Circular RNAs in Viral Infection and Antiviral Treatment
Abstract
1. Introduction
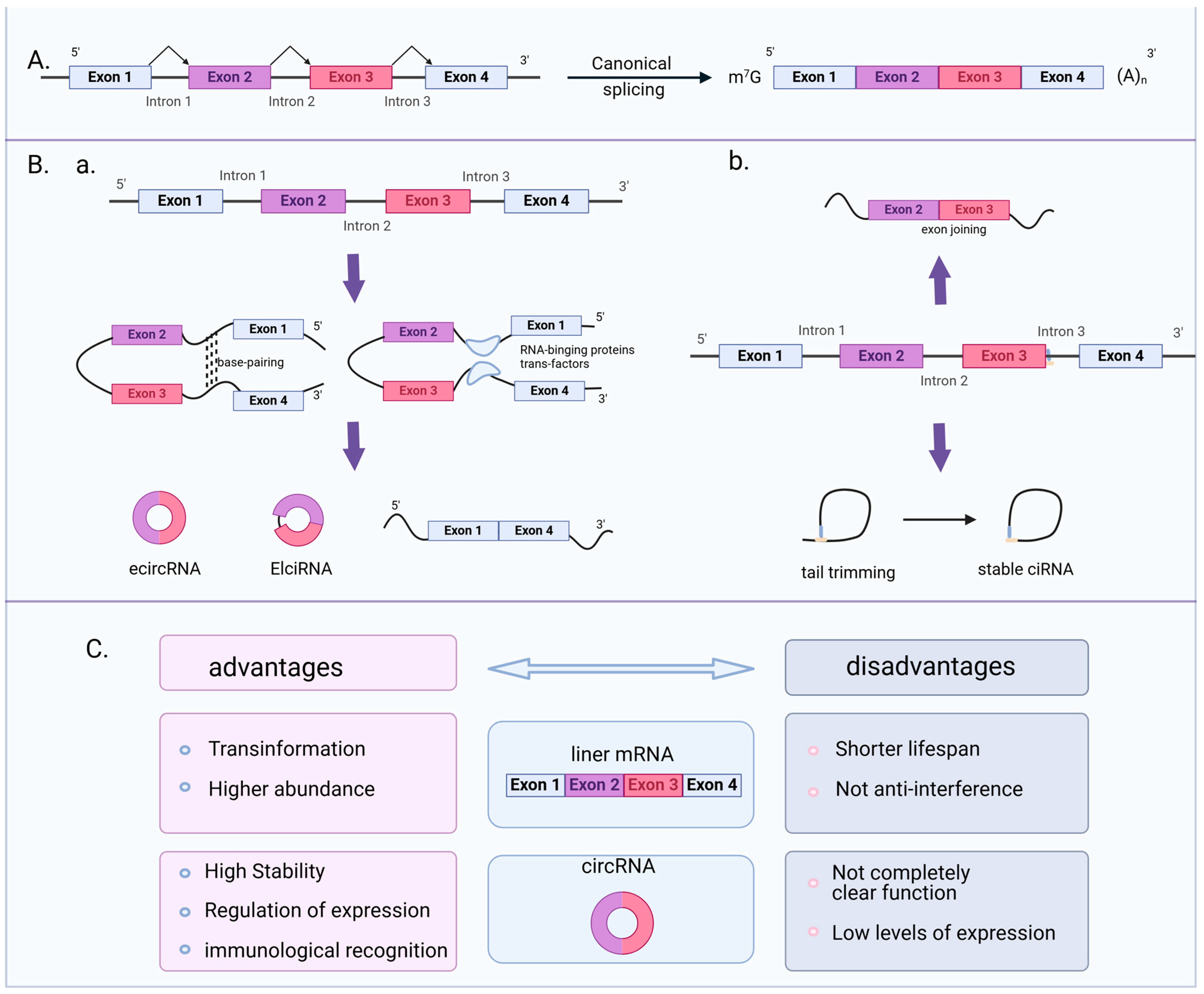
2. Circular RNA Biogenesis and Classification
3. Biological Functions of circRNAs During Viral Infections
3.1. Role as a Molecular Sponge for miRNA
3.2. Regulation of the Viral Infection Process via Protein Interactions
3.3. Proteins Translation
4. CircRNAs and the Host Immune System
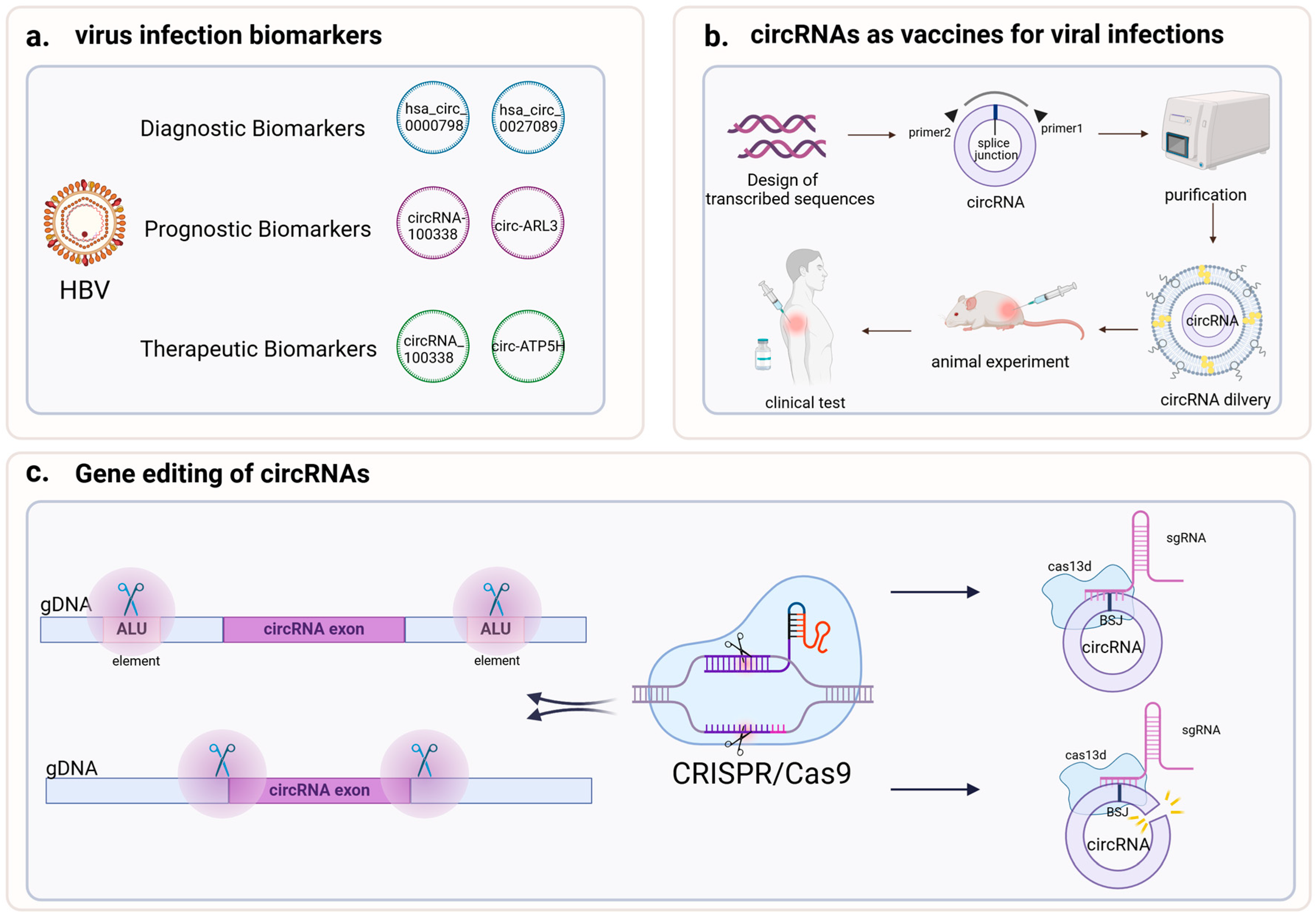
5. Application of circRNAs to Treat Viral Infections
5.1. CircRNAs as a Biomarker of Viral Infections
5.2. CircRNAs and Antiviral Vaccines
5.3. CircRNAs and Gene Editing
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sanger, H.L.; Klotz, G.; Riesner, D.; Gross, H.J.; Kleinschmidt, A.K. Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc. Natl. Acad. Sci. USA 1976, 73, 3852–3856. [Google Scholar] [CrossRef] [PubMed]
- Cocquerelle, C.; Mascrez, B.; Hétuin, D.; Bailleul, B. Mis-splicing yields circular RNA molecules. FASEB J. 1993, 7, 155–160. [Google Scholar] [CrossRef] [PubMed]
- Chen, R.; Wang, S.K.; Belk, J.A.; Amaya, L.; Li, Z.; Cardenas, A.; Abe, B.T.; Chen, C.K.; Wender, P.A.; Chang, H.Y. Engineering circular RNA for enhanced protein production. Nat. Biotechnol. 2023, 41, 262–272. [Google Scholar] [CrossRef]
- Jeck, W.R.; Sorrentino, J.A.; Wang, K.; Slevin, M.K.; Burd, C.E.; Liu, J.; Marzluff, W.F.; Sharpless, N.E. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 2013, 19, 141–157. [Google Scholar] [CrossRef]
- Chen, L.L. The expanding regulatory mechanisms and cellular functions of circular RNAs. Nat. Rev. Mol. Cell Biol. 2020, 21, 475–490. [Google Scholar] [CrossRef]
- Jarlstad Olesen, M.T.; Kristensen, L.S. Circular RNAs as microRNA sponges: Evidence and controversies. Essays Biochem. 2021, 65, 685–696. [Google Scholar] [CrossRef]
- Zhou, W.Y.; Cai, Z.R.; Liu, J.; Wang, D.S.; Ju, H.Q.; Xu, R.H. Circular RNA: Metabolism, functions and interactions with proteins. Mol. Cancer 2020, 19, 172. [Google Scholar] [CrossRef] [PubMed]
- Liang, Z.X.; Liu, H.S.; Xiong, L.; Yang, X.; Wang, F.W.; Zeng, Z.W.; He, X.W.; Wu, X.R.; Lan, P. A novel NF-κB regulator encoded by circPLCE1 inhibits colorectal carcinoma progression by promoting RPS3 ubiquitin-dependent degradation. Mol. Cancer 2021, 20, 103. [Google Scholar] [CrossRef]
- Shao, T.; Pan, Y.H.; Xiong, X.D. Circular RNA: An important player with multiple facets to regulate its parental gene expression. Mol. Ther. Nucleic Acids 2021, 23, 369–376. [Google Scholar] [CrossRef]
- Liu, C.X.; Chen, L.L. Circular RNAs: Characterization, cellular roles, and applications. Cell 2022, 185, 2016–2034. [Google Scholar] [CrossRef]
- Zhang, P.F.; Gao, C.; Huang, X.Y.; Lu, J.C.; Guo, X.J.; Shi, G.M.; Cai, J.B.; Ke, A.W. Cancer cell-derived exosomal circUHRF1 induces natural killer cell exhaustion and may cause resistance to anti-PD1 therapy in hepatocellular carcinoma. Mol. Cancer 2020, 19, 110. [Google Scholar] [CrossRef] [PubMed]
- Pan, X.; Huang, B.; Ma, Q.; Ren, J.; Liu, Y.; Wang, C.; Zhang, D.; Fu, J.; Ran, L.; Yu, T.; et al. Circular RNA circ-TNPO3 inhibits clear cell renal cell carcinoma metastasis by binding to IGF2BP2 and destabilizing SERPINH1 mRNA. Clin. Transl. Med. 2022, 12, e994. [Google Scholar] [CrossRef] [PubMed]
- Yu, T.; Ran, L.; Zhao, H.; Yin, P.; Li, W.; Lin, J.; Mao, H.; Cai, D.; Ma, Q.; Pan, X.; et al. Circular RNA circ-TNPO3 suppresses metastasis of GC by acting as a protein decoy for IGF2BP3 to regulate the expression of MYC and SNAIL. Mol. Ther. Nucleic Acids 2021, 26, 649–664. [Google Scholar] [CrossRef]
- Xie, M.; Yu, T.; Jing, X.; Ma, L.; Fan, Y.; Yang, F.; Ma, P.; Jiang, H.; Wu, X.; Shu, Y.; et al. Exosomal circSHKBP1 promotes gastric cancer progression via regulating the miR-582-3p/HUR/VEGF axis and suppressing HSP90 degradation. Mol. Cancer 2020, 19, 112. [Google Scholar] [CrossRef]
- Liang, G.; Ling, Y.; Mehrpour, M.; Saw, P.E.; Liu, Z.; Tan, W.; Tian, Z.; Zhong, W.; Lin, W.; Luo, Q.; et al. Autophagy-associated circRNA circCDYL augments autophagy and promotes breast cancer progression. Mol. Cancer 2020, 19, 65. [Google Scholar] [CrossRef]
- Wang, J.; Zhao, X.; Wang, Y.; Ren, F.; Sun, D.; Yan, Y.; Kong, X.; Bu, J.; Liu, M.; Xu, S. circRNA-002178 act as a ceRNA to promote PDL1/PD1 expression in lung adenocarcinoma. Cell Death Dis. 2020, 11, 32. [Google Scholar] [CrossRef]
- Wang, F.; Xu, X.; Zhang, N.; Chen, Z. Identification and integrated analysis of hepatocellular carcinoma-related circular RNA signature. Ann. Transl. Med. 2020, 8, 294. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Zhan, L.; Huang, K.; Wang, X. The functions and clinical significance of circRNAs in hematological malignancies. J. Hematol. Oncol. 2020, 13, 138. [Google Scholar] [CrossRef]
- Dong, X.; Bai, Y.; Liao, Z.; Gritsch, D.; Liu, X.; Wang, T.; Borges-Monroy, R.; Ehrlich, A.; Serrano, G.E.; Feany, M.B.; et al. Circular RNAs in the human brain are tailored to neuron identity and neuropsychiatric disease. Nat. Commun. 2023, 14, 5327. [Google Scholar] [CrossRef]
- Saaoud, F.; Drummer, I.V.C.; Shao, Y.; Sun, Y.; Lu, Y.; Xu, K.; Ni, D.; Jiang, X.; Wang, H.; Yang, X. Circular RNAs are a novel type of non-coding RNAs in ROS regulation, cardiovascular metabolic inflammations and cancers. Pharmacol. Ther. 2021, 220, 107715. [Google Scholar] [CrossRef]
- Hoque, P.; Romero, B.; Akins, R.E.; Batish, M. Exploring the Multifaceted Biologically Relevant Roles of circRNAs: From Regulation, Translation to Biomarkers. Cells 2023, 12, 2813. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Xue, Q.; Cheng, C.; Wang, Y.; Wang, X.; Chang, J.; Miao, C. Circular RNA in autoimmune diseases: Special emphasis on regulation mechanism in RA and SLE. J. Pharm. Pharmacol. 2023, 75, 370–384. [Google Scholar] [CrossRef]
- Koonin, E.V.; Dolja, V.V.; Krupovic, M. The logic of virus evolution. Cell Host Microbe 2022, 30, 917–929. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Liang, Z.; Wang, C.; Shen, Z.; Sun, S.; Gong, C.; Hu, X. Viral Circular RNAs and Their Possible Roles in Virus-Host Interaction. Front. Immunol. 2022, 13, 939768. [Google Scholar] [CrossRef]
- Qiu, H.; Yang, B.; Chen, Y.; Zhu, Q.; Wen, F.; Peng, M.; Wang, G.; Guo, G.; Chen, B.; Maarouf, M.; et al. Influenza A Virus-Induced circRNA circMerTK Negatively Regulates Innate Antiviral Responses. Microbiol. Spectr. 2023, 11, e0363722. [Google Scholar] [CrossRef] [PubMed]
- Yu, T.; Ding, Y.; Zhang, Y.; Liu, Y.; Li, Y.; Lei, J.; Zhou, J.; Song, S.; Hu, B. Circular RNA GATAD2A promotes H1N1 replication through inhibiting autophagy. Vet. Microbiol. 2019, 231, 238–245. [Google Scholar] [CrossRef]
- Chen, M.; Kang, L.; Zhang, T.; Zheng, J.; Chen, D.; Shao, D.; Li, Z.; Li, B.; Wei, J.; Qiu, Y.; et al. Circular RNA network plays a potential antiviral role in the early stage of JEV infection in mouse brain. Front. Microbiol. 2023, 14, 1165378. [Google Scholar] [CrossRef]
- Kazemi, S.; Mirzaei, R.; Karampoor, S.; Hosseini-Fard, S.R.; Ahmadyousefi, Y.; Soltanian, A.R.; Keramat, F.; Saidijam, M.; Alikhani, M.Y. Circular RNAs in tuberculosis: From mechanism of action to potential diagnostic biomarker. Microb. Pathog. 2023, 185, 106459. [Google Scholar] [CrossRef]
- Li, S.; Li, X.; Xue, W.; Zhang, L.; Yang, L.Z.; Cao, S.M.; Lei, Y.N.; Liu, C.X.; Guo, S.K.; Shan, L.; et al. Screening for functional circular RNAs using the CRISPR-Cas13 system. Nat. Methods 2021, 18, 51–59. [Google Scholar] [CrossRef]
- Sberna, G.; Maggi, F.; Amendola, A. Virus-Encoded Circular RNAs: Role and Significance in Viral Infections. Int. J. Mol. Sci. 2023, 24, 15. [Google Scholar] [CrossRef]
- Wang, W.; Sun, L.; Huang, M.T.; Quan, Y.; Jiang, T.; Miao, Z.; Zhang, Q. Regulatory circular RNAs in viral diseases: Applications in diagnosis and therapy. RNA Biol. 2023, 20, 847–858. [Google Scholar] [CrossRef] [PubMed]
- He, A.T.; Liu, J.; Li, F.; Yang, B.B. Targeting circular RNAs as a therapeutic approach: Current strategies and challenges. Signal Transduct. Target. Ther. 2021, 6, 185. [Google Scholar] [CrossRef]
- Gu, A.; Jaijyan, D.K.; Yang, S.; Zeng, M.; Pei, S.; Zhu, H. Functions of Circular RNA in Human Diseases and Illnesses. Noncoding RNA 2023, 9, 38. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Zheng, J.; Zhang, B.; Tang, X.; Cun, Y.; Wu, T.; Xu, Y.; Ma, T.; Cheng, J.; Yu, Z.; et al. Identification and characterization of virus-encoded circular RNAs in host cells. Microb. Genom. 2022, 8, mgen000848. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Zhou, H.; Liu, M.; Jaijyan, D.; Cruz-Cosme, R.; Ramasamy, S.; Subbian, S.; Liu, D.; Xu, J.; Niu, X.; et al. SARS-CoV-2, SARS-CoV, and MERS-CoV encode circular RNAs of spliceosome-independent origin. J. Med. Virol. 2022, 94, 3203–3222. [Google Scholar] [CrossRef]
- Huang, Y.; Abdelgawad, A.; Turchinovich, A.; Queen, S.; Abreu, C.M.; Zhu, X.; Batish, M.; Zheng, L.; Witwer, K.W. RNA landscapes of brain and brain-derived extracellular vesicles in simian immunodeficiency virus (SIV) infection and central nervous system pathology. J. Infect. Dis. 2023, 229, 1295–1305. [Google Scholar] [CrossRef]
- Liao, R.; Liu, L.; Zhou, J.; Wei, X.; Huang, P. Current Molecular Biology and Therapeutic Strategy Status and Prospects for circRNAs in HBV-Associated Hepatocellular Carcinoma. Front. Oncol. 2021, 11, 697747. [Google Scholar] [CrossRef]
- Hong, X.; Liu, N.; Liang, Y.; He, Q.; Yang, X.; Lei, Y.; Zhang, P.; Zhao, Y.; He, S.; Wang, Y.; et al. Circular RNA CRIM1 functions as a ceRNA to promote nasopharyngeal carcinoma metastasis and docetaxel chemoresistance through upregulating FOXQ1. Mol. Cancer 2020, 19, 33. [Google Scholar] [CrossRef]
- Li, Y.; Ashraf, U.; Chen, Z.; Zhou, D.; Imran, M.; Ye, J.; Chen, H.; Cao, S. Genome-wide profiling of host-encoded circular RNAs highlights their potential role during the Japanese encephalitis virus-induced neuroinflammatory response. BMC Genom. 2020, 21, 409. [Google Scholar] [CrossRef]
- Hu, Y.; Yu, Y.; Yang, R.; Wang, R.; Pu, D.; Wang, Y.; Fan, J.; Zhang, Y.; Song, J. The neuropathological mechanism of EV-A71 infection attributes to inflammatory pryoptosis and viral replication via activating the hsa_circ_0045431/hsa_miR_584/NLRP3 regulatory axis. Virus Res. 2023, 335, 199195. [Google Scholar] [CrossRef]
- Kang, L.; Xie, H.; Ye, H.; Jeyarajan, A.J.; Warner, C.A.; Huang, Y.; Shi, Y.; Li, Y.; Yang, C.; Xu, M.; et al. Hsa_circ_0007321 regulates Zika virus replication through miR-492/NFKBID/NF-κB signaling pathway. J. Virol. 2023, 97, e0123223. [Google Scholar] [CrossRef] [PubMed]
- Huang, A.; Zheng, H.; Wu, Z.; Chen, M.; Huang, Y. Circular RNA-protein interactions: Functions, mechanisms, and identification. Theranostics 2020, 10, 3503–3517. [Google Scholar] [CrossRef] [PubMed]
- Min, J.; Li, Y.; Li, X.; Wang, M.; Li, H.; Bi, Y.; Xu, P.; Liu, W.; Ye, X.; Li, J. The circRNA circVAMP3 restricts influenza A virus replication by interfering with NP and NS1 proteins. PLoS Pathog. 2023, 19, e1011577. [Google Scholar] [CrossRef]
- Tagawa, T.; Oh, D.; Dremel, S.; Mahesh, G.; Koparde, V.N.; Duncan, G.; Andresson, T.; Ziegelbauer, J.M. A virus-induced circular RNA maintains latent infection of Kaposi’s sarcoma herpesvirus. Proc. Natl. Acad. Sci. USA 2023, 120, e2212864120. [Google Scholar] [CrossRef]
- Mo, Y.; Wang, Y.; Zhang, S.; Xiong, F.; Yan, Q.; Jiang, X.; Deng, X.; Wang, Y.; Fan, C.; Tang, L.; et al. Circular RNA circRNF13 inhibits proliferation and metastasis of nasopharyngeal carcinoma via SUMO2. Mol. Cancer 2021, 20, 112. [Google Scholar] [CrossRef] [PubMed]
- Fan, X.; Yang, Y.; Chen, C.; Wang, Z. Pervasive translation of circular RNAs driven by short IRES-like elements. Nat. Commun. 2022, 13, 3751. [Google Scholar] [CrossRef]
- Yuan, W.; Zhang, X.; Cong, H. Advances in the protein-encoding functions of circular RNAs associated with cancer (Review). Oncol. Rep. 2023, 50, 160. [Google Scholar] [CrossRef]
- Wang, Y.; Wu, C.; Du, Y.; Li, Z.; Li, M.; Hou, P.; Shen, Z.; Chu, S.; Zheng, J.; Bai, J. Expanding uncapped translation and emerging function of circular RNA in carcinomas and noncarcinomas. Mol. Cancer 2022, 21, 13. [Google Scholar] [CrossRef]
- Wang, R.C.; Lee, E.E.; Zhao, J.; Kim, J. Assessment of the Abundance and Potential Function of Human Papillomavirus Type 16 Circular E7 RNA. mBio 2022, 13, e0041122. [Google Scholar] [CrossRef]
- Zhao, J.; Lee, E.E.; Kim, J.; Yang, R.; Chamseddin, B.; Ni, C.; Gusho, E.; Xie, Y.; Chiang, C.M.; Buszczak, M.; et al. Author Correction: Transforming activity of an oncoprotein-encoding circular RNA from human papillomavirus. Nat. Commun. 2022, 13, 2889. [Google Scholar] [CrossRef]
- Chen, Y.G.; Kim, M.V.; Chen, X.; Batista, P.J.; Aoyama, S.; Wilusz, J.E.; Iwasaki, A.; Chang, H.Y. Sensing Self and Foreign Circular RNAs by Intron Identity. Mol. Cell 2017, 67, 228–238.e5. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.G.; Chen, R.; Ahmad, S.; Verma, R.; Kasturi, S.P.; Amaya, L.; Broughton, J.P.; Kim, J.; Cadena, C.; Pulendran, B.; et al. N6-Methyladenosine Modification Controls Circular RNA Immunity. Mol. Cell 2019, 76, 96–109.e9. [Google Scholar] [CrossRef] [PubMed]
- Yan, L.; Chen, Y.G. Circular RNAs in Immune Response and Viral Infection. Trends Biochem. Sci. 2020, 45, 1022–1034. [Google Scholar] [CrossRef]
- Zheng, Z.M. Circular RNAs and RNase L in PKR activation and virus infection. Cell Biosci. 2019, 9, 43. [Google Scholar] [CrossRef]
- Liu, C.X.; Li, X.; Nan, F.; Jiang, S.; Gao, X.; Guo, S.K.; Xue, W.; Cui, Y.; Dong, K.; Ding, H.; et al. Structure and Degradation of Circular RNAs Regulate PKR Activation in Innate Immunity. Cell 2019, 177, 865–880.e21. [Google Scholar] [CrossRef]
- Wilusz, J.E. Circle the Wagons: Circular RNAs Control Innate Immunity. Cell 2019, 177, 797–799. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.X.; Guo, S.K.; Nan, F.; Xu, Y.F.; Yang, L.; Chen, L.L. RNA circles with minimized immunogenicity as potent PKR inhibitors. Mol. Cell 2022, 82, 420–434.e6. [Google Scholar] [CrossRef]
- Li, X.; Liu, C.X.; Xue, W.; Zhang, Y.; Jiang, S.; Yin, Q.F.; Wei, J.; Yao, R.W.; Yang, L.; Chen, L.L. Coordinated circRNA Biogenesis and Function with NF90/NF110 in Viral Infection. Mol. Cell 2017, 67, 214–227.e7. [Google Scholar] [CrossRef]
- Maarouf, M.; Wang, L.; Wang, Y.; Rai, K.R.; Chen, Y.; Fang, M.; Chen, J.L. Functional Involvement of circRNAs in the Innate Immune Responses to Viral Infection. Viruses 2023, 15, 1697. [Google Scholar] [CrossRef]
- Santer, L.; Bär, C.; Thum, T. Circular RNAs: A Novel Class of Functional RNA Molecules with a Therapeutic Perspective. Mol. Ther. 2019, 27, 1350–1363. [Google Scholar] [CrossRef]
- Barbagallo, D.; Palermo, C.I.; Barbagallo, C.; Battaglia, R.; Caponnetto, A.; Spina, V.; Ragusa, M.; Di Pietro, C.; Scalia, G.; Purrello, M. Competing endogenous RNA network mediated by circ_3205 in SARS-CoV-2 infected cells. Cell. Mol. Life Sci. 2022, 79, 75. [Google Scholar] [CrossRef] [PubMed]
- Oudit, G.Y.; Wang, K.; Viveiros, A.; Kellner, M.J.; Penninger, J.M. Angiotensin-converting enzyme 2-at the heart of the COVID-19 pandemic. Cell 2023, 186, 906–922. [Google Scholar] [CrossRef] [PubMed]
- Bohan, D.; Van Ert, H.; Ruggio, N.; Rogers, K.J.; Badreddine, M.; Aguilar Briseño, J.A.; Elliff, J.M.; Rojas Chavez, R.A.; Gao, B.; Stokowy, T.; et al. Phosphatidylserine receptors enhance SARS-CoV-2 infection. PLoS Pathog. 2021, 17, e1009743. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Jiang, J.; Qian, H.; Yan, Y.; Xu, W. Exosomal circRNA: Emerging insights into cancer progression and clinical application potential. J. Hematol. Oncol. 2023, 16, 67. [Google Scholar] [CrossRef]
- Saadh, M.J.; Abedi Kiasari, B.; Shahrtash, S.A.; Arias-Gonzáles, J.L.; Chaitanya, M.; Cotrina-Aliaga, J.C.; Kadham, M.J.; Sârbu, I.; Akhavan-Sigari, R. Exosomal non-coding RNAs’ role in immune regulation and potential therapeutic applications. Pathol. Res. Pract. 2023, 247, 154522. [Google Scholar] [CrossRef]
- Lou, Y.Y.; Wang, Q.D.; Lu, Y.T.; Tu, M.Y.; Xu, X.; Xia, Y.; Peng, Y.; Lai, M.M.; Zheng, X.Q. Differential circRNA expression profiles in latent human cytomegalovirus infection and validation using clinical samples. Physiol. Genom. 2019, 51, 51–58. [Google Scholar] [CrossRef]
- Wang, Y.; Li, M.; Pan, C.; Huang, H.; Hu, X.; Liu, J. Hsa_circ_0007637 Facilitates Nasopharyngeal Carcinoma Progression by Sponging miR-636/TPD52 Axis. Cancer Manag. Res. 2021, 13, 9439–9452. [Google Scholar] [CrossRef]
- Qu, Z.; Meng, F.; Shi, J.; Deng, G.; Zeng, X.; Ge, J.; Li, Y.; Liu, L.; Chen, P.; Jiang, Y.; et al. A Novel Intronic Circular RNA Antagonizes Influenza Virus by Absorbing a microRNA That Degrades CREBBP and Accelerating IFN-β Production. mBio 2021, 12, e0101721. [Google Scholar] [CrossRef]
- Qin, Y.; Lin, L.; Yang, S.; Dai, Z.; Zhang, C.; Huang, J.; Deng, F.; Yue, X.; Ren, L.; Fei, Y.; et al. Circular RNA circ_0076631 promotes coxsackievirus B3 infection through modulating viral translation by sponging miR-214-3p. Front. Microbiol. 2022, 13, 975223. [Google Scholar] [CrossRef]
- Li, J.; Teng, P.; Yang, F.; Ou, X.; Zhang, J.; Chen, W. Bioinformatics and Screening of a Circular RNA-microRNA-mRNA Regulatory Network Induced by Coxsackievirus Group B5 in Human Rhabdomyosarcoma Cells. Int. J. Mol. Sci. 2022, 23, 4628. [Google Scholar] [CrossRef]
- Bhardwaj, V.; Singh, A.; Choudhary, A.; Dalavi, R.; Ralte, L.; Chawngthu, R.L.; Senthil Kumar, N.; Vijay, N.; Chande, A. HIV-1 Vpr induces ciTRAN to prevent transcriptional repression of the provirus. Sci. Adv. 2023, 9, eadh9170. [Google Scholar] [CrossRef] [PubMed]
- Aborehab, N.M.; Kandeil, M.A.; Sabry, D.; Rabie, R.; Ibrahim, I.T. Circular SERPINA3 and its target microRNA-944 as potential biomarkers in hepatitis C virus-induced hepatocellular carcinoma in Egyptian population. Noncoding RNA Res. 2023, 8, 401–412. [Google Scholar] [CrossRef] [PubMed]
- Yang, D.; Sun, K.; Huang, F.; Fan, H.; Shi, T.; Chen, X.; Lu, G. Whole blood circular RNA hsa_circ_0002171 serves as a potential diagnostic biomarker for human adenovirus pneumonia in children. Braz. J. Med. Biol. Res. 2022, 55, e12347. [Google Scholar] [CrossRef]
- Wang, Y.; Pei, L.; Yue, Z.; Jia, M.; Wang, H.; Cao, L.L. The Potential of Serum Exosomal hsa_circ_0028861 as the Novel Diagnostic Biomarker of HBV-Derived Hepatocellular Cancer. Front. Genet. 2021, 12, 703205. [Google Scholar] [CrossRef]
- Zhu, K.; Zhan, H.; Peng, Y.; Yang, L.; Gao, Q.; Jia, H.; Dai, Z.; Tang, Z.; Fan, J.; Zhou, J. Plasma hsa_circ_0027089 is a diagnostic biomarker for hepatitis B virus-related hepatocellular carcinoma. Carcinogenesis 2020, 41, 296–302. [Google Scholar] [CrossRef]
- Li, Y.; Li, R.; Cheng, D.; Fu, X.; Fu, L.; Peng, S. The potential of CircRNA1002 as a biomarker in hepatitis B virus-related hepatocellular carcinoma. PeerJ 2022, 10, e13640. [Google Scholar] [CrossRef]
- Kang, M.K.; Kim, G.; Park, J.G.; Jang, S.Y.; Lee, H.W.; Tak, W.Y.; Kweon, Y.O.; Park, S.Y.; Lee, Y.R.; Hur, K. Tissue Circular RNA_0004018 and 0003570 as Novel Prognostic Biomarkers for Hepatitis B-Related Hepatocellular Carcinoma. Genes 2023, 14, 1963. [Google Scholar] [CrossRef] [PubMed]
- Rao, X.; Lai, L.; Li, X.; Wang, L.; Li, A.; Yang, Q. N(6) -methyladenosine modification of circular RNA circ-ARL3 facilitates Hepatitis B virus-associated hepatocellular carcinoma via sponging miR-1305. IUBMB Life 2021, 73, 408–417. [Google Scholar] [CrossRef]
- Wang, M.; Gu, B.; Yao, G.; Li, P.; Wang, K. Circular RNA Expression Profiles and the Pro-tumorigenic Function of CircRNA_10156 in Hepatitis B Virus-Related Liver Cancer. Int. J. Med. Sci. 2020, 17, 1351–1365. [Google Scholar] [CrossRef]
- Ye, X.; Zhu, B.; Han, J.; Huang, J.; Wu, Y. Circ-0036602 Acts As a Sponge of MiR-34a-5p and MiR-431-5p to Promote Cervical Cancer Proliferation and Invasion. J. Genom. 2022, 10, 16–25. [Google Scholar] [CrossRef]
- Cheng, X.; Shen, C.; Liao, Z. High Expression of Circular RNA-Mitochondrial tRNA Translation Optimization 1 Assists the Diagnosis of High-Risk Human Papillomavirus Infection in Cervical Cancer. J. Low. Genit. Tract. Dis. 2022, 26, 212–218. [Google Scholar] [CrossRef] [PubMed]
- Ge, J.; Wang, J.; Xiong, F.; Jiang, X.; Zhu, K.; Wang, Y.; Mo, Y.; Gong, Z.; Zhang, S.; He, Y.; et al. Epstein-Barr Virus-Encoded Circular RNA CircBART2.2 Promotes Immune Escape of Nasopharyngeal Carcinoma by Regulating PD-L1. Cancer Res. 2021, 81, 5074–5088. [Google Scholar] [CrossRef]
- Gong, L.P.; Chen, J.N.; Dong, M.; Xiao, Z.D.; Feng, Z.Y.; Pan, Y.H.; Zhang, Y.; Du, Y.; Zhang, J.Y.; Bi, Y.H.; et al. Epstein-Barr virus-derived circular RNA LMP2A induces stemness in EBV-associated gastric cancer. EMBO Rep. 2020, 21, e49689. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Liu, Y.; Duan, H.; Fan, X.; Wang, Y.; Song, J.; Han, J.; Yang, M.; Lu, L.; Nie, G. Identification of differentially expressed circular RNAs in human nasopharyngeal carcinoma. Cancer Biomark. 2020, 29, 483–492. [Google Scholar] [CrossRef] [PubMed]
- Abere, B.; Zhou, H.; Li, J.; Cao, S.; Toptan, T.; Grundhoff, A.; Fischer, N.; Moore, P.S.; Chang, Y. Merkel Cell Polyomavirus Encodes Circular RNAs (circRNAs) Enabling a Dynamic circRNA/microRNA/mRNA Regulatory Network. mBio 2020, 11, e03059-20. [Google Scholar] [CrossRef]
- Sun, W.; Zhou, S.; Peng, L.; Liu, Y.; Cheng, D.; Wang, Y.; Ni, C. CircZNF609 regulates pulmonary fibrosis via miR-145-5p/KLF4 axis and its translation function. Cell. Mol. Biol. Lett. 2023, 28, 105. [Google Scholar] [CrossRef]
- Song, Z.; Lin, J.; Su, R.; Ji, Y.; Jia, R.; Li, S.; Shan, G.; Huang, C. eIF3j inhibits translation of a subset of circular RNAs in eukaryotic cells. Nucleic Acids Res. 2022, 50, 11529–11549. [Google Scholar] [CrossRef]
- Abe, N.; Kodama, A.; Abe, H. Preparation of Circular RNA In Vitro. Methods Mol. Biol. 2018, 1724, 181–192. [Google Scholar] [CrossRef]
- Liu, X.; Zhang, Y.; Zhou, S.; Dain, L.; Mei, L.; Zhu, G. Circular RNA: An emerging frontier in RNA therapeutic targets, RNA therapeutics, and mRNA vaccines. J. Control. Release 2022, 348, 84–94. [Google Scholar] [CrossRef]
- Amaya, L.; Grigoryan, L.; Li, Z.; Lee, A.; Wender, P.A.; Pulendran, B.; Chang, H.Y. Circular RNA vaccine induces potent T cell responses. Proc. Natl. Acad. Sci. USA 2023, 120, e2302191120. [Google Scholar] [CrossRef]
- Qu, L.; Yi, Z.; Shen, Y.; Lin, L.; Chen, F.; Xu, Y.; Wu, Z.; Tang, H.; Zhang, X.; Tian, F.; et al. Circular RNA vaccines against SARS-CoV-2 and emerging variants. Cell 2022, 185, 1728–1744.e16. [Google Scholar] [CrossRef] [PubMed]
- Wan, J.; Wang, Z.; Wang, L.; Wu, L.; Zhang, C.; Zhou, M.; Fu, Z.F.; Zhao, L. Circular RNA vaccines with long-term lymph node-targeting delivery stability after lyophilization induce potent and persistent immune responses. mBio 2024, 15, e0177523. [Google Scholar] [CrossRef] [PubMed]
- Wesselhoeft, R.A.; Kowalski, P.S.; Parker-Hale, F.C.; Huang, Y.; Bisaria, N.; Anderson, D.G. RNA Circularization Diminishes Immunogenicity and Can Extend Translation Duration In Vivo. Mol. Cell 2019, 74, 508–520.e4. [Google Scholar] [CrossRef] [PubMed]
- Niu, D.; Wu, Y.; Lian, J. Circular RNA vaccine in disease prevention and treatment. Signal Transduct. Target. Ther. 2023, 8, 341. [Google Scholar] [CrossRef] [PubMed]
- Song, P.; Zhang, P.; Qin, K.; Su, F.; Gao, K.; Liu, X.; Li, Z. CRISPR/Cas13a induced exponential amplification for highly sensitive and specific detection of circular RNA. Talanta 2022, 246, 123521. [Google Scholar] [CrossRef]
- Wang, J.H.; Shi, C.W.; Lu, Y.Y.; Zeng, Y.; Cheng, M.Y.; Wang, R.Y.; Sun, Y.; Jiang, Y.L.; Yang, W.T.; Zhao, D.D.; et al. MicroRNA and circRNA Expression Analysis in a Zbtb1 Gene Knockout Monoclonal EL4 Cell Line. Front. Cell. Infect. Microbiol. 2021, 11, 706919. [Google Scholar] [CrossRef]
- Zhang, Y.; Nguyen, T.M.; Zhang, X.O.; Wang, L.; Phan, T.; Clohessy, J.G.; Pandolfi, P.P. Optimized RNA-targeting CRISPR/Cas13d technology outperforms shRNA in identifying functional circRNAs. Genome Biol. 2021, 22, 41. [Google Scholar] [CrossRef]
- Yin, X.; Luo, H.; Zhou, H.; Zhang, Z.; Lan, Y.; Feng, Z.; Chen, W.; Zheng, H. A rapid isothermal CRISPR-Cas13a diagnostic test for genital herpes simplex virus infection. iScience 2024, 27, 108581. [Google Scholar] [CrossRef]
- Li, X.; Su, B.; Yang, L.; Kou, Z.; Wu, H.; Zhang, T.; Liu, L.; Han, Y.; Niu, M.; Sun, Y.; et al. Highly sensitive and rapid point-of-care testing for HIV-1 infection based on CRISPR-Cas13a system. BMC Infect. Dis. 2023, 23, 627. [Google Scholar] [CrossRef]
- Tian, Y.; Fan, Z.; Zhang, X.; Xu, L.; Cao, Y.; Pan, Z.; Mo, Y.; Gao, Y.; Zheng, S.; Huang, J.; et al. CRISPR/Cas13a-Assisted accurate and portable hepatitis D virus RNA detection. Emerg. Microbes Infect. 2023, 12, 2276337. [Google Scholar] [CrossRef]
- Li, M.; He, Q.; Li, T.; Wan, W.; Zhou, H. Development and evaluation of a CRISPR-Cas13a system-based diagnostic for hepatitis E virus. Clin. Chem. Lab. Med. 2023, 62, 1237–1247. [Google Scholar] [CrossRef] [PubMed]

| CircRNA Name | Mechanism of Action | Impact on HBV-HCC | Reference |
|---|---|---|---|
| circRNA_101764 | Interacts with miR-181 | May promote the development of HBV-HCC | |
| circRNA_100338 | Interacts with miR-141-3p | Closely correlated with metastatic progression of HBV-HCC | |
| circ-ARL3 | Acts as a sponge for miR-1305 | Promotes proliferation and invasion of HBV-HCC cells | [37] |
| circ-ATP5H | Sponges miR-138-5p | Promotes HBV-HCC development | |
| HBV_circ_1 | Interacts with CDK1 | Promotes proliferation, migration, inhibits apoptosis of HBV-HCC cells |
| Virus | CircRNA | Test Method | Different Diseases | Mechanism | Effect | Reference |
|---|---|---|---|---|---|---|
| human cytomegalovirus | hsa_circ_0001445 hsa_circ_0001206 | unknown | Severe or even fatal diseases in newborns with immunodeficiency and adults with immunodeficiency | circRNA/miRNA/mRNA analysis | circRNA participates in the regulation of host cell secretion pathways, cell cycle, and apoptosis | [66] |
| Epstein–Barr virus | hsa_circ_0007637 | qRT-PCR | Nasopharyngeal carcinoma | hsa_circ_0007637/miR-636/TPD52 | High hsa_circ_0007637 expression predicted a poor outcome for NPC patients | [67] |
| influenza A virus | circVAMP3 | Deep RNA sequencing | Influenza | Reduces the interaction between NP and polymerase alkalinity 1, polymerase alkalinity 2, or vRNA to interfere with the activity of viral ribonucleoprotein complexes | Directly inhibition of virus replication | [43] |
| influenza A virus | circMerTK | RNA sequencing | Influenza | circMerTK affects IFN-β signal generation and downstream signal transduction | Enhances replication of IAV | [25] |
| influenza Virus | circRNA AIVR | Reverse transcription-quantitative PCR | Influenza | circRNA AIVR/miR-330-3p/recombinant CREB binding protein | Increases IFN-β production and advances the understanding of the roles of circRNAs in the cellular innate antiviral response. | [68] |
| coxsackievirus | circ_0076631 | Real-time quantitative PCR | Viral myocarditis and dilated cardiomyopathy | circ_0076631/miR-214-3p/cvb3 | Suppression of viral translation | [69] |
| coxsackievirus group B5 | novel_circ_0002006 novel_circ_0001066 | Real-time quantitative PCR | Hand, foot and mouth disease | Two novel circRNA might act as a molecular sponge for miRNA through the IFN-I pathway and NF-κB pathway | Inhibition of CVB5 replication | [70] |
| Japanese encephalitis virus | circStrbp | RNA sequencing | Japanese encephalitis | circStrbp/miR709/CX3CR1 | ceRNA pathway impacts JEV infection in vivo and in vitro | [27] |
| Japanese encephalitis virus | circ_0000220 | Illumina RNA-sequencing | Japanese encephalitis virus-induced neuroinflammatory response | circ_0000220-miR-326-3p-BCL3/MK2/TRIM25 | Knockdown of circ_0000220 or overexpression of miR-326-3p leads to a decrease in JEV-induced inflammatory cytokine production | [39] |
| enterovirus 71 | hsa_circ_0045431 | Quantitative real-time PCR | Hand foot and mouth disease | sa_circ_0045431/hsa_miR_584/NLRP3 | Promotes inflammatory necrosis and virus replication | [40] |
| zika virus | hsa_circ_0007321 | RNA sequencing | Congenital Zika syndrome | miR-492/NFKBID/NF-κB | Downregulates circRNA and inhibits NF-κB pathway. Promotes the replication of ZIKV. | [41] |
| human immunodeficiency virus | ciTRAN | RNA nanopore sequencing | Acquired immunodeficiency syndrome | HIV-1 hijacks ciTRAN to exclude serine/arginine-rich splicing factor 1 (SRSF1) from viral transcriptional complexes | Promotes effective viral transcription | [71] |
| hepatitis C virus | circular SERPINA3 | Real-time qPCR | Hepatocellular carcinoma | circSERPINA3/miR-944/MDM2 | Promotes the metastasis and oxidative stress of liver cancer cases | [72] |
| human adenovirus | hsa_circ_0002171 | Unknown | Adenovirus pneumonia | circRNA mRNA regulatory network | hsa_circ_0002171 has significant value in diagnosing highly pathogenic pneumonia and severe highly pathogenic pneumonia | [73] |
| hepatitis B virus | hsa_circ_0028861 | Microarray analysis | HBV-derived hepatocellular cancer | circRNA/miRNA/mRNA and downstream signaling pathway analysis | The combination of hsa_circ:0028861 and AFP shows better diagnostic ability | [74] |
| hepatitis B virus | circRNA_101764 circRNA_100338 circ-ARL3 circ-ATP5H | circRNA microarray and qRT-PCR | Hepatocellular carcinoma | circRNA_101764/miR-181/PI3K | Plays an important role in the cellular network during the development of HBV-HCC liver cancer | [37] |
| hepatitis B virus | hsa_circ_0027089 | Quantitative reverse transcription polymerase chain reaction | Hepatitis B virus-related hepatocellular carcinoma | circRNA_100338/miR141-3p/MTSS1 | Regulates the growth and metastasis of HCC cells | [75] |
| hepatitis B virus | circRNA1002 | Quantitative reverse transcription PCR | Hepatocellular carcinoma | circ-ARL3/miR-1305/WNT2 | circRNA1002 is involved in the progression of HCC, providing an improved early detection method for HCC | [76] |
| hepatitis B virus | hsa_circ_0003570 hsa_circ_0004018 | Real-time quantitative polymerase chain reaction | Hepatitis B virus-associated hepatocellular carcinoma | circ-ATP5H/miR138-5p/TNFAIP3 | Related to cell growth, tumor progression, invasiveness, and metastasis | [77] |
| hepatitis B virus | circ-ARL3 | circRNA microarray | Hepatocellular carcinoma | hsa_circ_0027089/miR-15b-3p/OIP5 | high circ-ARL3 was positively correlated with malignant clinical features and poor prognosis | [78] |
| hepatitis B virus | circRNA_10156 | High-throughput RNA sequencing | Hepatitis B virus-related liver cancer | Enrichment of circRNA1002-related genes under GO conditions related to hormone pathways and cell–cell interactions | circRNA-10156 may be a promising therapeutic target for liver cancer treatment | [79] |
| human papilloma virus | circ0036602 | qRT-PCR | Cervical cancer | circRNA/miRNA/mRNA | Promotes growth of CC cells | [80] |
| human papilloma virus | circE7 | Reverse transcription-quantitative polymerase chain reaction | Cervical cancer | circ-ARL3/miR-1305/cancer gene | circE7 reduces E7 protein levels and inhibits cancer cell growth in vitro and in tumor xenografts | [50] |
| high-risk human papillomavirus | Circular RNA-mitochondrial tRNA translation optimization 1 (circMTO1) | Reverse transcription-quantitative polymerase chain reaction | Cervical cancer | Upregulation of miR-149-3p by consumption of ircRNA-10156, reduction in Akt1 expression, and inhibition of liver cancer cell proliferation | circMTO1 associated with clinical stage, tumor differentiation, lymph node metastasis, invasion depth, and independently linked with HR-HPV infection in CC. | [81] |
| Epstein–Barr virus | circBART2.2 | Quantitative reverse transcription polymerase chain reaction | Nasopharyngeal carcinoma | Alters HMGB1 expression by sponging miR-34-5p and miR-431-5p | Crucial for regulating PD-L1 and subsequent immune escape in nasopharyngeal carcinoma | [82] |
| Epstein–Barr virus | ebv-circLMP2A | Reverse transcription-quantitative polymerase chain reaction and real-time PCR | EBV-associated gastric cancer | circE7 is localized in the cytoplasm through N6 methyladenosine (m6A) modification and translated to produce E7 oncoprotein | High expression of circRNA plays a crucial role in inducing and maintaining stemness phenotype, and is significantly associated with metastasis and poor prognosis | [83] |
| Epstein–Barr virus | circEAF2 | Quantitative real-time PCR | Lymphoma | Serum miR-199a was downregulated in HR-HPV-positive CC patients and inversely correlated with circMTO1. | circEAF2 is a potential prognostic biomarker | [83] |
| Epstein–Barr virus | hsa_circ_0007637 | High-throughput RNA sequencing (RNA-Seq) | Nasopharyngeal carcinoma | circBART2.2 activates transcription factors IRF3 and NF by binding to the helicase domain of RIG-I- κ B promoting the transcription of PD-L1 | hsa_circ_0007637 expression distinguished NPC tissues from paired healthy tissues and NPC cell lines (HNE1 6-10B, 5-8F, CNE-2, and so on) from a normal epithelial (NP460) cell line. | [84] |
| merkel cell polyomavirus | circMCV-T | RNA enzyme R resistance RNA sequencing | Merkel cell carcinoma | ebv-circLMP2A/miR-3908/TRIM59/p53 | Functional regulation of early transcriptional expression in regions important for virus replication and long-term persistence of the upper body | [85] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yin, X.; Li, H.; Zhou, Y. Circular RNAs in Viral Infection and Antiviral Treatment. Cells 2024, 13, 2033. https://doi.org/10.3390/cells13232033
Yin X, Li H, Zhou Y. Circular RNAs in Viral Infection and Antiviral Treatment. Cells. 2024; 13(23):2033. https://doi.org/10.3390/cells13232033
Chicago/Turabian StyleYin, Xiaocai, Hongjun Li, and Yan Zhou. 2024. "Circular RNAs in Viral Infection and Antiviral Treatment" Cells 13, no. 23: 2033. https://doi.org/10.3390/cells13232033
APA StyleYin, X., Li, H., & Zhou, Y. (2024). Circular RNAs in Viral Infection and Antiviral Treatment. Cells, 13(23), 2033. https://doi.org/10.3390/cells13232033






