messeguer, X. Identification of patterns in biological sequences at the ALGGEN server: PROMO and MALGEN. Nucleic Acids Res. 2003, 31, 3651–3653. [Google Scholar] [CrossRef]
- Gazon, H.; Lemasson, I.; Polakowski, N.; Cesaire, R.; Matsuoka, M.; Barbeau, B.; Mesnard, J.M.; Peloponese, J.M. Human T-cell leukemia virus type 1 (HTLV-1) bZIP factor requires cellular transcription factor JunD to upregulate HTLV-1 antisense transcription from the 3′ long terminal repeat. J. Virol. 2012, 86, 9070–9078. [Google Scholar] [CrossRef]
- Santoni, F.A.; Guerra, J.; Luban, J. HERV-H RNA is abundant in human embryonic stem cells and a precise marker for pluripotency. Retrovirology 2012, 9, 111. [Google Scholar] [CrossRef] [PubMed]
- Curty, G.; Marston, J.L.; de Mulder Rougvie, M.; Leal, F.E.; Nixon, D.F.; Soares, M.A. Human Endogenous Retrovirus K in Cancer: A Potential Biomarker and Immunotherapeutic Target. Viruses 2020, 12, 726. [Google Scholar] [CrossRef] [PubMed]
- Mareschi, K.; Montanari, P.; Rassu, M.; Galliano, I.; Daprà, V.; Adamini, A.; Castiglia, S.; Fagioli, F.; Bergallo, M. Human Endogenous Retrovirus-H and K Expression in Human Mesenchymal Stem Cells as Potential Markers of Stemness. Intervirology 2019, 62, 9–14. [Google Scholar] [CrossRef]
- Rezaei, S.D.; Hayward, J.A.; Norden, S.; Pedersen, J.; Mills, J.; Hearps, A.C.; Tachedjian, G. HERV-K Gag RNA and Protein Levels Are Elevated in Malignant Regions of the Prostate in Males with Prostate Cancer. Viruses 2021, 13, 449. [Google Scholar] [CrossRef] [PubMed]
- Dervan, E.; Bhattacharyya, D.D.; McAuliffe, J.D.; Khan, F.H.; Glynn, S.A. Ancient Adversary–HERV-K (HML-2) in Cancer. Front. Oncol. 2021, 11, 658489. [Google Scholar] [CrossRef]
- Agoni, L.; Guha, C.; Lenz, J. Detection of Human Endogenous Retrovirus K (HERV-K) Transcripts in Human Prostate Cancer Cell Lines. Front. Oncol. 2013, 3, 180. [Google Scholar] [CrossRef]
- Manca, M.A.; Solinas, T.; Simula, E.R.; Noli, M.; Ruberto, S.; Madonia, M.; Sechi, L.A. HERV-K and HERV-H Env Proteins Induce a Humoral Response in Prostate Cancer Patients. Pathogens 2022, 11, 95. [Google Scholar] [CrossRef]
- Vincendeau, M.; Göttesdorfer, I.; Schreml, J.M.; Wetie AG, N.; Mayer, J.; Greenwood, A.D.; Helfer, M.; Kramer, S.; Seifarth, W.; Leib-Mösch, C.; et al. Modulation of human endogenous retrovirus (HERV) transcription during persistent and de novo HIV-1 infection. Retrovirology 2015, 12, 27. [Google Scholar] [CrossRef]
- Yang, C.; Guo, X.; Li, J.; Han, J.; Jia, L.; Wen, H.L.; Sun, C.; Wang, X.; Zhang, B.; Li, L.; et al. Significant Upregulation of HERV-K (HML-2) Transcription Levels in Human Lung Cancer and Cancer Cells. Front. Microbiol. 2022, 13, 850444. [Google Scholar] [CrossRef]
- Chang, N.-T.; Yang, W.K.; Huang, H.-C.; Yeh, K.-W.; Wu, C.-W. The transcriptional activity of HERV-I LTR is negatively regulated by its cis-elements and wild-type p53 tumor suppressor protein. J. Biomed. Sci. 2007, 14, 211–222. [Google Scholar] [CrossRef]
- Toufaily, C.; Landry, S.; Leib-Mosch, C.; Rassart, E.; Barbeau, B. Activation of LTRs from different human endogenous retrovirus (HERV) families by the HTLV-1 tax protein and T-cell activators. Viruses 2011, 3, 2146–2159. [Google Scholar] [CrossRef]
- Duc Dodon, M.; Mesnard, J.M.; Barbeau, B. Adult T-cell leukemia induced by HTLV-1: Before and after HBZ. Med. Sci. 2010, 26, 391–396. [Google Scholar]
- Giam, C.Z.; Jeang, K.T. HTLV-1 Tax and adult T-cell leukemia. Front. Biosci. 2007, 12, 1496–1507. [Google Scholar] [CrossRef] [PubMed]
- Peloponese, J.M.; Kinjo, T.; Jeang, K.T. Human T-cell leukemia virus type 1 Tax and cellular transformation. Int. J. Hematol. 2007, 86, 101–106. [Google Scholar] [CrossRef] [PubMed]
- Belrose, G.; Gazon, H.; Meniane, J.-C.; Olindo, S.; Mesnard, J.-M.; Peloponese, J.-M.; Cesaire, R. Effects of valproate on Tax and HBZ expression in ex vivo cultured ATL cells. Retrovirology 2014, 11, P39. [Google Scholar] [CrossRef]
- Balada, E.; Vilardell-Tarres, M.; Ordi-Ros, J. Implication of human endogenous retroviruses in the development of autoimmune diseases. Int. Rev. Immunol. 2010, 29, 351–370. [Google Scholar] [CrossRef]
- Katoh, I.; Kurata, S. Association of endogenous retroviruses and long terminal repeats with human disorders. Front. Oncol. 2013, 3, 234. [Google Scholar] [CrossRef]
- Mameli, G.; Erre, G.L.; Caggiu, E.; Mura, S.; Cossu, D.; Bo, M.; Cadoni, M.L.; Piras, A.; Mundula, N.; Sechi, L.A.; et al. Identification of a HERV-K env surface peptide highly recognized in Rheumatoid Arthritis (RA) patients: A cross-sectional case-control study. Clin. Exp. Immunol. 2017, 189, 127–131. [Google Scholar] [CrossRef]
- Perzova, R.; Graziano, E.; Sanghi, S.; Welch, C.; Benz, P.; Abbott, L.; Lalone, D.; Glaser, J.; Loughran, T.; Poiesz, B.J.; et al. Increased seroreactivity to HERV-K10 peptides in patients with HTLV myelopathy. Virol. J. 2013, 10, 360. [Google Scholar] [CrossRef]
- Gessain, A.; Barin, F.; Vernant, J.C.; Gout, O.; Maurs, L.; Calender, A.; de The, G. Antibodies to human T-lymphotropic virus type-I in patients with tropical spastic paraparesis. Lancet 1985, 2, 407–410. [Google Scholar] [CrossRef]
- Hjelle, B.; Appenzeller, O.; Mills, R.; Alexander, S.; Torrez-Martinez, N.; Jahnke, R.; Ross, G. Chronic neurodegenerative disease associated with HTLV-II infection. Lancet 1992, 339, 645–646. [Google Scholar] [CrossRef] [PubMed]
- Perzova, R.; Graziano, E.; Sanghi, S.; Welch, C.; Benz, P.; Abbott, L.; Lalone, D.; Glaser, J.; Loughran, T.; Poiesz, B.J.; et al. Increased seroreactivity to human T cell lymphoma/leukemia virus-related endogenous sequence-1 Gag peptides in patients with human T cell lymphoma/leukemia virus myelopathy. Aids Res. Hum. Retroviruses 2015, 31, 242–249. [Google Scholar] [CrossRef]
- Jones, R.B.; Leal, F.E.; Hasenkrug, A.M.; Segurado, A.C.; Nixon, D.F.; Ostrowski, M.A. Human endogenous retrovirus K(HML-2) Gag and Env specific T-cell responses are not detected in HTLV-I-infected subjects using standard peptide screening methods. J. Negat. Results Biomed. 2013, 12, 3. [Google Scholar] [CrossRef]
- Burbelo, P.D.; Lebovitz, E.E.; Notkins, A.L. Luciferase immunoprecipitation systems for measuring antibodies in autoimmune and infectious diseases. Transl. Res. J. Lab. Clin. Med. 2015, 165, 325–335. [Google Scholar] [CrossRef]
- Eun, K.; Ham, S.W.; Kim, H. Cancer stem cell heterogeneity: Origin and new perspectives on CSC targeting. BMB Rep. 2017, 50, 117–125. [Google Scholar] [CrossRef]
- Sancho, P.; Barneda, D.; Heeschen, C. Hallmarks of cancer stem cell metabolism. Br. J. Cancer 2016, 114, 1305–1312. [Google Scholar] [CrossRef] [PubMed]
- Jayabal, P.; Ma, X.; Shiio, Y. EZH2 suppresses endogenous retroviruses and an interferon response in cancers. Genes Cancer 2021, 12, 96–105. [Google Scholar] [CrossRef]
- Ratner, L. Epigenetic Regulation of Human T-Cell Leukemia Virus Gene Expression. Microorganisms 2021, 10, 84. [Google Scholar] [CrossRef] [PubMed]
- Yamagishi, M.; Fujikawa, D.; Watanabe, T.; Uchimaru, K. HTLV-1-Mediated Epigenetic Pathway to Adult T-Cell Leukemia–Lymphoma. Front. Microbiol. 2018, 9, 1686. [Google Scholar] [CrossRef]
- Toyoda, K.; Yasunaga, J.I.; Shichijo, T.; Arima, Y.; Tsujita, K.; Tanaka, A.; Salah, T.; Zhang, W.; Hussein, O.; Matsuoka, M.; et al. HTLV-1 bZIP Factor-Induced Reprogramming of Lactate Metabolism and Epigenetic Status Promote Leukemic Cell Expansion. Blood Cancer Discov. 2023, 4, 374–393. [Google Scholar] [CrossRef]
- Dolci, M.; Favero, C.; Toumi, W.; Favi, E.; Tarantini, L.; Signorini, L.; Basile, G.; Bollati, V.; D’Alessandro, S.; Delbue, S.; et al. Human Endogenous Retroviruses Long Terminal Repeat Methylation, Transcription, and Protein Expression in Human Colon Cancer. Front. Oncol. 2020, 10, 569015. [Google Scholar] [CrossRef] [PubMed]
- Tavakolian, S.; Iranshahi, M.; Faghihloo, E. The Evaluation of HERV-K np9, rec, gag Expression in Isolated Human Peripheral Blood Mononuclear Cell (PBMC) of Gastric and Colon Cancer. Adv. Biomed. Res. 2023, 12, 131. [Google Scholar] [CrossRef] [PubMed]
- Kajanne, R.; Miettinen, P.; Tenhunen, M.; Leppa, S. Transcription factor AP-1 promotes growth and radioresistance in prostate cancer cells. Int. J. Oncol. 2009, 35, 1175–1182. [Google Scholar] [PubMed]
- Ishida, T.; Obata, Y.; Ohara, N.; Matsushita, H.; Sato, S.; Uenaka, A.; Saika, T.; Miyamura, T.; Chayama, K.; Nakayama, E.; et al. Identification of the HERV-K gag antigen in prostate cancer by SEREX using autologous patient serum and its immunogenicity. Cancer Immun. 2008, 8, 15. [Google Scholar]
- Milde-Langosch, K. The Fos family of transcription factors and their role in tumourigenesis. Eur. J. Cancer 2005, 41, 2449–2461. [Google Scholar] [CrossRef]
- Vaiopoulos, A.G.; Papachroni, K.K.; Papavassiliou, A.G. Colon carcinogenesis: Learning from NF-kappaB and AP-1. Int. J. Biochem. Cell Biol. 2010, 42, 1061–1065. [Google Scholar] [CrossRef]
- Zhang, W.; Hart, J.; McLeod, H.L.; Wang, H.L. Differential expression of the AP-1 transcription factor family members in human colorectal epithelial and neuroendocrine neoplasms. Am. J. Clin. Pathol. 2005, 124, 11–19. [Google Scholar] [CrossRef]
- Pérot, P.; Mullins, C.S.; Naville, M.; Bressan, C.; Hühns, M.; Gock, M.; Kühn, F.; Volff, J.-N.; Trillet-Lenoir, V.; Mallet, F.; et al. Expression of young HERV-H loci in the course of colorectal carcinoma and correlation with molecular subtypes. Oncotarget 2015, 6, 40095–40111. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tram, J.; Marty, L.; Mourouvin, C.; Abrantes, M.; Jaafari, I.; Césaire, R.; Hélias, P.; Barbeau, B.; Mesnard, J.-M.; Baccini, V.; et al. The Oncoprotein Fra-2 Drives the Activation of Human Endogenous Retrovirus Env Expression in Adult T-Cell Leukemia/Lymphoma (ATLL) Patients. Cells 2024, 13, 1517. https://doi.org/10.3390/cells13181517
Tram J, Marty L, Mourouvin C, Abrantes M, Jaafari I, Césaire R, Hélias P, Barbeau B, Mesnard J-M, Baccini V, et al. The Oncoprotein Fra-2 Drives the Activation of Human Endogenous Retrovirus Env Expression in Adult T-Cell Leukemia/Lymphoma (ATLL) Patients. Cells. 2024; 13(18):1517. https://doi.org/10.3390/cells13181517
Chicago/Turabian StyleTram, Julie, Laetitia Marty, Célima Mourouvin, Magali Abrantes, Ilham Jaafari, Raymond Césaire, Philippe Hélias, Benoit Barbeau, Jean-Michel Mesnard, Véronique Baccini, and et al. 2024. "The Oncoprotein Fra-2 Drives the Activation of Human Endogenous Retrovirus Env Expression in Adult T-Cell Leukemia/Lymphoma (ATLL) Patients" Cells 13, no. 18: 1517. https://doi.org/10.3390/cells13181517






