The Roles of Cystatin B in the Brain and Pathophysiological Mechanisms of Progressive Myoclonic Epilepsy Type 1
Abstract
1. Introduction
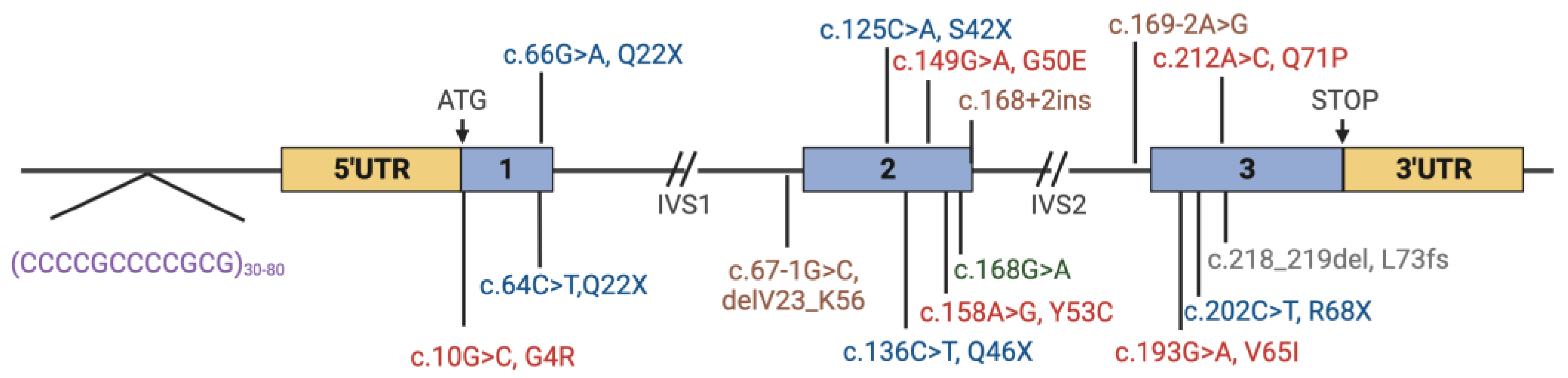
| Exon | gDNA (hg38) | cDNA | Protein (Reference) |
|---|---|---|---|
| 1 | g.43776260C>G | c.10G>C | p.Gly4Arg [14,15] |
| 1 | g.43776204C>T | c.66G>A | p.Gln22Ter [13] |
| intron 1 | g.43774760C>G | c.67-1G>C | p.Gln22Ter [11,16,17,18] |
| 2 | g.43774701G>T | c.125C>A | p.Ser42Ter [19] |
| 2 | g.43774690G>A | c.136C>T | p.Gln46Ter [11] |
| 2 | g.43774677C>T | c.149G>A | p.Gly50Glu [20] |
| 2 | g.43774668T>C | c.158A>G | p.Try53Cys [21] |
| 2 | g.43774658C>T | c.168G>A | p.Lys56= [22] |
| 2 intron | g.43774642_43774659del | c.168+2_168+19del | p.Val23_Lys56delVal57fs*28 [11,20] |
| 2 intron | g.43774637_43774656delinsTT | c.168+2_168+21delinsAA | Splice Site [11] |
| 2 intron | g.43774332T>C | c.169-2A>G | Splice site [14] |
| 3 | g.43774306C>T | c.193G>A | p.Val65Ile [21] |
| 3 | g.43774297G>A | c.202C>T | p.Arg68ter [16,17,23,24] |
| 3 | g.43774287T>G | c.212A>C | p.Gln71Pro [24] |
| 3 | g.43774281dup | c.218dup | p.His75Ser_fs*2 [25] |
| 3 | g. 43774284_43774285del | c.218_219del | p.Leu73Pro_fs*3 [17,18,26] |
| 5′ UTR_1 | g.43776444_43776479ins(500_900) | [11,17,20,22,24,27,28] | |
| 5′ UTR_1 | g.43776468_43776479del | [18] | |
| 5′ UTR_1 | g.43776444_43776479insN(1500_1600) | [29] |
2. Clinical Picture
Neurological Findings
3. Molecular Genetics of EPM1
4. The CSTB Protein
4.1. CSTB Expression and Localization in the Brain
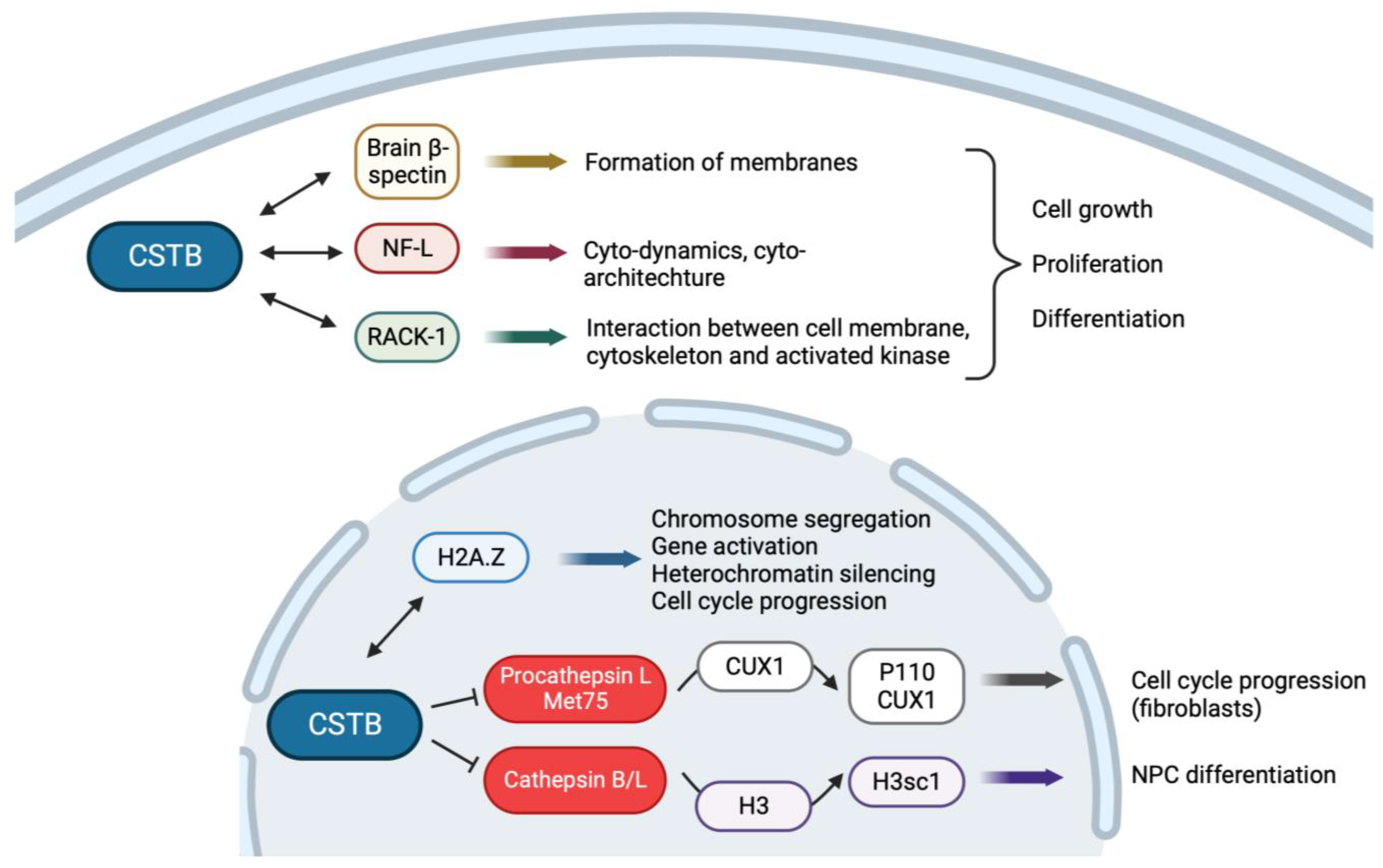
4.2. CSTB Plays a Role in Brain Development
4.3. CSTB Regulates Cell Cycle
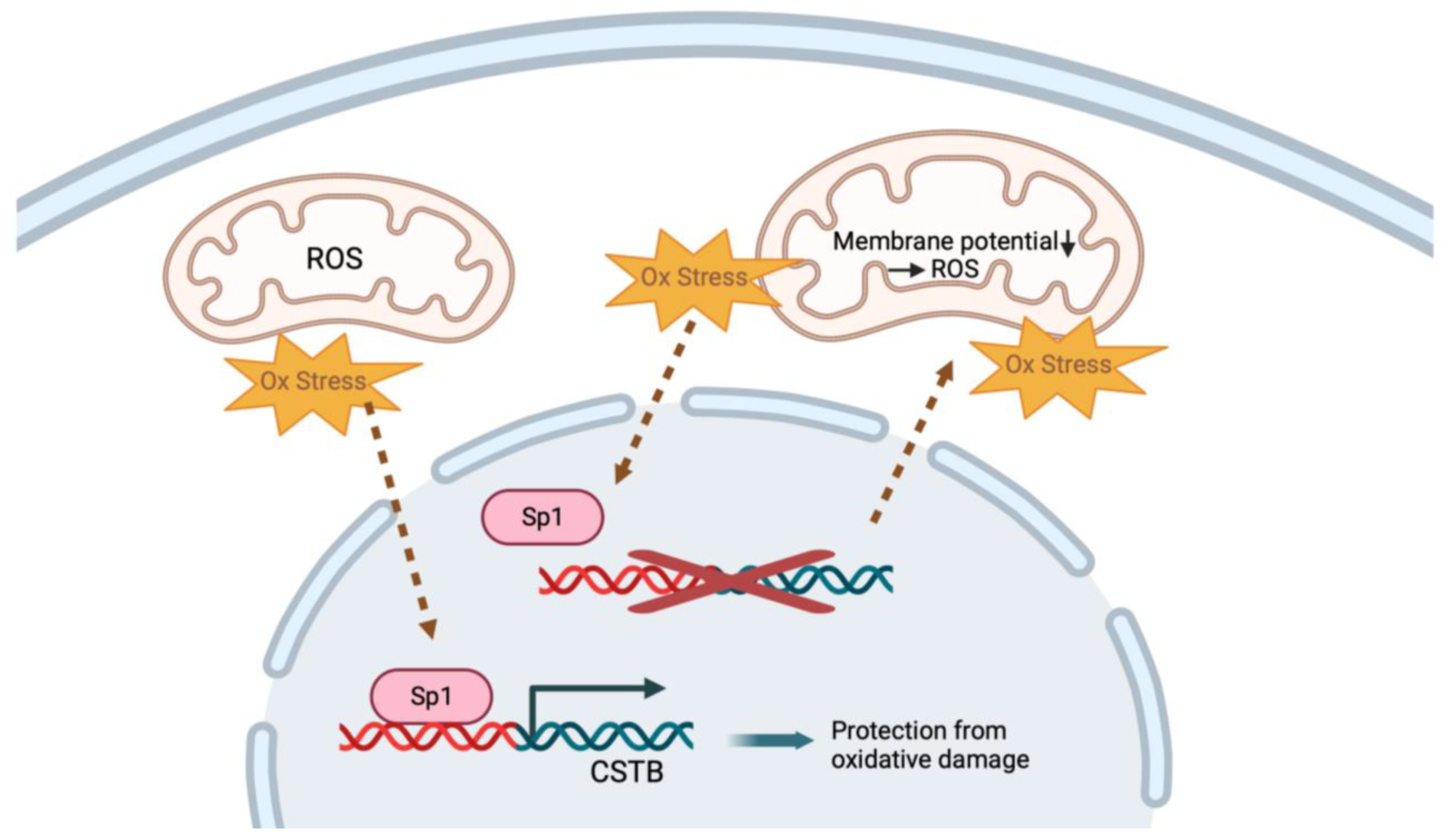
4.4. CSTB Regulates Mitochondria and Protects against Oxidative Stress
4.5. CSTB in Neuroinflammation
4.6. CSTB in GABAergic Neurons
4.7. CSTB in Synaptosomes
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Lehesjoki, A.E.; Koskiniemi, M.; Pandolfo, M.; Antonelli, A.; Kyllerman, M.; Wahlström, J.; Nergårdh, A.; Burmeister, M.; Sistonen, P.; Norio, R. Linkage studies in progressive myoclonus epilepsy: Unverricht-Lundborg and Lafora’s diseases. Neurology 1992, 42, 1545–1550. [Google Scholar] [CrossRef] [PubMed]
- Orsini, A.; Valetto, A.; Bertini, V.; Esposito, M.; Carli, N.; Minassian, B.A.; Bonuccelli, A.; Peroni, D.; Michelucci, R.; Striano, P. The best evidence for progressive myoclonic epilepsy: A pathway to precision therapy. Seizure 2019, 71, 247–257. [Google Scholar] [CrossRef]
- Sipilä, J.O.T.; Hyppönen, J.; Kytö, V.; Kälviäinen, R. Unverricht-Lundborg disease (EPM1) in Finland: A nationwide population-based study. Neurology 2020, 95, e3117–e3123. [Google Scholar] [CrossRef] [PubMed]
- Kälviäinen, R.; Khyuppenen, J.; Koskenkorva, P.; Eriksson, K.; Vanninen, R.; Mervaala, E. Clinical picture of EPM1-Unverricht-Lundborg disease. Epilepsia 2008, 49, 549–556. [Google Scholar] [CrossRef] [PubMed]
- Alakurtti, K.; Weber, E.; Rinne, R.; Theil, G.; de Haan, G.J.; Lindhout, D.; Salmikangas, P.; Saukko, P.; Lahtinen, U.; Lehesjoki, A.E. Loss of lysosomal association of cystatin B proteins representing progressive myoclonus epilepsy, EPM1, mutations. Eur. J. Hum. Genet. 2005, 13, 208–215. [Google Scholar] [CrossRef] [PubMed]
- Brännvall, K.; Hjelm, H.; Korhonen, L.; Lahtinen, U.; Lehesjoki, A.E.; Lindholm, D. Cystatin-B is expressed by neural stem cells and by differentiated neurons and astrocytes. Biochem. Biophys. Res. Commun. 2003, 308, 369–374. [Google Scholar] [CrossRef]
- Di Giaimo, R.; Riccio, M.; Santi, S.; Galeotti, C.; Ambrosetti, D.C.; Melli, M. New insights into the molecular basis of progressive myoclonus epilepsy: A multiprotein complex with cystatin B. Hum. Mol. Genet. 2002, 11, 2941–2950. [Google Scholar] [CrossRef]
- Gorski, K.; Spoljaric, A.; Nyman, T.A.; Kaila, K.; Battersby, B.J.; Lehesjoki, A.E. Quantitative Changes in the Mitochondrial Proteome of Cerebellar Synaptosomes From Preclinical Cystatin B-Deficient Mice. Front. Mol. Neurosci. 2020, 13, 570640. [Google Scholar] [CrossRef]
- Di Matteo, F.; Pipicelli, F.; Kyrousi, C.; Tovecci, I.; Penna, E.; Crispino, M.; Chambery, A.; Russo, R.; Ayo-Martin, A.C.; Giordano, M.; et al. Cystatin B is essential for proliferation and interneuron migration in individuals with EPM1 epilepsy. EMBO Mol. Med. 2020, 12, e11419. [Google Scholar] [CrossRef]
- Okuneva, O.; Körber, I.; Li, Z.; Tian, L.; Joensuu, T.; Kopra, O.; Lehesjoki, A.E. Abnormal microglial activation in the Cstb(-/-) mouse, a model for progressive myoclonus epilepsy, EPM1. Glia 2015, 63, 400–411. [Google Scholar] [CrossRef]
- Canafoglia, L.; Gennaro, E.; Capovilla, G.; Gobbi, G.; Boni, A.; Beccaria, F.; Viri, M.; Michelucci, R.; Agazzi, P.; Assereto, S.; et al. Electroclinical presentation and genotype-phenotype relationships in patients with Unverricht-Lundborg disease carrying compound heterozygous CSTB point and indel mutations. Epilepsia 2012, 53, 2120–2127. [Google Scholar] [CrossRef]
- Joensuu, T.; Lehesjoki, A.E.; Kopra, O. Molecular background of EPM1-Unverricht-Lundborg disease. Epilepsia 2008, 49, 557–563. [Google Scholar] [CrossRef] [PubMed]
- Pinto, E.; Freitas, J.; Duarte, A.J.; Ribeiro, I.; Ribeiro, D.; Lima, J.L.; Chaves, J.; Amaral, O. Unverricht-Lundborg disease: Homozygosity for a new splicing mutation in the cystatin B gene. Epilepsy Res. 2012, 99, 187–190. [Google Scholar] [CrossRef] [PubMed]
- Lalioti, M.D.; Scott, H.S.; Buresi, C.; Rossier, C.; Bottani, A.; Morris, M.A.; Malafosse, A.; Antonarakis, S.E. Dodecamer repeat expansion in cystatin B gene in progressive myoclonus epilepsy. Nature 1997, 386, 847–851. [Google Scholar] [CrossRef] [PubMed]
- Tarailo-Graovac, M.; Shyr, C.; Ross, C.J.; Horvath, G.A.; Salvarinova, R.; Ye, X.C.; Zhang, L.H.; Bhavsar, A.P.; Lee, J.J.; Drögemöller, B.I.; et al. Exome Sequencing and the Management of Neurometabolic Disorders. N. Engl. J. Med. 2016, 374, 2246–2255. [Google Scholar] [CrossRef] [PubMed]
- Pennacchio, L.A.; Lehesjoki, A.E.; Stone, N.E.; Willour, V.L.; Virtaneva, K.; Miao, J.; D’Amato, E.; Ramirez, L.; Faham, M.; Koskiniemi, M.; et al. Mutations in the gene encoding cystatin B in progressive myoclonus epilepsy (EPM1). Science 1996, 271, 1731–1734. [Google Scholar] [CrossRef] [PubMed]
- Lafrenière, R.G.; Rochefort, D.L.; Chrétien, N.; Rommens, J.M.; Cochius, J.I.; Kälviäinen, R.; Nousiainen, U.; Patry, G.; Farrell, K.; Söderfeldt, B.; et al. Unstable insertion in the 5’ flanking region of the cystatin B gene is the most common mutation in progressive myoclonus epilepsy type 1, EPM1. Nat. Genet. 1997, 15, 298–302. [Google Scholar] [CrossRef]
- Lalioti, M.D.; Mirotsou, M.; Buresi, C.; Peitsch, M.C.; Rossier, C.; Ouazzani, R.; Baldy-Moulinier, M.; Bottani, A.; Malafosse, A.; Antonarakis, S.E. Identification of mutations in cystatin B, the gene responsible for the Unverricht-Lundborg type of progressive myoclonus epilepsy (EPM1). Am. J. Hum. Genet. 1997, 60, 342–351. [Google Scholar]
- Lehesjoki, A.E.; Kälviäinen, R. Progressive Myoclonic Epilepsy Type 1. 2004 Jun 24 [Updated 2 July 2020]. In GeneReviews® [Internet]; Adam, M.P., Feldman, J., Mirzaa, G.M., Eds.; University of Washington: Seattle, WA, USA, 1993–2024. Available online: https://www.ncbi.nlm.nih.gov/books/NBK1142 (accessed on 1 November 2023).
- Joensuu, T.; Kuronen, M.; Alakurtti, K.; Tegelberg, S.; Hakala, P.; Aalto, A.; Huopaniemi, L.; Aula, N.; Michellucci, R.; Eriksson, K.; et al. Cystatin B: Mutation detection, alternative splicing and expression in progressive myclonus epilepsy of Unverricht-Lundborg type (EPM1) patients. Eur. J. Hum. Genet. 2007, 15, 185–193. [Google Scholar] [CrossRef]
- Nykamp, K.; Anderson, M.; Powers, M.; Garcia, J.; Herrera, B.; Ho, Y.Y.; Kobayashi, Y.; Patil, N.; Thusberg, J.; Westbrook, M.; et al. Correction: Sherloc: A comprehensive refinement of the ACMG-AMP variant classification criteria. Genet. Med. 2020, 22, 240. [Google Scholar] [CrossRef]
- Kagitani-Shimono, K.; Imai, K.; Okamoto, N.; Ono, J.; Okada, S. Unverricht-Lundborg disease with cystatin B gene abnormalities. Pediatr. Neurol. 2002, 26, 55–60. [Google Scholar] [CrossRef] [PubMed]
- Bobbili, D.R.; Lal, D.; May, P.; Reinthaler, E.M.; Jabbari, K.; Thiele, H.; Nothnagel, M.; Jurkowski, W.; Feucht, M.; Nürnberg, P.; et al. Exome-wide analysis of mutational burden in patients with typical and atypical Rolandic epilepsy. Eur. J. Hum. Genet. 2018, 26, 258–264. [Google Scholar] [CrossRef]
- de Haan, G.J.; Halley, D.J.; Doelman, J.C.; Geesink, H.H.; Augustijn, P.B.; Jager-Jongkind, A.D.; Majoie, M.; Bader, A.J.; Leliefeld-Ten Doeschate, L.A.; Deelen, W.H.; et al. Univerricht-Lundborg disease: Underdiagnosed in the Netherlands. Epilepsia 2004, 45, 1061–1063. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, A.; Marshall, C.R.; Blaser, S.; Ray, P.N.; Yoon, G. Severe neurodegeneration, progressive cerebral volume loss and diffuse hypomyelination associated with a homozygous frameshift mutation in CSTB. Eur. J. Hum. Genet. 2017, 25, 775–778. [Google Scholar] [CrossRef] [PubMed]
- Bespalova, I.N.; Adkins, S.; Pranzatelli, M.; Burmeister, M. Novel cystatin B mutation and diagnostic PCR assay in an Unverricht-Lundborg progressive myoclonus epilepsy patient. Am. J. Med. Genet. 1997, 74, 467–471. [Google Scholar] [CrossRef]
- Mazarib, A.; Xiong, L.; Neufeld, M.Y.; Birnbaum, M.; Korczyn, A.D.; Pandolfo, M.; Berkovic, S.F. Unverricht-Lundborg disease in a five-generation Arab family: Instability of dodecamer repeats. Neurology 2001, 57, 1050–1054. [Google Scholar] [CrossRef]
- Larson, G.P.; Ding, S.; Lafrenière, R.G.; Rouleau, G.A.; Krontiris, T.G. Instability of the EPM1 minisatellite. Hum. Mol. Genet. 1999, 8, 1985–1988. [Google Scholar] [CrossRef][Green Version]
- Nokelainen, P.; Heiskala, H.; Lehesjoki, A.E.; Kaski, M. A patient with 2 different repeat expansion mutations. Arch. Neurol. 2000, 57, 1199–1203. [Google Scholar] [CrossRef]
- Sipilä, J.O.T.; Kälviäinen, R. Comorbidities in patients with Unverricht-Lundborg disease (EPM1). Acta Neurol. Scand. 2022, 146, 690–693. [Google Scholar] [CrossRef]
- Eldridge, R.; Iivanainen, M.; Stern, R.; Koerber, T.; Wilder, B.J. “Baltic” myoclonus epilepsy: Hereditary disorder of childhood made worse by phenytoin. Lancet 1983, 2, 838–842. [Google Scholar] [CrossRef] [PubMed]
- Kyllerman, M.; Sommerfelt, K.; Hedström, A.; Wennergren, G.; Holmgren, D. Clinical and neurophysiological development of Unverricht-Lundborg disease in four Swedish siblings. Epilepsia 1991, 32, 900–909. [Google Scholar] [CrossRef] [PubMed]
- Canafoglia, L.; Ciano, C.; Panzica, F.; Scaioli, V.; Zucca, C.; Agazzi, P.; Visani, E.; Avanzini, G.; Franceschetti, S. Sensorimotor cortex excitability in Unverricht-Lundborg disease and Lafora body disease. Neurology 2004, 63, 2309–2315. [Google Scholar] [CrossRef] [PubMed]
- Joensuu, T.; Tegelberg, S.; Reinmaa, E.; Segerstråle, M.; Hakala, P.; Pehkonen, H.; Korpi, E.R.; Tyynelä, J.; Taira, T.; Hovatta, I.; et al. Gene expression alterations in the cerebellum and granule neurons of Cstb(-/-) mouse are associated with early synaptic changes and inflammation. PLoS ONE 2014, 9, e89321. [Google Scholar] [CrossRef]
- Norio, R.; Koskiniemi, M. Progressive myoclonus epilepsy: Genetic and nosological aspects with special reference to 107 Finnish patients. Clin. Genet. 1979, 15, 382–398. [Google Scholar] [CrossRef] [PubMed]
- Mascalchi, M.; Michelucci, R.; Cosottini, M.; Tessa, C.; Lolli, F.; Riguzzi, P.; Lehesjoki, A.E.; Tosetti, M.; Villari, N.; Tassinari, C.A. Brainstem involvement in Unverricht-Lundborg disease (EPM1): An MRI and (1)H MRS study. Neurology 2002, 58, 1686–1689. [Google Scholar] [CrossRef]
- Silvennoinen, K.; Säisänen, L.; Hyppönen, J.; Rissanen, S.M.; Karjalainen, P.A.; D’Ambrosio, S.; Jimenez-Jimenez, D.; Zagaglia, S.; Rothwell, J.C.; Balestrini, S.; et al. Short- and long-interval intracortical inhibition in EPM1 is related to genotype. Epilepsia 2023, 64, 208–217. [Google Scholar] [CrossRef] [PubMed]
- Virtaneva, K.; D’Amato, E.; Miao, J.; Koskiniemi, M.; Norio, R.; Avanzini, G.; Franceschetti, S.; Michelucci, R.; Tassinari, C.A.; Omer, S.; et al. Unstable minisatellite expansion causing recessively inherited myoclonus epilepsy, EPM1. Nat. Genet. 1997, 15, 393–396. [Google Scholar] [CrossRef]
- Hyppönen, J.; Äikiä, M.; Joensuu, T.; Julkunen, P.; Danner, N.; Koskenkorva, P.; Vanninen, R.; Lehesjoki, A.E.; Mervaala, E.; Kälviäinen, R. Refining the phenotype of Unverricht-Lundborg disease (EPM1): A population-wide Finnish study. Neurology 2015, 84, 1529–1536. [Google Scholar] [CrossRef]
- Jerala, R.; Trstenjak, M.; Lenarcic, B.; Turk, V. Cloning a synthetic gene for human stefin B and its expression in E. coli. FEBS Lett. 1988, 239, 41–44. [Google Scholar] [CrossRef]
- Cipollini, E.; Riccio, M.; Di Giaimo, R.; Dal Piaz, F.; Pulice, G.; Catania, S.; Caldarelli, I.; Dembic, M.; Santi, S.; Melli, M. Cystatin B and its EPM1 mutants are polymeric and aggregate prone in vivo. Biochim. Biophys. Acta 2008, 1783, 312–322. [Google Scholar] [CrossRef]
- Žerovnik, E. Human stefin B: From its structure, folding, and aggregation to its function in health and disease. Front. Mol. Neurosci. 2022, 15, 1009976. [Google Scholar] [CrossRef] [PubMed]
- Kopitar-Jerala, N. The Role of Stefin B in Neuro-inflammation. Front. Cell. Neurosci. 2015, 9, 458. [Google Scholar] [CrossRef]
- Riccio, M.; Santi, S.; Dembic, M.; Di Giaimo, R.; Cipollini, E.; Costantino-Ceccarini, E.; Ambrosetti, D.; Maraldi, N.M.; Melli, M. Cell-specific expression of the epm1 (cystatin B) gene in developing rat cerebellum. Neurobiol. Dis. 2005, 20, 104–114. [Google Scholar] [CrossRef] [PubMed]
- D’Amato, E.; Kokaia, Z.; Nanobashvili, A.; Reeben, M.; Lehesjoki, A.E.; Saarma, M.; Lindvall, O. Seizures induce widespread upregulation of cystatin B, the gene mutated in progressive myoclonus epilepsy, in rat forebrain neurons. Eur. J. Neurosci. 2000, 12, 1687–1695. [Google Scholar] [CrossRef] [PubMed]
- Pennacchio, L.A.; Bouley, D.M.; Higgins, K.M.; Scott, M.P.; Noebels, J.L.; Myers, R.M. Progressive ataxia, myoclonic epilepsy and cerebellar apoptosis in cystatin B-deficient mice. Nat. Genet. 1998, 20, 251–258. [Google Scholar] [CrossRef]
- Penna, E.; Cerciello, A.; Chambery, A.; Russo, R.; Cernilogar, F.M.; Pedone, E.M.; Perrone-Capano, C.; Cappello, S.; Di Giaimo, R.; Crispino, M. Cystatin B Involvement in Synapse Physiology of Rodent Brains and Human Cerebral Organoids. Front. Mol. Neurosci. 2019, 12, 195. [Google Scholar] [CrossRef]
- Riccio, M.; Di Giaimo, R.; Pianetti, S.; Palmieri, P.P.; Melli, M.; Santi, S. Nuclear localization of cystatin B, the cathepsin inhibitor implicated in myoclonus epilepsy (EPM1). Exp. Cell Res. 2001, 262, 84–94. [Google Scholar] [CrossRef]
- Macioce, P.; Gandolfi, N.; Leung, C.L.; Chin, S.S.; Malchiodi-Albedi, F.; Ceccarini, M.; Petrucci, T.C.; Liem, R.K. Characterization of NF-L and betaIISigma1-spectrin interaction in live cells. Exp. Cell Res. 1999, 250, 142–154. [Google Scholar] [CrossRef]
- Liliental, J.; Chang, D.D. Rack1, a receptor for activated protein kinase C, interacts with integrin beta subunit. J. Biol. Chem. 1998, 273, 2379–2383. [Google Scholar] [CrossRef]
- Nardelli, E.; Buonanno, F.; Onnis, L.; Rizzuto, N. Progressive myoclonic epilepsy: Anatomo-clinical study of a sporadic case with a marked cerebellar symptomatology (author’s transl). Riv. Patol. Nerv. Ment. 1975, 96, 221–232. [Google Scholar]
- Sierra-Torre, V.; Plaza-Zabala, A.; Bonifazi, P.; Abiega, O.; Díaz-Aparicio, I.; Tegelberg, S.; Lehesjoki, A.E.; Valero, J.; Sierra, A. Microglial phagocytosis dysfunction in the dentate gyrus is related to local neuronal activity in a genetic model of epilepsy. Epilepsia 2020, 61, 2593–2608. [Google Scholar] [CrossRef]
- Kaasik, A.; Kuum, M.; Aonurm, A.; Kalda, A.; Vaarmann, A.; Zharkovsky, A. Seizures, ataxia, and neuronal loss in cystatin B heterozygous mice. Epilepsia 2007, 48, 752–757. [Google Scholar] [CrossRef] [PubMed]
- Manninen, O.; Koskenkorva, P.; Lehtimäki, K.K.; Hyppönen, J.; Könönen, M.; Laitinen, T.; Kalimo, H.; Kopra, O.; Kälviäinen, R.; Gröhn, O.; et al. White matter degeneration with Unverricht-Lundborg progressive myoclonus epilepsy: A translational diffusion-tensor imaging study in patients and cystatin B-deficient mice. Radiology 2013, 269, 232–239. [Google Scholar] [CrossRef] [PubMed]
- Tegelberg, S.; Kopra, O.; Joensuu, T.; Cooper, J.D.; Lehesjoki, A.E. Early microglial activation precedes neuronal loss in the brain of the Cstb-/- mouse model of progressive myoclonus epilepsy, EPM1. J. Neuropathol. Exp. Neurol. 2012, 71, 40–53. [Google Scholar] [CrossRef]
- Buzzi, A.; Chikhladze, M.; Falcicchia, C.; Paradiso, B.; Lanza, G.; Soukupova, M.; Marti, M.; Morari, M.; Franceschetti, S.; Simonato, M. Loss of cortical GABA terminals in Unverricht-Lundborg disease. Neurobiol. Dis. 2012, 47, 216–224. [Google Scholar] [CrossRef]
- Čeru, S.; Konjar, Š.; Maher, K.; Repnik, U.; Križaj, I.; Benčina, M.; Renko, M.; Nepveu, A.; Žerovnik, E.; Turk, B.; et al. Stefin B interacts with histones and cathepsin L in the nucleus. J. Biol. Chem. 2010, 285, 10078–10086. [Google Scholar] [CrossRef]
- Dhillon, N.; Oki, M.; Szyjka, S.J.; Aparicio, O.M.; Kamakaka, R.T. H2A.Z functions to regulate progression through the cell cycle. Mol. Cell. Biol. 2006, 26, 489–501. [Google Scholar] [CrossRef] [PubMed]
- Jin, C.; Felsenfeld, G. Nucleosome stability mediated by histone variants H3.3 and H2A.Z. Genes Dev. 2007, 21, 1519–1529. [Google Scholar] [CrossRef]
- Abrahamson, M.; Alvarez-Fernandez, M.; Nathanson, C.M. Cystatins. Biochem. Soc. Symp. 2003, 70, 179–199. [Google Scholar] [CrossRef]
- Goulet, B.; Baruch, A.; Moon, N.S.; Poirier, M.; Sansregret, L.L.; Erickson, A.; Bogyo, M.; Nepveu, A. A cathepsin L isoform that is devoid of a signal peptide localizes to the nucleus in S phase and processes the CDP/Cux transcription factor. Mol. Cell 2004, 14, 207–219. [Google Scholar] [CrossRef]
- Sansregret, L.; Goulet, B.; Harada, R.; Wilson, B.; Leduy, L.; Bertoglio, J.; Nepveu, A. The p110 isoform of the CDP/Cux transcription factor accelerates entry into S phase. Mol. Cell. Biol. 2006, 26, 2441–2455. [Google Scholar] [CrossRef] [PubMed]
- Adams-Cioaba, M.A.; Krupa, J.C.; Xu, C.; Mort, J.S.; Min, J. Structural basis for the recognition and cleavage of histone H3 by cathepsin L. Nat. Commun. 2011, 2, 197. [Google Scholar] [CrossRef] [PubMed]
- Duarte, L.F.; Young, A.R.; Wang, Z.; Wu, H.A.; Panda, T.; Kou, Y.; Kapoor, A.; Hasson, D.; Mills, N.R.; Ma’ayan, A.; et al. Histone H3.3 and its proteolytically processed form drive a cellular senescence programme. Nat. Commun. 2014, 5, 5210. [Google Scholar] [CrossRef] [PubMed]
- Duncan, E.M.; Muratore-Schroeder, T.L.; Cook, R.G.; Garcia, B.A.; Shabanowitz, J.; Hunt, D.F.; Allis, C.D. Cathepsin L proteolytically processes histone H3 during mouse embryonic stem cell differentiation. Cell 2008, 135, 284–294. [Google Scholar] [CrossRef] [PubMed]
- Daura, E.; Tegelberg, S.; Yoshihara, M.; Jackson, C.; Simonetti, F.; Aksentjeff, K.; Ezer, S.; Hakala, P.; Katayama, S.; Kere, J.; et al. Cystatin B-deficiency triggers ectopic histone H3 tail cleavage during neurogenesis. Neurobiol. Dis. 2021, 156, 105418. [Google Scholar] [CrossRef] [PubMed]
- Alakurtti, K.; Virtaneva, K.; Joensuu, T.; Palvimo, J.J.; Lehesjoki, A.E. Characterization of the cystatin B gene promoter harboring the dodecamer repeat expanded in progressive myoclonus epilepsy, EPM1. Gene 2000, 242, 65–73. [Google Scholar] [CrossRef] [PubMed]
- Lehtinen, M.K.; Tegelberg, S.; Schipper, H.; Su, H.; Zukor, H.; Manninen, O.; Kopra, O.; Joensuu, T.; Hakala, P.; Bonni, A.; et al. Cystatin B deficiency sensitizes neurons to oxidative stress in progressive myoclonus epilepsy, EPM1. J. Neurosci. 2009, 29, 5910–5915. [Google Scholar] [CrossRef]
- Gorski, K.; Jackson, C.B.; Nyman, T.A.; Rezov, V.; Battersby, B.J.; Lehesjoki, A.E. Progressive mitochondrial dysfunction in cerebellar synaptosomes of cystatin B-deficient mice. Front. Mol. Neurosci. 2023, 16, 1175851. [Google Scholar] [CrossRef]
- Nayak, D.; Roth, T.L.; McGavern, D.B. Microglia development and function. Annu. Rev. Immunol. 2014, 32, 367–402. [Google Scholar] [CrossRef]
- Fraser, D.A.; Pisalyaput, K.; Tenner, A.J. C1q enhances microglial clearance of apoptotic neurons and neuronal blebs, and modulates subsequent inflammatory cytokine production. J. Neurochem. 2010, 112, 733–743. [Google Scholar] [CrossRef]
- Lieuallen, K.; Pennacchio, L.A.; Park, M.; Myers, R.M.; Lennon, G.G. Cystatin B-deficient mice have increased expression of apoptosis and glial activation genes. Hum. Mol. Genet. 2001, 10, 1867–1871. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Yin, L.; Chen, Z. Neuroprotective role of fibronectin in neuron-glial extrasynaptic transmission. Neural Regen. Res. 2013, 8, 376–382. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Wang, K.K. Glial fibrillary acidic protein: From intermediate filament assembly and gliosis to neurobiomarker. Trends Neurosci. 2015, 38, 364–374. [Google Scholar] [CrossRef] [PubMed]
- Han, J.; Lee, J.D.; Bibbs, L.; Ulevitch, R.J. A MAP kinase targeted by endotoxin and hyperosmolarity in mammalian cells. Science 1994, 265, 808–811. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Trombley, P.Q.; van den Pol, A.N. GABA receptors precede glutamate receptors in hypothalamic development; differential regulation by astrocytes. J. Neurophysiol. 1995, 74, 1473–1484. [Google Scholar] [CrossRef]
- Wu, C.; Sun, D. GABA receptors in brain development, function, and injury. Metab. Brain Dis. 2015, 30, 367–379. [Google Scholar] [CrossRef]
- Lobb, C.J.; Wilson, C.J.; Paladini, C.A. A dynamic role for GABA receptors on the firing pattern of midbrain dopaminergic neurons. J. Neurophysiol. 2010, 104, 403–413. [Google Scholar] [CrossRef]
- Franceschetti, S.; Sancini, G.; Buzzi, A.; Zucchini, S.; Paradiso, B.; Magnaghi, G.; Frassoni, C.; Chikhladze, M.; Avanzini, G.; Simonato, M. A pathogenetic hypothesis of Unverricht-Lundborg disease onset and progression. Neurobiol. Dis. 2007, 25, 675–685. [Google Scholar] [CrossRef]
- Okada, Y.; Yamazaki, H.; Sekine-Aizawa, Y.; Hirokawa, N. The neuron-specific kinesin superfamily protein KIF1A is a unique monomeric motor for anterograde axonal transport of synaptic vesicle precursors. Cell 1995, 81, 769–780. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Singh, S.; Hämäläinen, R.H. The Roles of Cystatin B in the Brain and Pathophysiological Mechanisms of Progressive Myoclonic Epilepsy Type 1. Cells 2024, 13, 170. https://doi.org/10.3390/cells13020170
Singh S, Hämäläinen RH. The Roles of Cystatin B in the Brain and Pathophysiological Mechanisms of Progressive Myoclonic Epilepsy Type 1. Cells. 2024; 13(2):170. https://doi.org/10.3390/cells13020170
Chicago/Turabian StyleSingh, Shekhar, and Riikka H. Hämäläinen. 2024. "The Roles of Cystatin B in the Brain and Pathophysiological Mechanisms of Progressive Myoclonic Epilepsy Type 1" Cells 13, no. 2: 170. https://doi.org/10.3390/cells13020170
APA StyleSingh, S., & Hämäläinen, R. H. (2024). The Roles of Cystatin B in the Brain and Pathophysiological Mechanisms of Progressive Myoclonic Epilepsy Type 1. Cells, 13(2), 170. https://doi.org/10.3390/cells13020170







