Coronavirus Receptor Expression Profiles in Human Mast Cells, Basophils, and Eosinophils
Abstract
1. Introduction
2. Materials and Methods
2.1. Reagents
2.2. Isolation of Primary MC, BA, and EO
2.3. Cell Lines
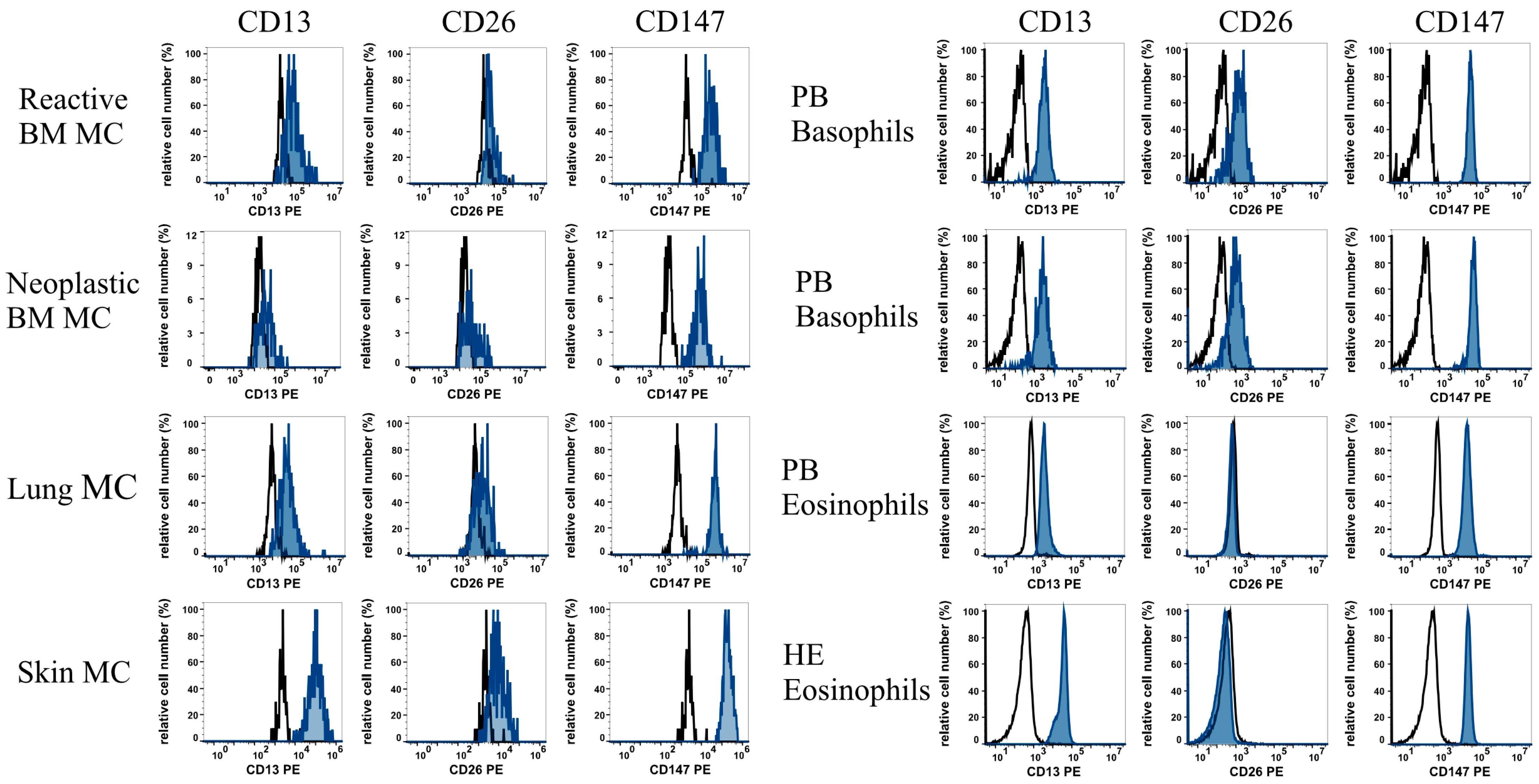
2.4. Evaluation of Surface Expression of CoV-R on Cell Lines and Primary Leukocytes Using Flow Cytometry
2.5. Effects of Various Drugs on Expression of CoV-R on MC, BA, and EO
2.6. Quantitative PCR (qPCR)
2.7. Statistical Analysis
3. Results
3.1. MC, BA, and EO Display Distinct Profiles of Cell Surface CoV-R
3.2. Expression of CoV-R in MC, BA, and EO Cell Lines
3.3. Effects of Pharmacologic Inhibitors on Expression of CoV-R on MC, BA, and EO
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Morris, S.B.; Schwartz, N.G.; Patel, P.; Abbo, L.; Beauchamps, L.; Balan, S.; Lee, E.H.; Paneth-Pollak, R.; Geevarughese, A.; Lash, M.K.; et al. Case Series of Multisystem Inflammatory Syndrome in Adults Associated with SARS-CoV-2 Infection—United Kingdom and United States, March–August 2020. MMWR Morb. Mortal. Wkly. Rep. 2020, 69, 1450–1456. [Google Scholar] [CrossRef] [PubMed]
- Pollard, C.A.; Morran, M.P.; Nestor-Kalinoski, A.L. The COVID-19 pandemic: A global health crisis. Physiol. Genom. 2020, 52, 549–557. [Google Scholar] [CrossRef] [PubMed]
- Montenegro, F.; Unigarro, L.; Paredes, G.; Moya, T.; Romero, A.; Torres, L.; López, J.C.; González, F.E.J.; Del Pozo, G.; López-Cortés, A.; et al. Acute respiratory distress syndrome (ARDS) caused by the novel coronavirus disease (COVID-19): A practical comprehensive literature review. Expert. Rev. Respir. Med. 2021, 15, 183–195. [Google Scholar] [CrossRef] [PubMed]
- Gustine, J.N.; Jones, D. Immunopathology of Hyperinflammation in COVID-19. Am. J. Pathol. 2021, 191, 4–17. [Google Scholar] [CrossRef] [PubMed]
- Alon, R.; Sportiello, M.; Kozlovski, S.; Kumar, A.; Reilly, E.C.; Zarbock, A.; Garbi, N.; Topham, D.J. Leukocyte trafficking to the lungs and beyond: Lessons from influenza for COVID-19. Nat. Rev. Immunol. 2021, 21, 49–64. [Google Scholar] [CrossRef]
- Degauque, N.; Haziot, A.; Brouard, S.; Mooney, N. Endothelial cell, myeloid, and adaptive immune responses in SARS-CoV-2 infection. FASEB J. 2021, 35, e21577. [Google Scholar] [CrossRef] [PubMed]
- Darif, D.; Hammi, I.; Kihel, A.; El Idrissi Saik, I.; Guessous, F.; Akarid, K. The pro-inflammatory cytokines in COVID-19 pathogenesis: What goes wrong? Microb. Pathog. 2021, 153, 104799. [Google Scholar] [CrossRef]
- Leisman, D.E.; Ronner, L.; Pinotti, R.; Taylor, M.D.; Sinha, P.; Calfee, C.S.; Hirayama, A.V.; Mastroiani, F.; Turtle, C.J.; Harhay, M.O.; et al. Cytokine elevation in severe and critical COVID-19: A rapid systematic review, meta-analysis, and comparison with other inflammatory syndromes. Lancet Respir. Med. 2020, 8, 1233–1244. [Google Scholar] [CrossRef] [PubMed]
- Meidaninikjeh, S.; Sabouni, N.; Marzouni, H.Z.; Bengar, S.; Khalili, A.; Jafari, R. Monocytes and macrophages in COVID-19: Friends and foes. Life Sci. 2021, 269, 119010. [Google Scholar] [CrossRef]
- Choudhary, S.; Sharma, K.; Silakari, O. The interplay between inflammatory pathways and COVID-19: A critical review on pathogenesis and therapeutic options. Microb. Pathog. 2021, 150, 104673. [Google Scholar] [CrossRef]
- Indari, O.; Jakhmola, S.; Manivannan, E.; Jha, H.C. An Update on Antiviral Therapy Against SARS-CoV-2: How Far Have We Come? Front. Pharmacol. 2021, 12, 632677. [Google Scholar] [CrossRef] [PubMed]
- Vivekanandhan, K.; Shanmugam, P.; Barabadi, H.; Arumugam, V.; Daniel Raj Daniel Paul Raj, D.; Sivasubramanian, M.; Ramasamy, S.; Anand, K.; Boomi, P.; Chandrasekaran, B.; et al. Emerging Therapeutic Approaches to Combat COVID-19: Present Status and Future Perspectives. Front. Mol. Biosci. 2021, 8, 604447. [Google Scholar] [CrossRef] [PubMed]
- Welte, T.; Ambrose, L.J.; Sibbring, G.C.; Sheikh, S.; Müllerová, H.; Sabir, I. Current evidence for COVID-19 therapies: A systematic literature review. Eur. Respir. Rev. 2021, 30, 200384. [Google Scholar] [CrossRef] [PubMed]
- Abeldaño Zuñiga, R.A.; Coca, S.M.; Abeldaño, G.F.; González-Villoria, R.A.M. Clinical effectiveness of drugs in hospitalized patients with COVID-19: A systematic review and meta-analysis. Ther. Adv. Respir. Dis. 2021, 15, 17534666211007214. [Google Scholar] [CrossRef] [PubMed]
- Abdala-Valencia, H.; Coden, M.E.; Chiarella, S.E.; Jacobsen, E.A.; Bochner, B.S.; Lee, J.J.; Berdnikovs, S. Shaping eosinophil identity in the tissue contexts of development, homeostasis, and disease. J. Leukoc. Biol. 2018, 104, 95–108. [Google Scholar] [CrossRef]
- Galli, S.J.; Tsai, M. Mast cells in allergy and infection: Versatile effector and regulatory cells in innate and adaptive immunity. Eur. J. Immunol. 2010, 40, 1843–1851. [Google Scholar] [CrossRef]
- Gibbs, B.F. Human basophils as effectors and immunomodulators of allergic inflammation and innate immunity. Clin. Exp. Med. 2005, 5, 43–49. [Google Scholar] [CrossRef]
- McBrien, C.N.; Menzies-Gow, A. The Biology of Eosinophils and Their Role in Asthma. Front. Med. 2017, 4, 93. [Google Scholar] [CrossRef]
- Theoharides, T.C.; Valent, P.; Akin, C. Mast Cells, Mastocytosis, and Related Disorders. N. Engl. J. Med. 2015, 373, 1885–1886. [Google Scholar] [CrossRef]
- Valent, P.; Akin, C.; Hartmann, K.; Nilsson, G.; Reiter, A.; Hermine, O.; Sotlar, K.; Sperr, W.R.; Escribano, L.; George, T.I.; et al. Mast cells as a unique hematopoietic lineage and cell system: From Paul Ehrlich’s visions to precision medicine concepts. Theranostics 2020, 10, 10743–10768. [Google Scholar] [CrossRef]
- Valent, P.; Bettelheim, P. Cell surface structures on human basophils and mast cells: Biochemical and functional characterization. Adv. Immunol. 1992, 52, 333–423. [Google Scholar] [CrossRef] [PubMed]
- Mayerhofer, M.; Aichberger, K.J.; Florian, S.; Valent, P. Recognition sites for microbes and components of the immune system on human mast cells: Relationship to CD antigens and implications for host defense. Int. J. Immunopathol. Pharmacol. 2007, 20, 421–434. [Google Scholar] [CrossRef] [PubMed]
- Akoto, C.; Davies, D.E.; Swindle, E.J. Mast cells are permissive for rhinovirus replication: Potential implications for asthma exacerbations. Clin. Exp. Allergy 2017, 47, 351–360. [Google Scholar] [CrossRef] [PubMed]
- Murphy-Schafer, A.R.; Paust, S. Divergent Mast Cell Responses Modulate Antiviral Immunity During Influenza Virus Infection. Front. Cell. Infect. Microbiol. 2021, 11, 580679. [Google Scholar] [CrossRef] [PubMed]
- Phipps, S.; Lam, C.E.; Mahalingam, S.; Newhouse, M.; Ramirez, R.; Rosenberg, H.F.; Foster, P.S.; Matthaei, K.I. Eosinophils contribute to innate antiviral immunity and promote clearance of respiratory syncytial virus. Blood 2007, 110, 1578–1586. [Google Scholar] [CrossRef] [PubMed]
- Rathore, A.P.; St John, A.L. Protective and pathogenic roles for mast cells during viral infections. Curr. Opin. Immunol. 2020, 66, 74–81. [Google Scholar] [CrossRef]
- Sundstrom, J.B.; Ellis, J.E.; Hair, G.A.; Kirshenbaum, A.S.; Metcalfe, D.D.; Yi, H.; Cardona, A.C.; Lindsay, M.K.; Ansari, A.A. Human tissue mast cells are an inducible reservoir of persistent HIV infection. Blood 2007, 109, 5293–5300. [Google Scholar] [CrossRef] [PubMed]
- St John, A.L.; Rathore, A.P.; Yap, H.; Ng, M.L.; Metcalfe, D.D.; Vasudevan, S.G.; Abraham, S.N. Immune surveillance by mast cells during dengue infection promotes natural killer (NK) and NKT-cell recruitment and viral clearance. Proc. Natl. Acad. Sci. USA 2011, 108, 9190–9195. [Google Scholar] [CrossRef]
- Gadanec, L.K.; McSweeney, K.R.; Qaradakhi, T.; Ali, B.; Zulli, A.; Apostolopoulos, V. Can SARS-CoV-2 Virus Use Multiple Receptors to Enter Host Cells? Int. J. Mol. Sci. 2021, 22, 992. [Google Scholar] [CrossRef]
- Matusiak, M.; Schürch, C.M. Expression of SARS-CoV-2 entry receptors in the respiratory tract of healthy individuals, smokers and asthmatics. Respir. Res. 2020, 21, 252. [Google Scholar] [CrossRef]
- Radzikowska, U.; Ding, M.; Tan, G.; Zhakparov, D.; Peng, Y.; Wawrzyniak, P.; Wang, M.; Li, S.; Morita, H.; Altunbulakli, C.; et al. Distribution of ACE2, CD147, CD26, and other SARS-CoV-2 associated molecules in tissues and immune cells in health and in asthma, COPD, obesity, hypertension, and COVID-19 risk factors. Allergy 2020, 75, 2829–2845. [Google Scholar] [CrossRef] [PubMed]
- Yeager, C.L.; Ashmun, R.A.; Williams, R.K.; Cardellichio, C.B.; Shapiro, L.H.; Look, A.T.; Holmes, K.V. Human aminopeptidase N is a receptor for human coronavirus 229E. Nature 1992, 357, 420–422. [Google Scholar] [CrossRef]
- Zhou, J.; Li, C.; Zhao, G.; Chu, H.; Wang, D.; Yan, H.H.; Poon, V.K.; Wen, L.; Wong, B.H.; Zhao, X.; et al. Human intestinal tract serves as an alternative infection route for Middle East respiratory syndrome coronavirus. Sci. Adv. 2017, 3, eaao4966. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Chen, W.; Zhang, Z.; Deng, Y.; Lian, J.Q.; Du, P.; Wei, D.; Zhang, Y.; Sun, X.X.; Gong, L.; et al. CD147-spike protein is a novel route for SARS-CoV-2 infection to host cells. Signal Transduct. Target. Ther. 2020, 5, 283. [Google Scholar] [CrossRef] [PubMed]
- Gschwandtner, M.; Paulitschke, V.; Mildner, M.; Brunner, P.M.; Hacker, S.; Eisenwort, G.; Sperr, W.R.; Valent, P.; Gerner, C.; Tschachler, E. Proteome analysis identifies L1CAM/CD171 and DPP4/CD26 as novel markers of human skin mast cells. Allergy 2017, 72, 85–97. [Google Scholar] [CrossRef]
- Schulman, E.S.; MacGlashan, D.W., Jr.; Peters, S.P.; Schleimer, R.P.; Newball, H.H.; Lichtenstein, L.M. Human lung mast cells: Purification and characterization. J. Immunol. 1982, 129, 2662–2667. [Google Scholar] [CrossRef]
- Valent, P.; Ashman, L.K.; Hinterberger, W.; Eckersberger, F.; Majdic, O.; Lechner, K.; Bettelheim, P. Mast cell typing: Demonstration of a distinct hematopoietic cell type and evidence for immunophenotypic relationship to mononuclear phagocytes. Blood 1989, 73, 1778–1785. [Google Scholar] [CrossRef]
- Kishi, K. A new leukemia cell line with Philadelphia chromosome characterized as basophil precursors. Leuk. Res. 1985, 9, 381–390. [Google Scholar] [CrossRef]
- Butterfield, J.H.; Weiler, D.; Dewald, G.; Gleich, G.J. Establishment of an immature mast cell line from a patient with mast cell leukemia. Leuk. Res. 1988, 12, 345–355. [Google Scholar] [CrossRef]
- Akin, C.; Brockow, K.; D’Ambrosio, C.; Kirshenbaum, A.S.; Ma, Y.; Longley, B.J.; Metcalfe, D.D. Effects of tyrosine kinase inhibitor STI571 on human mast cells bearing wild-type or mutated c-kit. Exp. Hematol. 2003, 31, 686–692. [Google Scholar] [CrossRef]
- Gleixner, K.V.; Mayerhofer, M.; Aichberger, K.J.; Derdak, S.; Sonneck, K.; Böhm, A.; Gruze, A.; Samorapoompichit, P.; Manley, P.W.; Fabbro, D.; et al. PKC412 inhibits in vitro growth of neoplastic human mast cells expressing the D816V-mutated variant of KIT: Comparison with AMN107, imatinib, and cladribine (2CdA) and evaluation of cooperative drug effects. Blood 2006, 107, 752–759. [Google Scholar] [CrossRef]
- Saleh, R.; Wedeh, G.; Herrmann, H.; Bibi, S.; Cerny-Reiterer, S.; Sadovnik, I.; Blatt, K.; Hadzijusufovic, E.; Jeanningros, S.; Blanc, C.; et al. A new human mast cell line expressing a functional IgE receptor converts to tumorigenic growth by KIT D816V transfection. Blood 2014, 124, 111–120. [Google Scholar] [CrossRef] [PubMed]
- Hoermann, G.; Blatt, K.; Greiner, G.; Putz, E.M.; Berger, A.; Herrmann, H.; Cerny-Reiterer, S.; Gleixner, K.V.; Walz, C.; Hoetzenecker, K.; et al. CD52 is a molecular target in advanced systemic mastocytosis. FASEB J. 2014, 28, 3540–3551. [Google Scholar] [CrossRef] [PubMed]
- Smiljkovic, D.; Herrmann, H.; Sadovnik, I.; Gamperl, S.; Berger, D.; Stefanzl, G.; Eisenwort, G.; Hoermann, G.; Kopanja, S.; Dorofeeva, Y.; et al. Expression and regulation of Siglec-6 (CD327) on human mast cells and basophils. J. Allergy Clin. Immunol. 2023, 151, 202–211. [Google Scholar] [CrossRef] [PubMed]
- Sadovnik, I.; Ivanov, D.; Smiljkovic, D.; Stefanzl, G.; Degenfeld-Schonburg, L.; Herndlhofer, S.; Eisenwort, G.; Hauswirth, A.W.; Sliwa, T.; Keil, F.; et al. Identification of CD203c as a New Basophil-Specific Flow-Marker in Ph(+) Chronic Myeloid Leukemia. Cells 2022, 12, 3. [Google Scholar] [CrossRef] [PubMed]
- Herrmann, H.; Cerny-Reiterer, S.; Gleixner, K.V.; Blatt, K.; Herndlhofer, S.; Rabitsch, W.; Jäger, E.; Mitterbauer-Hohendanner, G.; Streubel, B.; Selzer, E.; et al. CD34(+)/CD38(−) stem cells in chronic myeloid leukemia express Siglec-3 (CD33) and are responsive to the CD33-targeting drug gemtuzumab/ozogamicin. Haematologica 2012, 97, 219–226. [Google Scholar] [CrossRef]
- Herrmann, H.; Sadovnik, I.; Cerny-Reiterer, S.; Rülicke, T.; Stefanzl, G.; Willmann, M.; Hoermann, G.; Bilban, M.; Blatt, K.; Herndlhofer, S.; et al. Dipeptidylpeptidase IV (CD26) defines leukemic stem cells (LSC) in chronic myeloid leukemia. Blood 2014, 123, 3951–3962. [Google Scholar] [CrossRef] [PubMed]
- Krysko, O.; Bourne, J.H.; Kondakova, E.; Galova, E.A.; Whitworth, K.; Newby, M.L.; Bachert, C.; Hill, H.; Crispin, M.; Stamataki, Z.; et al. Severity of SARS-CoV-2 infection is associated with high numbers of alveolar mast cells and their degranulation. Front. Immunol. 2022, 13, 968981. [Google Scholar] [CrossRef]
- Gebremeskel, S.; Schanin, J.; Coyle, K.M.; Butuci, M.; Luu, T.; Brock, E.C.; Xu, A.; Wong, A.; Leung, J.; Korver, W.; et al. Mast Cell and Eosinophil Activation Are Associated With COVID-19 and TLR-Mediated Viral Inflammation: Implications for an Anti-Siglec-8 Antibody. Front. Immunol. 2021, 12, 650331. [Google Scholar] [CrossRef]
- Schaller, T.; Märkl, B.; Claus, R.; Sholl, L.; Hornick, J.L.; Giannetti, M.P.; Schweizer, L.; Mann, M.; Castells, M. Mast cells in lung damage of COVID-19 autopsies: A descriptive study. Allergy 2022, 77, 2237–2239. [Google Scholar] [CrossRef]
- Budnevsky, A.V.; Avdeev, S.N.; Kosanovic, D.; Shishkina, V.V.; Filin, A.A.; Esaulenko, D.I.; Ovsyannikov, E.S.; Samoylenko, T.V.; Redkin, A.N.; Suvorova, O.A.; et al. Role of mast cells in the pathogenesis of severe lung damage in COVID-19 patients. Respir. Res. 2022, 23, 371. [Google Scholar] [CrossRef]
- Murdaca, G.; Di Gioacchino, M.; Greco, M.; Borro, M.; Paladin, F.; Petrarca, C.; Gangemi, S. Basophils and Mast Cells in COVID-19 Pathogenesis. Cells 2021, 10, 2754. [Google Scholar] [CrossRef]
- Wang, R.; Simoneau, C.R.; Kulsuptrakul, J.; Bouhaddou, M.; Travisano, K.A.; Hayashi, J.M.; Carlson-Stevermer, J.; Zengel, J.R.; Richards, C.M.; Fozouni, P.; et al. Genetic Screens Identify Host Factors for SARS-CoV-2 and Common Cold Coronaviruses. Cell 2021, 184, 106–119. [Google Scholar] [CrossRef] [PubMed]
- Ashmun, R.A.; Look, A.T. Metalloprotease activity of CD13/aminopeptidase N on the surface of human myeloid cells. Blood 1990, 75, 462–469. [Google Scholar] [CrossRef] [PubMed]
- Dixon, J.; Kaklamanis, L.; Turley, H.; Hickson, I.D.; Leek, R.D.; Harris, A.L.; Gatter, K.C. Expression of aminopeptidase-n (CD 13) in normal tissues and malignant neoplasms of epithelial and lymphoid origin. J. Clin. Pathol. 1994, 47, 43–47. [Google Scholar] [CrossRef] [PubMed]
- Piedfer, M.; Dauzonne, D.; Tang, R.; N’Guyen, J.; Billard, C.; Bauvois, B. Aminopeptidase-N/CD13 is a potential proapoptotic target in human myeloid tumor cells. FASEB J. 2011, 25, 2831–2842. [Google Scholar] [CrossRef]
- He, X.; Feng, Z.; Ma, J.; Ling, S.; Cao, Y.; Gurung, B.; Wu, Y.; Katona, B.W.; O’Dwyer, K.P.; Siegel, D.L.; et al. Bispecific and split CAR T cells targeting CD13 and TIM3 eradicate acute myeloid leukemia. Blood 2020, 135, 713–723. [Google Scholar] [CrossRef]
- Williams, B.A.; Law, A.; Hunyadkurti, J.; Desilets, S.; Leyton, J.V.; Keating, A. Antibody Therapies for Acute Myeloid Leukemia: Unconjugated, Toxin-Conjugated, Radio-Conjugated and Multivalent Formats. J. Clin. Med. 2019, 8, 1261. [Google Scholar] [CrossRef]
- Bouchet, S.; Tang, R.; Fava, F.; Legrand, O.; Bauvois, B. The CNGRC-GG-D(KLAKLAK)2 peptide induces a caspase-independent, Ca2+-dependent death in human leukemic myeloid cells by targeting surface aminopeptidase N/CD13. Oncotarget 2016, 7, 19445–19467. [Google Scholar] [CrossRef]
- Chott, A.; Guenther, P.; Huebner, A.; Selzer, E.; Parwaresch, R.M.; Horny, H.P.; Valent, P. Morphologic and immunophenotypic properties of neoplastic cells in a case of mast cell sarcoma. Am. J. Surg. Pathol. 2003, 27, 1013–1019. [Google Scholar] [CrossRef]
- Dahl, C.; Hoffmann, H.J.; Saito, H.; Schiøtz, P.O. Human mast cells express receptors for IL-3, IL-5 and GM-CSF; A partial map of receptors on human mast cells cultured in vitro. Allergy 2004, 59, 1087–1096. [Google Scholar] [CrossRef] [PubMed]
- Hennersdorf, F.; Florian, S.; Jakob, A.; Baumgärtner, K.; Sonneck, K.; Nordheim, A.; Biedermann, T.; Valent, P.; Bühring, H.J. Identification of CD13, CD107a, and CD164 as novel basophil-activation markers and dissection of two response patterns in time kinetics of IgE-dependent upregulation. Cell Res. 2005, 15, 325–335. [Google Scholar] [CrossRef]
- Sonneck, K.; Baumgartner, C.; Rebuzzi, L.; Marth, K.; Chen, K.W.; Hauswirth, A.W.; Florian, S.; Vrtala, S.; Bühring, H.J.; Valenta, R.; et al. Recombinant allergens promote expression of aminopeptidase-n (CD13) on basophils in allergic patients. Int. J. Immunopathol. Pharmacol. 2008, 21, 11–21. [Google Scholar] [CrossRef] [PubMed]
- Braun, R.K.; Foerster, M.; Workalemahu, G.; Haefner, D.; Kroegel, C.; Walker, C. Differential regulation of aminopeptidase N (CD13) by transendothelial migration and cytokines on human eosinophils. Exp. Lung Res. 2003, 29, 59–77. [Google Scholar] [CrossRef] [PubMed]
- Füreder, W.; Agis, H.; Sperr, W.R.; Lechner, K.; Valent, P. The surface membrane antigen phenotype of human blood basophils. Allergy 1994, 49, 861–865. [Google Scholar] [CrossRef] [PubMed]
- Pushkarsky, T.; Zybarth, G.; Dubrovsky, L.; Yurchenko, V.; Tang, H.; Guo, H.; Toole, B.; Sherry, B.; Bukrinsky, M. CD147 facilitates HIV-1 infection by interacting with virus-associated cyclophilin A. Proc. Natl. Acad. Sci. USA 2001, 89, 6360–6365. [Google Scholar] [CrossRef]
- Watanabe, A.; Yoneda, M.; Ikeda, F.; Terao-Muto, Y.; Sato, H.; Kai, C. CD147/EMMPRIN acts as a functional entry receptor for measles virus on epithelial cells. J. Virol. 2010, 84, 4183–4193. [Google Scholar] [CrossRef]
- Fenizia, C.; Galbiati, S.; Vanetti, C.; Vago, R.; Clerici, M.; Tacchetti, C.; Daniele, T. SARS-CoV-2 Entry: At the Crossroads of CD147 and ACE2. Cells 2021, 10, 1434. [Google Scholar] [CrossRef]
- Schernthaner, G.H.; Hauswirth, A.W.; Baghestanian, M.; Agis, H.; Ghannadan, M.; Worda, C.; Krauth, M.T.; Printz, D.; Fritsch, G.; Sperr, W.R.; et al. Detection of differentiation- and activation-linked cell surface antigens on cultured mast cell progenitors. Allergy 2005, 60, 1248–1255. [Google Scholar] [CrossRef]
- Wimazal, F.; Ghannadan, M.; Müller, M.R.; End, A.; Willheim, M.; Meidlinger, P.; Schernthaner, G.H.; Jordan, J.H.; Hagen, W.; Agis, H.; et al. Expression of homing receptors and related molecules on human mast cells and basophils: A comparative analysis using multi-color flow cytometry and toluidine blue/immunofluorescence staining techniques. Tissue Antigens 1999, 54, 499–507. [Google Scholar] [CrossRef]
- Hoffmann, M.; Kleine-Weber, H.; Schroeder, S.; Krüger, N.; Herrler, T.; Erichsen, S.; Schiergens, T.S.; Herrler, G.; Wu, N.H.; Nitsche, A.; et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 2020, 181, 271–280. [Google Scholar] [CrossRef] [PubMed]
- Hatmal, M.M.; Alshaer, W.; Al-Hatamleh, M.A.I.; Hatmal, M.; Smadi, O.; Taha, M.O.; Oweida, A.J.; Boer, J.C.; Mohamud, R.; Plebanski, M. Comprehensive Structural and Molecular Comparison of Spike Proteins of SARS-CoV-2, SARS-CoV and MERS-CoV, and Their Interactions with ACE2. Cells 2020, 9, 2638. [Google Scholar] [CrossRef]
- Kim, H.Y.; Kang, H.G.; Kim, H.M.; Jeong, H.J. Expression of SARS-CoV-2 receptor angiotensin-converting enzyme 2 by activating protein-1 in human mast cells. Cell Immunol. 2023, 386, 104705. [Google Scholar] [CrossRef]
- Giannetti, M.P.; Weller, E.; Alvarez-Twose, I.; Torrado, I.; Bonadonna, P.; Zanotti, R.; Dwyer, D.F.; Foer, D.; Akin, C.; Hartmann, K.; et al. COVID-19 infection in patients with mast cell disorders including mastocytosis does not impact mast cell activation symptoms. J. Allergy Clin. Immunol. Pract. 2021, 9, 2083–2086. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Zhong, L.; Deng, J.; Peng, J.; Dan, H.; Zeng, X.; Li, T.; Chen, Q. High expression of ACE2 receptor of 2019-nCoV on the epithelial cells of oral mucosa. Int. J. Oral Sci. 2020, 12, 8. [Google Scholar] [CrossRef] [PubMed]
- Coleman, C.M.; Sisk, J.M.; Mingo, R.M.; Nelson, E.A.; White, J.M.; Frieman, M.B. Abelson Kinase Inhibitors Are Potent Inhibitors of Severe Acute Respiratory Syndrome Coronavirus and Middle East Respiratory Syndrome Coronavirus Fusion. J. Virol. 2016, 90, 8924–8933. [Google Scholar] [CrossRef]

| Antigen | CoV |
|---|---|
| ACE2 | SARS-CoV, SARS-CoV-2, hCoV-NL63 |
| ABL2 | SARS-CoV, MERS-CoV |
| CD13 = Aminopeptidase N | hCoV-229E |
| CD26 = Dipeptidylpeptidase IV | MERS-CoV |
| CD147 = Basigin | SARS-CoV-2 |
| 9-O-Acetylated Sialic Acid | hCoV-OC43, hCoV-HKU1 |
| Patient Number | Age (Years) | Sex (m/f) | Diagnosis | Sample PB or BM | Serum Tryptase (ng/mL) | Hb (g/dL) | WBC (G/L) | PLT (G/L) | % EO in PB | % MC in MNC a | % MC in BM Smears b | % MC in BM Histology |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| #1 | 61 | f | Pancytopenia * | BM | n.a. | 7.9 | 2.43 | 27 | 0 | 0.02% | <1 | n.a. |
| #2 | 32 | f | Chronic Kidney Disease and Idiopathic Hypereosinophilia | BM | 26.8 | 10.2 | 12.72 | 359 | 32 | 0.07% | <1 | n.a. |
| #3 | 45 | m | Chronic spontaneous Urticaria | BM | 5 | 13.1 | 7.74 | 230 | 9 | 0.006% | <1 | n.a. |
| #4 | 52 | m | Hodgkin Lymphoma | BM | n.a. | 12.3 | 10.05 | 330 | 4 | 0.003% | <1 | n.a. |
| #5 | 24 | f | Cutaneous Mastocytosis | BM | 10.2 | 14.3 | 7.10 | 210 | 4 | 0.02% | <1 | n.a. |
| #6 | 32 | f | Suspected HαT ** | BM | 151 | 12.6 | 8.27 | 336 | 11 | 0.0001% | <1 | n.a. |
| #7 | 30 | f | ISM | BM | 88.2 | 13.0 | 4.94 | 169 | 2 | 0.01% | <1 | 10 |
| #8 | 62 | m | SM-CMML | BM | 70.7 | 9.0 | 13.91 | 122 | 4 | 0.008% | <1 | 5–10 |
| #9 | 72 | m | SM-CMML | BM | 616 | 11.6 | 8.58 | 83 | 0 | 0.01% | <1 | 5 |
| #10 | 69 | f | Reactive Hypereosinophilia *** | PB | 8.9 | 12.7 | 12.76 | 159 | 62 | n.a. | n.a. | n.a. |
| #11 | 74 | m | MPN/MDS-Eo | PB | 5.8 | 9.4 | 34.27 | 24 | 36 | n.a. | n.a. | n.a. |
| #12 | 78 | f | Lymphocyte-Variant of Hypereosinophilic Syndrome | PB | 6.8 | 10.9 | 9.91 | 373 | 26 | n.a. | n.a. | n.a. |
| Cell Type | Surface Expression of Coronavirus Receptors | |||
|---|---|---|---|---|
| Cell Lines | CD13 | CD26 | CD147 | |
| HMC-1.1 | + | − | ++ | |
| HMC-1.2 | + | − | ++ | |
| MCPV-1.1 | + | − | ++ | |
| MCPV-1.2 | + | − | ++ | |
| MCPV-1.3 | + | − | ++ | |
| MCPV-1.4 | + | − | ++ | |
| ROSAKIT WT | + | − | +++ | |
| ROSAKIT D816V | + | − | +++ | |
| ROSAKIT K509I | +/− | − | +++ | |
| KU812 | + | − | +++ | |
| EOL-1 | + | + | +++ | |
| Primary cells | number of samples (n) | |||
| Reactive BM MC | n = 6 | +/− | +/− | +/++ |
| Lung MC | n = 4 | + | +/− | ++ |
| Skin MC | n = 3 | +/++ | +/− | ++ |
| Neoplastic BM MC | n = 3 | +/− | +/− | ++ |
| Basophils (HD PB) | n = 13 | +/++ | + | +++ |
| Eosinophils (HD PB) | n = 13 | + | − | ++ |
| Eosinophils (HE PB) | n = 3 | ++ | − | ++ |
| Cell Line | Expression of mRNA Specific for | |||||
|---|---|---|---|---|---|---|
| ACE2 | ABL1 | ABL2 | ANPEP (CD13) | DPPIV (CD26) | BSG (CD147) | |
| HMC-1.1 | – | + | + | + | – | + |
| HMC-1.2 | – | + | + | + | – | + |
| ROSAKIT WT | – | + | + | + | – | + |
| ROSAKIT D816V | – | + | + | –/+ | – | + |
| ROSAKIT K509I | – | + | + | – | – | + |
| MCPV-1.1 | – | + | + | + | – | + |
| MCPV-1.2 | – | + | + | + | – | + |
| MCPV-1.3 | – | + | + | + | – | + |
| MCPV-1.4 | – | + | + | + | – | + |
| KU812 | – | + | + | + | – | + |
| EOL-1 | – | + | + | – | + | + |
| Cell Line | CoV-R | Effects of Drugs on Expression of CoV-R | ||
|---|---|---|---|---|
| Hydroxychloroquine | Dexamethasone | Vitamin D | ||
| HMC-1.1 | CD13 | 69 ± 5.0 | 53 ± 14.7 * | 88 ± 2.8 |
| CD147 | 100 ± 6.6 | 63 ± 19.2 * | 103 ± 5.7 | |
| HMC-1.2 | CD13 | 104 ± 11.7 | 94 ± 3.9 | 80 ± 19.0 |
| CD147 | 100 ± 2.4 | 99 ± 2.4 | 102 ± 7.7 | |
| ROSAKIT WT | CD13 | 104 ± 2.5 | 95 ± 5.9 | 106 ± 7.9 |
| CD147 | 101 ± 11.4 | 87 ± 10.5 | 107 ± 4.8 | |
| ROSAKIT D816V | CD13 | 117 ± 15.6 | 110 ± 15.4 | 90 ± 19.8 |
| CD147 | 94 ± 14.5 | 90 ± 12.3 | 83 ± 35.5 | |
| ROSAKIT K509I | CD13 | 100 ± 8.3 | 98 ± 5.2 | 89 ± 13.2 |
| CD147 | 121 ± 27.1 | 108 ± 27.0 | 89 ± 41.8 | |
| MCPV-1.1 | CD13 | 98 ± 2.2 | 84 ± 4.2 | 101 ± 6.3 |
| CD147 | 91 ± 5.5 | 87 ± 9.4 | 101 ± 5.2 | |
| MCPV-1.2 | CD13 | 113 ± 9.4 | 91 ± 6.2 | 115 ± 26.4 |
| CD147 | 99 ± 4.5 | 106 ± 21.5 | 118 ± 14.0 | |
| MCPV-1.3 | CD13 | 100 ± 6.4 | 86 ± 7.3 | 113 ± 7.3 |
| CD147 | 83 ± 13.3 | 90 ± 8.2 | 96 ± 22.5 | |
| MCPV-1.4 | CD13 | 107 ± 3.7 | 89 ± 3.0 | 108 ± 12.0 |
| CD147 | 95 ± 8.4 | 90 ± 5.1 | 111 ± 11.3 | |
| KU812 | CD13 | 98 ± 5.8 | 98 ± 1.0 | 98 ± 4.2 |
| CD147 | 103 ± 3.6 | 105 ± 9.2 | 106 ± 10.9 | |
| EOL-1 | CD13 | 110 ± 8.5 | 92 ± 5.3 | 109 ± 12.1 |
| CD26 | 67 ± 18.0 * | 90 ± 5.0 | 81 ± 13.0 | |
| CD147 | 103 ± 5.2 | 106 ± 4.0 | 107 ± 0.9 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Degenfeld-Schonburg, L.; Sadovnik, I.; Smiljkovic, D.; Peter, B.; Stefanzl, G.; Gstoettner, C.; Jaksch, P.; Hoetzenecker, K.; Aigner, C.; Radtke, C.; et al. Coronavirus Receptor Expression Profiles in Human Mast Cells, Basophils, and Eosinophils. Cells 2024, 13, 173. https://doi.org/10.3390/cells13020173
Degenfeld-Schonburg L, Sadovnik I, Smiljkovic D, Peter B, Stefanzl G, Gstoettner C, Jaksch P, Hoetzenecker K, Aigner C, Radtke C, et al. Coronavirus Receptor Expression Profiles in Human Mast Cells, Basophils, and Eosinophils. Cells. 2024; 13(2):173. https://doi.org/10.3390/cells13020173
Chicago/Turabian StyleDegenfeld-Schonburg, Lina, Irina Sadovnik, Dubravka Smiljkovic, Barbara Peter, Gabriele Stefanzl, Clemens Gstoettner, Peter Jaksch, Konrad Hoetzenecker, Clemens Aigner, Christine Radtke, and et al. 2024. "Coronavirus Receptor Expression Profiles in Human Mast Cells, Basophils, and Eosinophils" Cells 13, no. 2: 173. https://doi.org/10.3390/cells13020173
APA StyleDegenfeld-Schonburg, L., Sadovnik, I., Smiljkovic, D., Peter, B., Stefanzl, G., Gstoettner, C., Jaksch, P., Hoetzenecker, K., Aigner, C., Radtke, C., Arock, M., Sperr, W. R., & Valent, P. (2024). Coronavirus Receptor Expression Profiles in Human Mast Cells, Basophils, and Eosinophils. Cells, 13(2), 173. https://doi.org/10.3390/cells13020173







