Synthetic Biology Meets Ca2+ Release-Activated Ca2+ Channel-Dependent Immunomodulation
Abstract
:1. Introduction—The Role of Ca2+ Release-Activated Ca2+ (CRAC) Channels in Immunology
2. CRAC Channel Working Mechanisms
3. Synthetic Biology, Application to CRAC Channels, and Impact on CRAC Channel-Dependent Downstream Signaling
3.1. Chemical Inducers of Proximity (CIPs)
3.2. Proteolysis-Based Signaling
3.3. Photosensory Domains
3.3.1. Light-Induced Oligomerization

3.3.2. Light-Induced Uncaging
3.3.3. Genetically Encoded Light-Sensitive CRAC Channel Components in the Immune Response
3.4. Unnatural Amino Acids
3.5. Upconversion Nanoparticles
3.6. Photosensitive Drugs
3.7. Engineered Immune Cells
3.8. Therapeutic Antibodies, Nanobodies, Antibody Mimetics
3.9. Conclusion and Perspectives
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| AARS | Aminoacyl-tRNA synthetase |
| AtCRY2 | Cryptochrome 2 derived from Arabidopsis thaliana |
| BACCS | Blue light-activated Ca2+ channel switch |
| BCR | B cell receptor |
| BG | Benzylguanine |
| Ca2+ | Calcium |
| CAD | CRAC activation domain |
| CAR | Chimeric antigen receptor |
| CC | coiled-coil |
| CIB1 | Cryptochrome-interacting basic helix-loop-helix 1 region |
| CIP | Chemical inducer |
| CRAC | Ca2+ release-activated Ca2+ |
| CREB | cAMP response element-binding protein |
| CRISPR-Cas9 | Clustered regularly interspaced short palindromic repeats–associated-9 nuclease |
| CRS | Cytokine release syndrome |
| CRY2 | Cryptochrome 2 |
| CTL | Cytotoxic T cells |
| CyP | Prolyl isomerase |
| DC | Dendritic cell |
| DAG | Diacylglycerol |
| DREADD | Designer receptor exclusively activated by designer drugs |
| EAE | Experimental autoimmune encephalomyelitis |
| EF-SAM | EF-sterile alpha motif |
| ER | Endoplasmic reticulum |
| ER–PM | Endoplasmic reticulum–plasma membrane |
| FcεRII | Fc-receptor for IgE |
| FcγR | Fc-receptor for IgG |
| FMN | Flavin mononucleotide |
| FKBP | FK506 binding protein |
| FRB | Ser/Thr phosphatase |
| GCE | Genetic code expansion |
| GNb | Nanobody against GFP |
| GoF | Gain-of-function |
| GPCR | G protein-coupled receptor |
| GvHD | Graft-versus-host disease |
| IBD | Inflammatory bowel disease |
| IFNγ | Interferon-γ |
| IL | Interleukin |
| iLID | Light-induced dimer |
| INNP5E | Inositol polyphosphate-5-phosphatase E |
| IP3 | Inositol-1,4,5-trisphosphate |
| LiCAR | Light-inducible CAR |
| LOCa | Light-gated Ca2+ channel |
| LoF | Loss-of-function |
| LOV2 | Light-oxygen-voltage-sensing 2 |
| NFAT | Nuclear factor of activated T cells |
| NF-κB | Nuclear factor kappa-light-chain-enhancer of activated B cells |
| NIR | Near-infrared |
| NK | Natural killer |
| OASF | Orai-activating small fragment |
| PCB | Phytocyanobilin |
| PACE | Protease-activated Orai activator |
| PAS | Period-ARNT-single-minded |
| PI | Phosphatidylinositol |
| PIF | Phytochrome-interacting factor |
| PI(4,5)P2 | Phosphatidylinositol-4,5-bisphosphate |
| PI4P | Phosphatidylinositol 4-phosphate |
| PHR | N-terminal photolyase homology region |
| PJ | Pseudojanin |
| PLCγ | Phospholipase Cγ |
| POI | Protein of interest |
| PM | Plasma membrane |
| PPV | Plum pox virus protease |
| RNAi | RNA interference |
| Sac1 | Phosphatidylinositol-3-phosphatase |
| SCID | Severe combined immunodeficiency |
| STIM | Stromal interaction molecule |
| SOAR | STIM1-Orai activating region |
| SOCE | Store-operated Ca2+ entry |
| SbMVp | Sunflower mild mosaic virus protease |
| TCR | T cell receptor |
| TEV | Tobacco etch virus |
| Th | T helper |
| TM | Transmembrane |
| TME | Tumor microenvironment |
| UAA | Unnatural amino acid |
| UCPN | Upconversion nanoparticles |
| UV8 | UV resistance locus 8 |
References
- Berridge, M.J.; Bootman, M.D.; Lipp, P. Calcium—A Life and Death Signal. Nature 1998, 395, 645–648. [Google Scholar] [CrossRef]
- Berridge, M.J.; Lipp, P.; Bootman, M.D. The Versatility and Universality of Calcium Signalling. Nat. Rev. Mol. Cell Biol. 2000, 1, 11–21. [Google Scholar] [CrossRef]
- Berridge, M.J.; Bootman, M.D.; Roderick, H.L. Calcium Signalling: Dynamics, Homeostasis and Remodelling. Nat. Rev. Mol. Cell Biol. 2003, 4, 517. [Google Scholar] [CrossRef]
- Luan, S.; Wang, C. Calcium Signaling Mechanisms Across Kingdoms. Annu. Rev. Cell Dev. Biol. 2021, 37, 311–340. [Google Scholar] [CrossRef]
- Berridge, M.J. The Inositol Trisphosphate/Calcium Signaling Pathway in Health and Disease. Physiol. Rev. 2016, 96, 1261–1296. [Google Scholar] [CrossRef]
- Putney, J.W. Store-Operated Calcium Entry: An Historical Overview. Adv. Exp. Med. Biol. 2017, 981, 205–214. [Google Scholar] [CrossRef]
- Lewis, R.S. Store-Operated Calcium Channels: New Perspectives on Mechanism and Function. Cold Spring Harb. Perspect. Biol. 2011, 3. [Google Scholar] [CrossRef]
- Parekh, A.B. On the Activation Mechanism of Store-Operated Calcium Channels. Pflug. Arch. 2006, 453, 303–311. [Google Scholar] [CrossRef]
- Prakriya, M.; Lewis, R.S. Store-Operated Calcium Channels. Physiol. Rev. 2015, 95, 1383–1436. [Google Scholar] [CrossRef]
- Feske, S.; Wulff, H.; Skolnik, E.Y. Ion Channels in Innate and Adaptive Immunity. Annu. Rev. Immunol. 2015, 33, 291–353. [Google Scholar] [CrossRef]
- Srikanth, S.; Woo, J.S.; Sun, Z.; Gwack, Y. Immunological Disorders: Regulation of Ca(2+) Signaling in T Lymphocytes. Adv. Exp. Med. Biol. 2017, 993, 397–424. [Google Scholar] [CrossRef]
- Shaw, P.J.; Qu, B.; Hoth, M.; Feske, S. Molecular Regulation of CRAC Channels and Their Role in Lymphocyte Function. Cell Mol. Life Sci. 2013, 70, 2637–2656. [Google Scholar] [CrossRef]
- Trebak, M.; Kinet, J.-P. Calcium Signalling in T Cells. Nat. Rev. Immunol. 2019, 19, 154–169. [Google Scholar] [CrossRef]
- Vaeth, M.; Kahlfuss, S.; Feske, S. CRAC Channels and Calcium Signaling in T Cell-Mediated Immunity. Trends Immunol. 2020, 41, 878–901. [Google Scholar] [CrossRef]
- Vaeth, M.; Feske, S. Ion Channelopathies of the Immune System. Curr. Opin. Immunol. 2018, 52, 39–50. [Google Scholar] [CrossRef] [PubMed]
- Clemens, R.A.; Lowell, C.A. CRAC Channel Regulation of Innate Immune Cells in Health and Disease. Cell Calcium 2019, 78, 56–65. [Google Scholar] [CrossRef]
- Parekh, A.B. Store-Operated CRAC Channels: Function in Health and Disease. Nat. Rev. Drug Discov. 2010, 9, 399–410. [Google Scholar] [CrossRef]
- Johnson, M.; Trebak, M. ORAI Channels in Cellular Remodeling of Cardiorespiratory Disease. Cell Calcium 2019, 79, 1–10. [Google Scholar] [CrossRef]
- Hoth, M. CRAC Channels, Calcium, and Cancer in Light of the Driver and Passenger Concept. Biochim. Biophys. Acta 2016, 1863, 1408–1417. [Google Scholar] [CrossRef]
- Chalmers, S.B.; Monteith, G.R. ORAI Channels and Cancer. Cell Calcium 2018, 74, 160–167. [Google Scholar] [CrossRef]
- Feske, S. CRAC Channels and Disease—From Human CRAC Channelopathies and Animal Models to Novel Drugs. Cell Calcium 2019, 80, 112–116. [Google Scholar] [CrossRef]
- Lacruz, R.S.; Feske, S. Diseases Caused by Mutations in ORAI1 and STIM1. Ann. N. Y Acad. Sci. 2015, 1356, 45–79. [Google Scholar] [CrossRef] [PubMed]
- Ravetch, J.V.; Bolland, S. IgG Fc Receptors. Annu. Rev. Immunol. 2001, 19, 275–290. [Google Scholar] [CrossRef] [PubMed]
- Lanier, L.L. NK CELL RECOGNITION. Annu. Rev. Immunol. 2005, 23, 225–274. [Google Scholar] [CrossRef] [PubMed]
- Gauld, S.B.; Dal Porto, J.M.; Cambier, J.C. B Cell Antigen Receptor Signaling: Roles in Cell Development and Disease. Science 2002, 296, 1641–1642. [Google Scholar] [CrossRef] [PubMed]
- Abraham, R.T.; Weiss, A. Jurkat T Cells and Development of the T-Cell Receptor Signalling Paradigm. Nat. Rev. Immunol. 2004, 4, 301–308. [Google Scholar] [CrossRef] [PubMed]
- Turner, H.; Kinet, J.P. Signalling through the High-Affinity IgE Receptor Fc EpsilonRI. Nature 1999, 402, 24–30. [Google Scholar] [CrossRef] [PubMed]
- Liou, J.; Kim, M.L.; Heo, W.D.; Jones, J.T.; Myers, J.W.; Ferrell, J.E., Jr.; Meyer, T. STIM Is a Ca2+ Sensor Essential for Ca2+-Store-Depletion-Triggered Ca2+ Influx. Curr. Biol. 2005, 15, 1235–1241. [Google Scholar] [CrossRef]
- Roos, J.; DiGregorio, P.J.; Yeromin, A.V.; Ohlsen, K.; Lioudyno, M.; Zhang, S.; Safrina, O.; Kozak, J.A.; Wagner, S.L.; Cahalan, M.D.; et al. STIM1, an Essential and Conserved Component of Store-Operated Ca2+ Channel Function. J. Cell Biol. 2005, 169, 435–445. [Google Scholar] [CrossRef]
- Picard, C.; McCarl, C.-A.; Papolos, A.; Khalil, S.; Lüthy, K.; Hivroz, C.; LeDeist, F.; Rieux-Laucat, F.; Rechavi, G.; Rao, A.; et al. STIM1 Mutation Associated with a Syndrome of Immunodeficiency and Autoimmunity. N. Engl. J. Med. 2009, 360, 1971–1980. [Google Scholar] [CrossRef]
- Zhang, S.L.; Yu, Y.; Roos, J.; Kozak, J.A.; Deerinck, T.J.; Ellisman, M.H.; Stauderman, K.A.; Cahalan, M.D. STIM1 Is a Ca2+ Sensor That Activates CRAC Channels and Migrates from the Ca2+ Store to the Plasma Membrane. Nature 2005, 437, 902–905. [Google Scholar] [CrossRef]
- Sallinger, M.; Grabmayr, H.; Humer, C.; Bonhenry, D.; Romanin, C.; Schindl, R.; Derler, I. Activation Mechanisms and Structural Dynamics of STIM Proteins. J. Physiol. 2023, in press. [Google Scholar] [CrossRef]
- Prakriya, M.; Feske, S.; Gwack, Y.; Srikanth, S.; Rao, A.; Hogan, P.G. Orai1 Is an Essential Pore Subunit of the CRAC Channel. Nature 2006, 443, 230–233. [Google Scholar] [CrossRef]
- Zhang, S.L.; Yeromin, A.V.; Zhang, X.H.-F.; Yu, Y.; Safrina, O.; Penna, A.; Roos, J.; Stauderman, K.A.; Cahalan, M.D. Genome-Wide RNAi Screen of Ca(2+) Influx Identifies Genes That Regulate Ca(2+) Release-Activated Ca(2+) Channel Activity. Proc. Natl. Acad. Sci. USA 2006, 103, 9357–9362. [Google Scholar] [CrossRef]
- Vig, M.; Peinelt, C.; Beck, A.; Koomoa, D.L.; Rabah, D.; Koblan-Huberson, M.; Kraft, S.; Turner, H.; Fleig, A.; Penner, R.; et al. CRACM1 Is a Plasma Membrane Protein Essential for Store-Operated Ca2+ Entry. Science 2006, 312, 1220–1223. [Google Scholar] [CrossRef] [PubMed]
- Prakriya, M.; Lewis, R.S. Regulation of CRAC Channel Activity by Recruitment of Silent Channels to a High Open-Probability Gating Mode. J. Gen. Physiol. 2006, 128, 373–386. [Google Scholar] [CrossRef] [PubMed]
- Feske, S. CRAC Channelopathies. Pflug. Arch. 2010, 460, 417–435. [Google Scholar] [CrossRef] [PubMed]
- Feske, S.; Gwack, Y.; Prakriya, M.; Srikanth, S.; Puppel, S.H.; Tanasa, B.; Hogan, P.G.; Lewis, R.S.; Daly, M.; Rao, A. A Mutation in Orai1 Causes Immune Deficiency by Abrogating CRAC Channel Function. Nature 2006, 441, 179–185. [Google Scholar] [CrossRef]
- Hogan, P.G.; Chen, L.; Nardone, J.; Rao, A. Transcriptional Regulation by Calcium, Calcineurin, and NFAT. Genes. Dev. 2003, 17, 2205–2232. [Google Scholar] [CrossRef]
- Lis, A.; Peinelt, C.; Beck, A.; Parvez, S.; Monteilh-Zoller, M.; Fleig, A.; Penner, R. CRACM1, CRACM2, and CRACM3 Are Store-Operated Ca2+ Channels with Distinct Functional Properties. Curr. Biol. 2007, 17, 794–800. [Google Scholar] [CrossRef]
- Shaw, P.J.; Feske, S. Physiological and Pathophysiological Functions of SOCE in the Immune System. Front. Biosci. 2012, 4, 2253–2268. [Google Scholar] [CrossRef]
- Froghi, S.; Grant, C.R.; Tandon, R.; Quaglia, A.; Davidson, B.; Fuller, B. New Insights on the Role of TRP Channels in Calcium Signalling and Immunomodulation: Review of Pathways and Implications for Clinical Practice. Clin. Rev. Allergy Immunol. 2021, 60, 271–292. [Google Scholar] [CrossRef]
- Immler, R.; Simon, S.I.; Sperandio, M. Calcium Signalling and Related Ion Channels in Neutrophil Recruitment and Function. Eur. J. Clin. Investig. 2018, 48 (Suppl. S2), e12964. [Google Scholar] [CrossRef]
- Steinckwich, N.; Schenten, V.; Melchior, C.; Bréchard, S.; Tschirhart, E.J. An Essential Role of STIM1, Orai1, and S100A8–A9 Proteins for Ca2+ Signaling and FcγR-Mediated Phagosomal Oxidative Activity. J. Immunol. 2011, 186, 2182–2191. [Google Scholar] [CrossRef]
- Nunes, P.; Demaurex, N. The Role of Calcium Signaling in Phagocytosis. J. Leukoc. Biol. 2010, 88, 57–68. [Google Scholar] [CrossRef]
- Zhang, B.; Ma, X.; Loor, J.J.; Jiang, Q.; Guo, H.; Zhang, W.; Li, M.; Lv, X.; Yin, Y.; Wen, J.; et al. Role of ORAI Calcium Release-Activated Calcium Modulator 1 (ORAI1) on Neutrophil Extracellular Trap Formation in Dairy Cows with Subclinical Hypocalcemia. J. Dairy. Sci. 2022, 105, 3394–3404. [Google Scholar] [CrossRef]
- Waldron, R.T.; Chen, Y.; Pham, H.; Go, A.; Su, H.; Hu, C.; Wen, L.; Husain, S.Z.; Sugar, C.A.; Roos, J.; et al. The Orai Ca2+ Channel Inhibitor CM4620 Targets Both Parenchymal and Immune Cells to Reduce Inflammation in Experimental Acute Pancreatitis. J. Physiol. 2019, 597, 3085–3105. [Google Scholar] [CrossRef] [PubMed]
- Grimes, D.; Johnson, R.; Pashos, M.; Cummings, C.; Kang, C.; Sampedro, G.R.; Tycksen, E.; McBride, H.J.; Sah, R.; Lowell, C.A.; et al. ORAI1 and ORAI2 Modulate Murine Neutrophil Calcium Signaling, Cellular Activation, and Host Defense. Proc. Natl. Acad. Sci. USA 2020, 117, 24403–24414. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Guo, H.; Yang, W.; Li, M.; Zou, Y.; Loor, J.J.; Xia, C.; Xu, C. Effects of ORAI Calcium Release-Activated Calcium Modulator 1 (ORAI1) on Neutrophil Activity in Dairy Cows with Subclinical Hypocalcemia1. J. Anim. Sci. 2019, 97, 3326–3336. [Google Scholar] [CrossRef] [PubMed]
- Vandier, C.; Velge-Roussel, F. Regulation of Human Dendritic Cell Immune Functions by Ion Channels. Curr. Opin. Immunol. 2018, 52, 27–31. [Google Scholar] [CrossRef] [PubMed]
- Shumilina, E.; Huber, S.M.; Lang, F. Ca2+ Signaling in the Regulation of Dendritic Cell Functions. Am. J. Physiol.-Cell Physiol. 2011, 300, C1205–C1214. [Google Scholar] [CrossRef]
- Félix, R.; Crottès, D.; Delalande, A.; Fauconnier, J.; Lebranchu, Y.; Le Guennec, J.-Y.; Velge-Roussel, F. The Orai-1 and STIM-1 Complex Controls Human Dendritic Cell Maturation. PLoS ONE 2013, 8, e61595. [Google Scholar] [CrossRef] [PubMed]
- Hsu, S.; O’Connell, P.J.; Klyachko, V.A.; Badminton, M.N.; Thomson, A.W.; Jackson, M.B.; Clapham, D.E.; Ahern, G.P. Fundamental Ca2+ Signaling Mechanisms in Mouse Dendritic Cells: CRAC Is the Major Ca2+ Entry Pathway. J. Immunol. 2001, 166, 6126–6133. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, N.; Suzuki, J.; Kobayashi, Y. Role of Calcium in Tumor Necrosis Factor Production by Activated Macrophages. J. Biochem. 1996, 120, 1190–1195. [Google Scholar] [CrossRef]
- Chen, B.-C.; Chou, C.-F.; Lin, W.-W. Pyrimidinoceptor-Mediated Potentiation of Inducible Nitric-Oxide Synthase Induction in J774 Macrophages. J. Biol. Chem. 1998, 273, 29754–29763. [Google Scholar] [CrossRef] [PubMed]
- Vaeth, M.; Zee, I.; Concepcion, A.R.; Maus, M.; Shaw, P.; Portal-Celhay, C.; Zahra, A.; Kozhaya, L.; Weidinger, C.; Philips, J.; et al. Ca2+ Signaling but Not Store-Operated Ca2+ Entry Is Required for the Function of Macrophages and Dendritic Cells. J. Immunol. 2015, 195, 1202–1217. [Google Scholar] [CrossRef] [PubMed]
- Wajdner, H.E.; Farrington, J.; Barnard, C.; Peachell, P.T.; Schnackenberg, C.G.; Marino, J.P.; Xu, X.; Affleck, K.; Begg, M.; Seward, E.P. Orai and TRPC Channel Characterization in Fc ε RI-mediated Calcium Signaling and Mediator Secretion in Human Mast Cells. Physiol. Rep. 2017, 5, e13166. [Google Scholar] [CrossRef] [PubMed]
- Di Capite, J.; Parekh, A.B. CRAC Channels and Ca2+ Signaling in Mast Cells. Immunol. Rev. 2009, 231, 45–58. [Google Scholar] [CrossRef] [PubMed]
- Zweifach, A. Target-Cell Contact Activates a Highly Selective Capacitative Calcium Entry Pathway in Cytotoxic T Lymphocytes. J. Cell Biol. 2000, 148, 603–614. [Google Scholar] [CrossRef]
- Baba, Y.; Nishida, K.; Fujii, Y.; Hirano, T.; Hikida, M.; Kurosaki, T. Essential Function for the Calcium Sensor STIM1 in Mast Cell Activation and Anaphylactic Responses. Nat. Immunol. 2008, 9, 81–88. [Google Scholar] [CrossRef]
- Vig, M.; DeHaven, W.I.; Bird, G.S.; Billingsley, J.M.; Wang, H.; Rao, P.E.; Hutchings, A.B.; Jouvin, M.-H.; Putney, J.W.; Kinet, J.-P. Defective Mast Cell Effector Functions in Mice Lacking the CRACM1 Pore Subunit of Store-Operated Calcium Release-Activated Calcium Channels. Nat. Immunol. 2008, 9, 89–96. [Google Scholar] [CrossRef] [PubMed]
- Hoth, M.; Penner, R. Depletion of Intracellular Calcium Stores Activates a Calcium Current in Mast Cells. Nature 1992, 355, 353–356. [Google Scholar] [CrossRef] [PubMed]
- Maul-Pavicic, A.; Chiang, S.C.C.; Rensing-Ehl, A.; Jessen, B.; Fauriat, C.; Wood, S.M.; Sjöqvist, S.; Hufnagel, M.; Schulze, I.; Bass, T.; et al. ORAI1-Mediated Calcium Influx Is Required for Human Cytotoxic Lymphocyte Degranulation and Target Cell Lysis. Proc. Natl. Acad. Sci. USA 2011, 108, 3324–3329. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Friedmann, K.S.; Lyrmann, H.; Zhou, Y.; Schoppmeyer, R.; Knörck, A.; Mang, S.; Hoxha, C.; Angenendt, A.; Backes, C.S.; et al. A Calcium Optimum for Cytotoxic T Lymphocyte and Natural Killer Cell Cytotoxicity. J. Physiol. 2018, 596, 2681–2698. [Google Scholar] [CrossRef] [PubMed]
- Kurosaki, T.; Shinohara, H.; Baba, Y. B Cell Signaling and Fate Decision. Annu. Rev. Immunol. 2010, 28, 21–55. [Google Scholar] [CrossRef] [PubMed]
- Feske, S.; Skolnik, E.Y.; Prakriya, M. Ion Channels and Transporters in Lymphocyte Function and Immunity. Nat. Rev. Immunol. 2012, 12, 532–547. [Google Scholar] [CrossRef] [PubMed]
- Aifantis, I.; Gounari, F.; Scorrano, L.; Borowski, C.; von Boehmer, H. Constitutive Pre-TCR Signaling Promotes Differentiation through Ca2+ Mobilization and Activation of NF-ΚB and NFAT. Nat. Immunol. 2001, 2, 403–409. [Google Scholar] [CrossRef]
- Nakayama, T.; Ueda, Y.; Yamada, H.; Shores, E.W.; Singer, A.; June, C.H. In Vivo Calcium Elevations in Thymocytes with T Cell Receptors That Are Specific for Self Ligands. Science 1992, 257, 96–99. [Google Scholar] [CrossRef]
- Melichar, H.J.; Ross, J.O.; Herzmark, P.; Hogquist, K.A.; Robey, E.A. Distinct Temporal Patterns of T Cell Receptor Signaling during Positive versus Negative Selection in Situ. Sci. Signal 2013, 6, ra92. [Google Scholar] [CrossRef]
- Daniels, M.A.; Teixeiro, E.; Gill, J.; Hausmann, B.; Roubaty, D.; Holmberg, K.; Werlen, G.; Holländer, G.A.; Gascoigne, N.R.J.; Palmer, E. Thymic Selection Threshold Defined by Compartmentalization of Ras/MAPK Signalling. Nature 2006, 444, 724–729. [Google Scholar] [CrossRef]
- Bhakta, N.R.; Oh, D.Y.; Lewis, R.S. Calcium Oscillations Regulate Thymocyte Motility during Positive Selection in the Three-Dimensional Thymic Environment. Nat. Immunol. 2005, 6, 143–151. [Google Scholar] [CrossRef]
- Oh-hora, M.; Rao, A. Calcium Signaling in Lymphocytes. Curr. Opin. Immunol. 2008, 20, 250–258. [Google Scholar] [CrossRef]
- Wei, S.H.; Safrina, O.; Yu, Y.; Garrod, K.R.; Cahalan, M.D.; Parker, I. Ca2+ Signals in CD4+ T Cells during Early Contacts with Antigen-Bearing Dendritic Cells in Lymph Node. J. Immunol. 2007, 179, 1586–1594. [Google Scholar] [CrossRef]
- Lioudyno, M.I.; Kozak, J.A.; Penna, A.; Safrina, O.; Zhang, S.L.; Sen, D.; Roos, J.; Stauderman, K.A.; Cahalan, M.D. Orai1 and STIM1 Move to the Immunological Synapse and Are Up-Regulated during T Cell Activation. Proc. Natl. Acad. Sci. USA 2008, 105, 2011–2016. [Google Scholar] [CrossRef]
- Barr, V.A.; Bernot, K.M.; Srikanth, S.; Gwack, Y.; Balagopalan, L.; Regan, C.K.; Helman, D.J.; Sommers, C.L.; Oh-hora, M.; Rao, A.; et al. Dynamic Movement of the Calcium Sensor STIM1 and the Calcium Channel Orai1 in Activated T-Cells: Puncta and Distal Caps. Mol. Biol. Cell 2008, 19, 2802–2817. [Google Scholar] [CrossRef]
- Quintana, A.; Pasche, M.; Junker, C.; Al-Ansary, D.; Rieger, H.; Kummerow, C.; Nuñez, L.; Villalobos, C.; Meraner, P.; Becherer, U.; et al. Calcium Microdomains at the Immunological Synapse: How ORAI Channels, Mitochondria and Calcium Pumps Generate Local Calcium Signals for Efficient T-Cell Activation. EMBO J. 2011, 30, 3895–3912. [Google Scholar] [CrossRef]
- Pipkin, M.E.; Lieberman, J. Delivering the Kiss of Death: Progress on Understanding How Perforin Works. Curr. Opin. Immunol. 2007, 19, 301–308. [Google Scholar] [CrossRef] [PubMed]
- Negulescu, P.A.; Krasieva, T.B.; Khan, A.; Kerschbaum, H.H.; Cahalan, M.D. Polarity of T Cell Shape, Motility, and Sensitivity to Antigen. Immunity 1996, 4, 421–430. [Google Scholar] [CrossRef] [PubMed]
- Orrenius, S.; Zhivotovsky, B.; Nicotera, P. Regulation of Cell Death: The Calcium–Apoptosis Link. Nat. Rev. Mol. Cell Biol. 2003, 4, 552–565. [Google Scholar] [CrossRef] [PubMed]
- Zhivotovsky, B.; Orrenius, S. Calcium and Cell Death Mechanisms: A Perspective from the Cell Death Community. Cell Calcium 2011, 50, 211–221. [Google Scholar] [CrossRef] [PubMed]
- Desvignes, L.; Weidinger, C.; Shaw, P.; Vaeth, M.; Ribierre, T.; Liu, M.; Fergus, T.; Kozhaya, L.; McVoy, L.; Unutmaz, D.; et al. STIM1 Controls T Cell-Mediated Immune Regulation and Inflammation in Chronic Infection. J. Clin. Invest. 2015, 125, 2347–2362. [Google Scholar] [CrossRef]
- Weidinger, C.; Shaw, P.J.; Feske, S. STIM1 and STIM2-Mediated Ca(2+) Influx Regulates Antitumour Immunity by CD8(+) T Cells. EMBO Mol. Med. 2013, 5, 1311–1321. [Google Scholar] [CrossRef]
- Kim, K.-D.; Srikanth, S.; Yee, M.-K.W.; Mock, D.C.; Lawson, G.W.; Gwack, Y. ORAI1 Deficiency Impairs Activated T Cell Death and Enhances T Cell Survival. J. Immunol. 2011, 187, 3620–3630. [Google Scholar] [CrossRef]
- Feske, S.; Giltnane, J.; Dolmetsch, R.; Staudt, L.M.; Rao, A. Gene Regulation Mediated by Calcium Signals in T Lymphocytes. Nat. Immunol. 2001, 2, 316–324. [Google Scholar] [CrossRef]
- Huppa, J.B.; Gleimer, M.; Sumen, C.; Davis, M.M. Continuous T Cell Receptor Signaling Required for Synapse Maintenance and Full Effector Potential. Nat. Immunol. 2003, 4, 749–755. [Google Scholar] [CrossRef]
- Waite, J.C.; Vardhana, S.; Shaw, P.J.; Jang, J.-E.; McCarl, C.-A.; Cameron, T.O.; Feske, S.; Dustin, M.L. Interference with Ca(2+) Release Activated Ca(2+) (CRAC) Channel Function Delays T-Cell Arrest in Vivo. Eur. J. Immunol. 2013, 43, 3343–3354. [Google Scholar] [CrossRef]
- Greenberg, M.L.; Yu, Y.; Leverrier, S.; Zhang, S.L.; Parker, I.; Cahalan, M.D. Orai1 Function Is Essential for T Cell Homing to Lymph Nodes. J. Immunol. 2013, 190, 3197–3206. [Google Scholar] [CrossRef] [PubMed]
- Vaeth, M.; Maus, M.; Klein-Hessling, S.; Freinkman, E.; Yang, J.; Eckstein, M.; Cameron, S.; Turvey, S.E.; Serfling, E.; Berberich-Siebelt, F.; et al. Store-Operated Ca2+ Entry Controls Clonal Expansion of T Cells through Metabolic Reprogramming. Immunity 2017, 47, 664–679.e6. [Google Scholar] [CrossRef] [PubMed]
- Feske, S. Calcium Signalling in Lymphocyte Activation and Disease. Nat. Rev. Immunol. 2007, 7, 690–702. [Google Scholar] [CrossRef]
- Feske, S.; Okamura, H.; Hogan, P.G.; Rao, A. Ca2+/Calcineurin Signalling in Cells of the Immune System. Biochem. Biophys. Res. Commun. 2003, 311, 1117–1132. [Google Scholar] [CrossRef] [PubMed]
- Macian, F. NFAT Proteins: Key Regulators of T-Cell Development and Function. Nat. Rev. Immunol. 2005, 5, 472–484. [Google Scholar] [CrossRef]
- Macián, F.; López-Rodríguez, C.; Rao, A. Partners in Transcription: NFAT and AP-1. Oncogene 2001, 20, 2476–2489. [Google Scholar] [CrossRef]
- Gwack, Y.; Feske, S.; Srikanth, S.; Hogan, P.G.; Rao, A. Signalling to Transcription: Store-Operated Ca2+ Entry and NFAT Activation in Lymphocytes. Cell Calcium 2007, 42, 145–156. [Google Scholar] [CrossRef]
- Srikanth, S.; Gwack, Y. Orai1-NFAT Signalling Pathway Triggered by T Cell Receptor Stimulation. Mol. Cells 2013, 35, 182–194. [Google Scholar] [CrossRef]
- Palkowitsch, L.; Marienfeld, U.; Brunner, C.; Eitelhuber, A.; Krappmann, D.; Marienfeld, R.B. The Ca2+-Dependent Phosphatase Calcineurin Controls the Formation of the Carma1-Bcl10-Malt1 Complex during T Cell Receptor-Induced NF-ΚB Activation. J. Biol. Chem. 2011, 286, 7522–7534. [Google Scholar] [CrossRef] [PubMed]
- Frischbutter, S.; Gabriel, C.; Bendfeldt, H.; Radbruch, A.; Baumgrass, R. Dephosphorylation of Bcl-10 by Calcineurin Is Essential for Canonical NF-κB Activation in Th Cells. Eur. J. Immunol. 2011, 41, 2349–2357. [Google Scholar] [CrossRef] [PubMed]
- Yeh, Y.-C.; Parekh, A.B. CRAC Channels and Ca2+-Dependent Gene Expression. In Calcium Entry Channels in Non-Excitable Cells; Series: Methods in Signal Transduction Series; CRC Press: Boca Raton, FL, USA; Taylor & Francis: Abingdon, UK, 2017; pp. 93–106. [Google Scholar]
- McCarl, C.-A.; Khalil, S.; Ma, J.; Oh-hora, M.; Yamashita, M.; Roether, J.; Kawasaki, T.; Jairaman, A.; Sasaki, Y.; Prakriya, M.; et al. Store-Operated Ca2+ Entry through ORAI1 Is Critical for T Cell-Mediated Autoimmunity and Allograft Rejection. J. Immunol. 2010, 185, 5845–5858. [Google Scholar] [CrossRef] [PubMed]
- Feske, S.; Draeger, R.; Peter, H.-H.; Eichmann, K.; Rao, A. The Duration of Nuclear Residence of NFAT Determines the Pattern of Cytokine Expression in Human SCID T Cells. J. Immunol. 2000, 165, 297–305. [Google Scholar] [CrossRef] [PubMed]
- Feske, S.; Müller, J.M.; Graf, D.; Kroczek, R.A.; Dräger, R.; Niemeyer, C.; Baeuerle, P.A.; Peter, H.H.; Schlesier, M. Severe Combined Immunodeficiency Due to Defective Binding of the Nuclear Factor of Activated T Cells in T Lymphocytes of Two Male Siblings. Eur. J. Immunol. 1996, 26, 2119–2126. [Google Scholar] [CrossRef] [PubMed]
- Endo, Y.; Noguchi, S.; Hara, Y.; Hayashi, Y.K.; Motomura, K.; Miyatake, S.; Murakami, N.; Tanaka, S.; Yamashita, S.; Kizu, R.; et al. Dominant Mutations in ORAI1 Cause Tubular Aggregate Myopathy with Hypocalcemia via Constitutive Activation of Store-Operated Ca(2)(+) Channels. Hum. Mol. Genet. 2015, 24, 637–648. [Google Scholar] [CrossRef]
- Garibaldi, M.; Fattori, F.; Riva, B.; Labasse, C.; Brochier, G.; Ottaviani, P.; Sacconi, S.; Vizzaccaro, E.; Laschena, F.; Romero, N.B.; et al. A Novel Gain-of-Function Mutation in ORAI1 Causes Late-Onset Tubular Aggregate Myopathy and Congenital Miosis. Clin. Genet. 2017, 91, 780–786. [Google Scholar] [CrossRef]
- Bohm, J.; Bulla, M.; Urquhart, J.E.; Malfatti, E.; Williams, S.G.; O’Sullivan, J.; Szlauer, A.; Koch, C.; Baranello, G.; Mora, M.; et al. ORAI1 Mutations with Distinct Channel Gating Defects in Tubular Aggregate Myopathy. Hum. Mutat. 2017, 38, 426–438. [Google Scholar] [CrossRef]
- Korzeniowski, M.K.; Manjarrés, I.M.; Varnai, P.; Balla, T. Activation of STIM1-Orai1 Involves an Intramolecular Switching Mechanism. Sci. Signal 2010, 3, ra82. [Google Scholar] [CrossRef]
- Schaballie, H.; Rodriguez, R.; Martin, E.; Moens, L.; Frans, G.; Lenoir, C.; Dutré, J.; Canioni, D.; Bossuyt, X.; Fischer, A.; et al. A Novel Hypomorphic Mutation in STIM1 Results in a Late-Onset Immunodeficiency. J. Allergy Clin. Immunol. 2015, 136, 816–819.e4. [Google Scholar] [CrossRef]
- Lian, J.; Cuk, M.; Kahlfuss, S.; Kozhaya, L.; Vaeth, M.; Rieux-Laucat, F.; Picard, C.; Benson, M.J.; Jakovcevic, A.; Bilic, K.; et al. ORAI1 Mutations Abolishing Store-Operated Ca2+ Entry Cause Anhidrotic Ectodermal Dysplasia with Immunodeficiency. J. Allergy Clin. Immunol. 2018, 142, 1297–1310.e11. [Google Scholar] [CrossRef]
- McCarl, C.-A.; Picard, C.; Khalil, S.; Kawasaki, T.; Röther, J.; Papolos, A.; Kutok, J.; Hivroz, C.; LeDeist, F.; Plogmann, K.; et al. ORAI1 Deficiency and Lack of Store-Operated Ca2+ Entry Cause Immunodeficiency, Myopathy, and Ectodermal Dysplasia. J. Allergy Clin. Immunol. 2009, 124, 1311–1318.e7. [Google Scholar] [CrossRef]
- Available online: https://cancergenome.nih.gov/Cancer Genome Atlas Network (accessed on 16 December 2023).
- Markello, T.; Chen, D.; Kwan, J.Y.; Horkayne-Szakaly, I.; Morrison, A.; Simakova, O.; Maric, I.; Lozier, J.; Cullinane, A.R.; Kilo, T.; et al. York Platelet Syndrome Is a CRAC Channelopathy Due to Gain-of-Function Mutations in STIM1. Mol. Genet. Metab. 2015, 114, 474–482. [Google Scholar] [CrossRef]
- Hedberg, C.; Niceta, M.; Fattori, F.; Lindvall, B.; Ciolfi, A.; D’Amico, A.; Tasca, G.; Petrini, S.; Tulinius, M.; Tartaglia, M.; et al. Childhood Onset Tubular Aggregate Myopathy Associated with de Novo STIM1 Mutations. J. Neurol. 2014, 261, 870–876. [Google Scholar] [CrossRef] [PubMed]
- Walter, M.C.; Rossius, M.; Zitzelsberger, M.; Vorgerd, M.; Müller-Felber, W.; Ertl-Wagner, B.; Zhang, Y.; Brinkmeier, H.; Senderek, J.; Schoser, B. 50 Years to Diagnosis: Autosomal Dominant Tubular Aggregate Myopathy Caused by a Novel STIM1 Mutation. Neuromuscul. Disord. 2015, 25, 577–584. [Google Scholar] [CrossRef] [PubMed]
- Böhm, J.; Chevessier, F.; Koch, C.; Peche, G.A.; Mora, M.; Morandi, L.; Pasanisi, B.; Moroni, I.; Tasca, G.; Fattori, F.; et al. Clinical, Histological and Genetic Characterisation of Patients with Tubular Aggregate Myopathy Caused by Mutations in STIM1. J. Med. Genet. 2014, 51, 824–833. [Google Scholar] [CrossRef] [PubMed]
- Böhm, J.; Chevessier, F.; De Paula, A.M.; Koch, C.; Attarian, S.; Feger, C.; Hantaï, D.; Laforêt, P.; Ghorab, K.; Vallat, J.-M.; et al. Constitutive Activation of the Calcium Sensor STIM1 Causes Tubular-Aggregate Myopathy. Am. J. Human. Genet. 2013, 92, 271–278. [Google Scholar] [CrossRef]
- Fuchs, S.; Rensing-Ehl, A.; Speckmann, C.; Bengsch, B.; Schmitt-Graeff, A.; Bondzio, I.; Maul-Pavicic, A.; Bass, T.; Vraetz, T.; Strahm, B.; et al. Antiviral and Regulatory T Cell Immunity in a Patient with Stromal Interaction Molecule 1 Deficiency. J. Immunol. 2012, 188, 1523–1533. [Google Scholar] [CrossRef]
- Byun, M.; Abhyankar, A.; Lelarge, V.; Plancoulaine, S.; Palanduz, A.; Telhan, L.; Boisson, B.; Picard, C.; Dewell, S.; Zhao, C.; et al. Whole-Exome Sequencing-Based Discovery of STIM1 Deficiency in a Child with Fatal Classic Kaposi Sarcoma. J. Exp. Med. 2010, 207, 2307–2312. [Google Scholar] [CrossRef] [PubMed]
- Vaeth, M.; Eckstein, M.; Shaw, P.J.; Kozhaya, L.; Yang, J.; Berberich-Siebelt, F.; Clancy, R.; Unutmaz, D.; Feske, S. Store-Operated Ca 2+ Entry in Follicular T Cells Controls Humoral Immune Responses and Autoimmunity. Immunity 2016, 44, 1350–1364. [Google Scholar] [CrossRef] [PubMed]
- Feske, S. Immunodeficiency Due to Defects in Store-operated Calcium Entry. Ann. N. Y Acad. Sci. 2011, 1238, 74–90. [Google Scholar] [CrossRef]
- Rubaiy, H.N. ORAI Calcium Channels: Regulation, Function, Pharmacology, and Therapeutic Targets. Pharmaceuticals 2023, 16, 162. [Google Scholar] [CrossRef] [PubMed]
- Figueiredo, I.A.D.; Ferreira, S.R.D.; Fernandes, J.M.; Silva, B.A.d.; Vasconcelos, L.H.C.; Cavalcante, F.d.A. A Review of the Pathophysiology and the Role of Ion Channels on Bronchial Asthma. Front. Pharmacol. 2023, 14, 1236550. [Google Scholar] [CrossRef]
- Cantonero, C.; Sanchez-Collado, J.; Gonzalez-Nuñez, M.A.; Salido, G.M.; Lopez, J.J.; Jardin, I.; Rosado, J.A. Store-Independent Orai1-Mediated Ca2+ Entry and Cancer. Cell Calcium 2019, 80, 1–7. [Google Scholar] [CrossRef]
- Kouba, S.; Buscaglia, P.; Guéguinou, M.; Ibrahim, S.; Félix, R.; Guibon, R.; Fromont, G.; Pigat, N.; Capiod, T.; Vandier, C.; et al. Pivotal Role of the ORAI3-STIM2 Complex in the Control of Mitotic Death and Prostate Cancer Cell Cycle Progression. Cell Calcium 2023, 115, 102794. [Google Scholar] [CrossRef]
- Sanchez-Collado, J.; Jardin, I.; López, J.J.; Ronco, V.; Salido, G.M.; Dubois, C.; Prevarskaya, N.; Rosado, J.A. Role of Orai3 in the Pathophysiology of Cancer. Int. J. Mol. Sci. 2021, 22, 11426. [Google Scholar] [CrossRef]
- Mignen, O.; Vannier, J.-P.; Schneider, P.; Renaudineau, Y.; Abdoul-Azize, S. Orai1 Ca2+ Channel Modulators as Therapeutic Tools for Treating Cancer: Emerging Evidence! Biochem. Pharmacol. 2024, 219, 115955. [Google Scholar] [CrossRef]
- Fiorio Pla, A.; Kondratska, K.; Prevarskaya, N. STIM and ORAI Proteins: Crucial Roles in Hallmarks of Cancer. Am. J. Physiol. Cell Physiol. 2016, 310, C509–C519. [Google Scholar] [CrossRef] [PubMed]
- Dubois, C.; Vanden Abeele, F.; Lehen’kyi, V.; Gkika, D.; Guarmit, B.; Lepage, G.; Slomianny, C.; Borowiec, A.S.; Bidaux, G.; Benahmed, M.; et al. Remodeling of Channel-Forming ORAI Proteins Determines an Oncogenic Switch in Prostate Cancer. Cancer Cell 2014, 26, 19–32. [Google Scholar] [CrossRef]
- Umemura, M.; Nakakaji, R.; Ishikawa, Y. Physiological Functions of Calcium Signaling via Orai1 in Cancer. J. Physiol. Sci. 2023, 73, 21. [Google Scholar] [CrossRef] [PubMed]
- Vashisht, A.; Trebak, M.; Motiani, R.K. STIM and Orai Proteins as Novel Targets for Cancer Therapy. A Review in the Theme: Cell and Molecular Processes in Cancer Metastasis. Am. J. Physiol. Cell Physiol. 2015, 309, C457–C469. [Google Scholar] [CrossRef] [PubMed]
- Tiffner, A.; Hopl, V.; Derler, I. CRAC and SK Channels: Their Molecular Mechanisms Associated with Cancer Cell Development. Cancers 2022, 15, 101. [Google Scholar] [CrossRef] [PubMed]
- Letizia, M.; Wang, Y.; Kaufmann, U.; Gerbeth, L.; Sand, A.; Brunkhorst, M.; Weidner, P.; Ziegler, J.F.; Böttcher, C.; Schlickeiser, S.; et al. Store-operated Calcium Entry Controls Innate and Adaptive Immune Cell Function in Inflammatory Bowel Disease. EMBO Mol. Med. 2022, 14, e15687. [Google Scholar] [CrossRef]
- Kaufmann, U.; Shaw, P.J.; Kozhaya, L.; Subramanian, R.; Gaida, K.; Unutmaz, D.; McBride, H.J.; Feske, S. Selective ORAI1 Inhibition Ameliorates Autoimmune Central Nervous System Inflammation by Suppressing Effector but Not Regulatory T Cell Function. J. Immunol. 2016, 196, 573–585. [Google Scholar] [CrossRef]
- Wu, B.; Woo, J.S.; Sun, Z.; Srikanth, S.; Gwack, Y. Ca2+ Signaling Augmented by ORAI1 Trafficking Regulates the Pathogenic State of Effector T Cells. J. Immunol. 2022, 208, 1329–1340. [Google Scholar] [CrossRef]
- Vaeth, M.; Yang, J.; Yamashita, M.; Zee, I.; Eckstein, M.; Knosp, C.; Kaufmann, U.; Karoly Jani, P.; Lacruz, R.S.; Flockerzi, V.; et al. ORAI2 Modulates Store-Operated Calcium Entry and T Cell-Mediated Immunity. Nat. Commun. 2017, 8, 14714. [Google Scholar] [CrossRef]
- Cox, J.H.; Hussell, S.; Søndergaard, H.; Roepstorff, K.; Bui, J.-V.; Deer, J.R.; Zhang, J.; Li, Z.-G.; Lamberth, K.; Kvist, P.H.; et al. Antibody-Mediated Targeting of the Orai1 Calcium Channel Inhibits T Cell Function. PLoS ONE 2013, 8, e82944. [Google Scholar] [CrossRef]
- Yuan, X.; Tang, B.; Chen, Y.; Zhou, L.; Deng, J.; Han, L.; Zhai, Y.; Zhou, Y.; Gill, D.L.; Lu, C.; et al. Celastrol Inhibits Store Operated Calcium Entry and Suppresses Psoriasis. Front. Pharmacol. 2023, 14, 1111798. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.-H.; Noyer, L.; Kahlfuss, S.; Raphael, D.; Tao, A.Y.; Kaufmann, U.; Zhu, J.; Mitchell-Flack, M.; Sidhu, I.; Zhou, F.; et al. Distinct Roles of ORAI1 in T Cell–Mediated Allergic Airway Inflammation and Immunity to Influenza A Virus Infection. Sci. Adv. 2022, 8, eabn6552. [Google Scholar] [CrossRef]
- Braun, A.; Gessner, J.E.; Varga-Szabo, D.; Syed, S.N.; Konrad, S.; Stegner, D.; Vögtle, T.; Schmidt, R.E.; Nieswandt, B. STIM1 Is Essential for Fcγ Receptor Activation and Autoimmune Inflammation. Blood 2009, 113, 1097–1104. [Google Scholar] [CrossRef] [PubMed]
- Rivet, C.A.; Hill, A.S.; Lu, H.; Kemp, M.L. Predicting Cytotoxic T-Cell Age from Multivariate Analysis of Static and Dynamic Biomarkers. Mol. Cell Proteom. 2011, 10, 3921. [Google Scholar] [CrossRef] [PubMed]
- Angenendt, A.; Steiner, R.; Knörck, A.; Schwär, G.; Konrad, M.; Krause, E.; Lis, A. Orai, STIM, and PMCA Contribute to Reduced Calcium Signal Generation in CD8+ T Cells of Elderly Mice. Aging 2020, 12, 3266–3286. [Google Scholar] [CrossRef] [PubMed]
- Khan, H.Y.; Mazahir, I.; Reddy, S.; Fazili, F.; Azmi, A.S. Roles of CRAC Channel in Cancer: Implications for Therapeutic Development. Expert. Rev. Precis. Med. Drug Dev. 2020, 5, 371–382. [Google Scholar] [CrossRef] [PubMed]
- Tanwar, J.; Arora, S.; Motiani, R.K. Orai3: Oncochannel with Therapeutic Potential. Cell Calcium 2020, 90, 102247. [Google Scholar] [CrossRef]
- Shapovalov, G.; Gordienko, D.; Prevarskaya, N. Store operated calcium channels in cancer progression. Int. Rev. Cell Mol. Biol. 2021, 363, 123–168. [Google Scholar] [CrossRef]
- Hammad, A.S.; Machaca, K. Store Operated Calcium Entry in Cell Migration and Cancer Metastasis. Cells 2021, 10, 1246. [Google Scholar] [CrossRef]
- Jardin, I.; Lopez, J.J.; Sanchez-Collado, J.; Gomez, L.J.; Salido, G.M.; Rosado, J.A. Store-Operated Calcium Entry and Its Implications in Cancer Stem Cells. Cells 2022, 11, 1332. [Google Scholar] [CrossRef]
- Villalobos, C.; Hernández-Morales, M.; Gutiérrez, L.G.; Núñez, L. TRPC1 and ORAI1 Channels in Colon Cancer. Cell Calcium 2019, 81, 59–66. [Google Scholar] [CrossRef]
- Backes, C.S.; Friedmann, K.S.; Mang, S.; Knörck, A.; Hoth, M.; Kummerow, C. Natural Killer Cells Induce Distinct Modes of Cancer Cell Death: Discrimination, Quantification, and Modulation of Apoptosis, Necrosis, and Mixed Forms. J. Biol. Chem. 2018, 293, 16348–16363. [Google Scholar] [CrossRef]
- Kaschek, L.; Zöphel, S.; Knörck, A.; Hoth, M. A Calcium Optimum for Cytotoxic T Lymphocyte and Natural Killer Cell Cytotoxicity. Semin. Cell Dev. Biol. 2021, 115, 10–18. [Google Scholar] [CrossRef]
- Rosado, J.A.; Diez, R.; Smani, T.; Jardín, I. STIM and Orai1 Variants in Store-Operated Calcium Entry. Front. Pharmacol. 2016, 6, 325. [Google Scholar] [CrossRef]
- Bogeski, I.; Al-Ansary, D.; Qu, B.; Niemeyer, B.A.; Hoth, M.; Peinelt, C. Pharmacology of ORAI Channels as a Tool to Understand Their Physiological Functions. Expert. Rev. Clin. Pharmacol. 2010, 3, 291–303. [Google Scholar] [CrossRef] [PubMed]
- Benson, J.C.; Trebak, M. Too Much of a Good Thing: The Case of SOCE in Cellular Apoptosis. Cell Calcium 2023, 111, 102716. [Google Scholar] [CrossRef] [PubMed]
- Hoth, M.; Niemeyer, B.A. The neglected CRAC proteins. Curr. Top. Membr. 2013, 71, 237–271. [Google Scholar] [CrossRef] [PubMed]
- Ong, H.L.; Subedi, K.P.; Son, G.-Y.; Liu, X.; Ambudkar, I.S. Tuning Store-Operated Calcium Entry to Modulate Ca2+-Dependent Physiological Processes. Biochim. Biophys. Acta (BBA) Mol. Cell Res. 2019, 1866, 1037–1045. [Google Scholar] [CrossRef] [PubMed]
- ČENDULA, R.; DRAGÚN, M.; GAŽOVÁ, A.; KYSELOVIČ, J.; HULMAN, M.; MÁŤUŠ, M. Changes in STIM Isoforms Expression and Gender-Specific Alterations in Orai Expression in Human Heart Failure. Physiol. Res. 2019, 68, S165–S172. [Google Scholar] [CrossRef] [PubMed]
- Moccia, F.; Zuccolo, E.; Poletto, V.; Turin, I.; Guerra, G.; Pedrazzoli, P.; Rosti, V.; Porta, C.; Montagna, D. Targeting Stim and Orai Proteins as an Alternative Approach in Anticancer Therapy. Curr. Med. Chem. 2016, 23, 3450–3480. [Google Scholar] [CrossRef] [PubMed]
- Grabmayr, H.; Romanin, C.; Fahrner, M. STIM Proteins: An Ever-Expanding Family. Int. J. Mol. Sci. 2020, 22, 378. [Google Scholar] [CrossRef]
- Lilliu, E.; Koenig, S.; Koenig, X.; Frieden, M. Store-Operated Calcium Entry in Skeletal Muscle: What Makes It Different? Cells 2021, 10, 2356. [Google Scholar] [CrossRef]
- Yoast, R.E.; Emrich, S.M.; Trebak, M. The Anatomy of Native CRAC Channel(s). Curr. Opin. Physiol. 2020, 17, 89–95. [Google Scholar] [CrossRef]
- Fahrner, M.; Grabmayr, H.; Romanin, C. Mechanism of STIM Activation. Curr. Opin. Physiol. 2020, 17, 74–79. [Google Scholar] [CrossRef] [PubMed]
- Butorac, C.; Krizova, A.; Derler, I. Review: Structure and Activation Mechanisms of CRAC Channels. Adv. Exp. Med. Biol. 2020, 1131, 547–604. [Google Scholar] [CrossRef] [PubMed]
- Yeung, P.S.; Yamashita, M.; Prakriya, M. Molecular Basis of Allosteric Orai1 Channel Activation by STIM1. J. Physiol. 2019, 598, 1707–1723. [Google Scholar] [CrossRef]
- Yeung, P.S.; Yamashita, M.; Prakriya, M. Pore Opening Mechanism of CRAC Channels. Cell Calcium 2016, 63, 14–19. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Cai, X.; Nwokonko, R.M.; Loktionova, N.A.; Wang, Y.; Gill, D.L. The STIM-Orai Coupling Interface and Gating of the Orai1 Channel. Cell Calcium 2017, 63, 8–13. [Google Scholar] [CrossRef]
- Humer, C.; Romanin, C.; Höglinger, C. Highlighting the Multifaceted Role of Orai1 N-Terminal- and Loop Regions for Proper CRAC Channel Functions. Cells 2022, 11, 371. [Google Scholar] [CrossRef]
- Qiu, R.; Lewis, R.S. Structural Features of STIM and Orai Underlying Store-Operated Calcium Entry. Curr. Opin. Cell Biol. 2019, 57, 90–98. [Google Scholar] [CrossRef]
- Frischauf, I.; Fahrner, M.; Jardín, I.; Romanin, C. The STIM1: Orai Interaction. Adv. Exp. Med. Biol. 2016, 898, 25–46. [Google Scholar] [CrossRef] [PubMed]
- Gudlur, A.; Hogan, P.G. The STIM-Orai Pathway: Orai, the Pore-Forming Subunit of the CRAC Channel. Adv. Exp. Med. Biol. 2017, 993, 39–57. [Google Scholar] [CrossRef] [PubMed]
- Tiffner, A.; Derler, I. Isoform-Specific Properties of Orai Homologues in Activation, Downstream Signaling, Physiology and Pathophysiology. Int. J. Mol. Sci. 2021, 22, 8020. [Google Scholar] [CrossRef]
- Sallinger, M.; Tiffner, A.; Schmidt, T.; Bonhenry, D.; Waldherr, L.; Frischauf, I.; Lunz, V.; Derler, I.; Schober, R.; Schindl, R. Luminal STIM1 Mutants That Cause Tubular Aggregate Myopathy Promote Autophagic Processes. Int. J. Mol. Sci. 2020, 21, 4410. [Google Scholar] [CrossRef] [PubMed]
- Schober, R.; Bonhenry, D.; Lunz, V.; Zhu, J.; Krizova, A.; Frischauf, I.; Fahrner, M.; Zhang, M.; Waldherr, L.; Schmidt, T.; et al. Sequential Activation of STIM1 Links Ca(2+) with Luminal Domain Unfolding. Sci. Signal 2019, 12, eaax3194. [Google Scholar] [CrossRef] [PubMed]
- Gudlur, A.; Zeraik, A.E.; Hirve, N.; Rajanikanth, V.; Bobkov, A.A.; Ma, G.; Zheng, S.; Wang, Y.; Zhou, Y.; Komives, E.A.; et al. Calcium Sensing by the STIM1 ER-Luminal Domain. Nat. Commun. 2018, 9, 4536. [Google Scholar] [CrossRef] [PubMed]
- Stathopulos, P.B.; Li, G.-Y.; Plevin, M.J.; Ames, J.B.; Ikura, M. Stored Ca2+ Depletion-Induced Oligomerization of Stromal Interaction Molecule 1 (STIM1) via the EF-SAM Region. J. Biol. Chem. 2006, 281, 35855–35862. [Google Scholar] [CrossRef] [PubMed]
- Gudlur, A.; Quintana, A.; Zhou, Y.; Hirve, N.; Mahapatra, S.; Hogan, P.G. STIM1 Triggers a Gating Rearrangement at the Extracellular Mouth of the ORAI1 Channel. Nat. Commun. 2014, 5, 5164. [Google Scholar] [CrossRef]
- Ma, G.; Wei, M.; He, L.; Liu, C.; Wu, B.; Zhang, S.L.; Jing, J.; Liang, X.; Senes, A.; Tan, P.; et al. Inside-out Ca(2+) Signalling Prompted by STIM1 Conformational Switch. Nat. Commun. 2015, 6, 7826. [Google Scholar] [CrossRef]
- Hirve, N.; Rajanikanth, V.; Hogan, P.G.; Gudlur, A. Coiled-Coil Formation Conveys a STIM1 Signal from ER Lumen to Cytoplasm. Cell Rep. 2018, 22, 72–83. [Google Scholar] [CrossRef] [PubMed]
- Fahrner, M.; Muik, M.; Schindl, R.; Butorac, C.; Stathopulos, P.; Zheng, L.; Jardin, I.; Ikura, M.; Romanin, C. A Coiled-Coil Clamp Controls Both Conformation and Clustering of Stromal Interaction Molecule 1 (STIM1). J. Biol. Chem. 2014, 289, 33231–33244. [Google Scholar] [CrossRef] [PubMed]
- van Dorp, S.; Qiu, R.; Choi, U.B.; Wu, M.M.; Yen, M.; Kirmiz, M.; Brunger, A.T.; Lewis, R.S. Conformational Dynamics of Auto-Inhibition in the ER Calcium Sensor STIM1. Elife 2021, 10, e66194. [Google Scholar] [CrossRef] [PubMed]
- Muik, M.; Fahrner, M.; Schindl, R.; Stathopulos, P.; Frischauf, I.; Derler, I.; Plenk, P.; Lackner, B.; Groschner, K.; Ikura, M.; et al. STIM1 Couples to ORAI1 via an Intramolecular Transition into an Extended Conformation. EMBO J. 2011, 30, 1678–1689. [Google Scholar] [CrossRef] [PubMed]
- Gudlur, A.; Zeraik, A.E.; Hirve, N.; Hogan, P.G. STIM Calcium Sensing and Conformational Change. J. Physiol. 2020, 598, 1695–1705. [Google Scholar] [CrossRef] [PubMed]
- Ma, G.; He, L.; Liu, S.; Xie, J.; Huang, Z.; Jing, J.; Lee, Y.T.; Wang, R.; Luo, H.; Han, W.; et al. Optogenetic Engineering to Probe the Molecular Choreography of STIM1-Mediated Cell Signaling. Nat. Commun. 2020, 11, 1039. [Google Scholar] [CrossRef]
- Muik, M.; Frischauf, I.; Derler, I.; Fahrner, M.; Bergsmann, J.; Eder, P.; Schindl, R.; Hesch, C.; Polzinger, B.; Fritsch, R.; et al. Dynamic Coupling of the Putative Coiled-Coil Domain of ORAI1 with STIM1 Mediates ORAI1 Channel Activation. J. Biol. Chem. 2008, 283, 8014–8022. [Google Scholar] [CrossRef]
- Frischauf, I.; Muik, M.; Derler, I.; Bergsmann, J.; Fahrner, M.; Schindl, R.; Groschner, K.; Romanin, C. Molecular Determinants of the Coupling between STIM1 and Orai Channels: Differential Activation of Orai1-3 Channels by a STIM1 Coiled-Coil Mutant. J. Biol. Chem. 2009, 284, 21696–21706. [Google Scholar] [CrossRef]
- Wang, X.; Wang, Y.; Zhou, Y.; Hendron, E.; Mancarella, S.; Andrake, M.D.; Rothberg, B.S.; Soboloff, J.; Gill, D.L. Distinct Orai-Coupling Domains in STIM1 and STIM2 Define the Orai-Activating Site. Nat. Commun. 2014, 5, 3183. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Jennette, M.R.; Ma, G.; Kazzaz, S.A.; Baraniak, J.H.; Nwokonko, R.M.; Groff, M.L.; Velasquez-Reynel, M.; Huang, Y.; Wang, Y.; et al. An Apical Phe-His Pair Defines the Orai1-Coupling Site and Its Occlusion within STIM1. Nat. Commun. 2023, 14, 6921. [Google Scholar] [CrossRef] [PubMed]
- Kawasaki, T.; Lange, I.; Feske, S. A Minimal Regulatory Domain in the C Terminus of STIM1 Binds to and Activates ORAI1 CRAC Channels. Biochem. Biophys. Res. Commun. 2009, 385, 49. [Google Scholar] [CrossRef] [PubMed]
- Luik, R.M.; Wang, B.; Prakriya, M.; Wu, M.M.; Lewis, R.S. Oligomerization of STIM1 Couples ER Calcium Depletion to CRAC Channel Activation. Nature 2008, 454, 538. [Google Scholar] [CrossRef] [PubMed]
- Yuan, J.P.; Zeng, W.; Dorwart, M.R.; Choi, Y.J.; Worley, P.F.; Muallem, S. SOAR and the Polybasic STIM1 Domains Gate and Regulate Orai Channels. Nat. Cell Biol. 2009, 11, 337–343. [Google Scholar] [CrossRef] [PubMed]
- Muik, M.; Fahrner, M.; Derler, I.; Schindl, R.; Bergsmann, J.; Frischauf, I.; Groschner, K.; Romanin, C. A Cytosolic Homomerization and a Modulatory Domain within STIM1 C Terminus Determine Coupling to ORAI1 Channels. J. Biol. Chem. 2009, 284, 8421–8426. [Google Scholar] [CrossRef] [PubMed]
- Derler, I.; Fahrner, M.; Muik, M.; Lackner, B.; Schindl, R.; Groschner, K.; Romanin, C. A Ca2(+ )Release-Activated Ca2(+) (CRAC) Modulatory Domain (CMD) within STIM1 Mediates Fast Ca2(+)-Dependent Inactivation of ORAI1 Channels. J. Biol. Chem. 2009, 284, 24933–24938. [Google Scholar] [CrossRef] [PubMed]
- Cohen, H.A.; Zomot, E.; Nataniel, T.; Militsin, R.; Palty, R. The SOAR of STIM1 Interacts with Plasma Membrane Lipids to Form ER-PM Contact Sites. Cell Rep. 2023, 42, 112238. [Google Scholar] [CrossRef]
- Ercan, E.; Momburg, F.; Engel, U.; Temmerman, K.; Nickel, W.; Seedorf, M. A Conserved, Lipid-Mediated Sorting Mechanism of Yeast Ist2 and Mammalian STIM Proteins to the Peripheral ER. Traffic 2009, 10, 1802–1818. [Google Scholar] [CrossRef]
- Liou, J.; Fivaz, M.; Inoue, T.; Meyer, T. Live-Cell Imaging Reveals Sequential Oligomerization and Local Plasma Membrane Targeting of Stromal Interaction Molecule 1 after Ca2+ Store Depletion. Proc. Natl. Acad. Sci. USA 2007, 104, 9301–9306. [Google Scholar] [CrossRef]
- Wu, M.M.; Buchanan, J.A.; Luik, R.M.; Lewis, R.S. Ca2+ Store Depletion Causes STIM1 to Accumulate in ER Regions Closely Associated with the Plasma Membrane. J. Cell Biol. 2006, 174, 803–813. [Google Scholar] [CrossRef] [PubMed]
- Hou, X.; Burstein, S.R.; Long, S.B. Structures Reveal Opening of the Store-Operated Calcium Channel Orai. Elife 2018, 7, e36758. [Google Scholar] [CrossRef] [PubMed]
- Hou, X.; Pedi, L.; Diver, M.M.; Long, S.B. Crystal Structure of the Calcium Release-Activated Calcium Channel Orai. Science 2012, 338, 1308–1313. [Google Scholar] [CrossRef]
- Hou, X.; Outhwaite, I.R.; Pedi, L.; Long, S.B. Cryo-EM Structure of the Calcium Release-Activated Calcium Channel Orai in an Open Conformation. BioRxiv 2020. [Google Scholar] [CrossRef]
- Liu, X.; Wu, G.; Yu, Y.; Chen, X.; Ji, R.; Lu, J.; Li, X.; Zhang, X.; Yang, X.; Shen, Y. Molecular Understanding of Calcium Permeation through the Open Orai Channel. PLoS Biol. 2019, 17, e3000096. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Cai, X.; Loktionova, N.A.; Wang, X.; Nwokonko, R.M.; Wang, X.; Wang, Y.; Rothberg, B.S.; Trebak, M.; Gill, D.L. The STIM1-Binding Site Nexus Remotely Controls Orai1 Channel Gating. Nat. Commun. 2016, 7, 13725. [Google Scholar] [CrossRef] [PubMed]
- Tiffner, A.; Maltan, L.; Weiß, S.; Derler, I. The Orai Pore Opening Mechanism. Int. J. Mol. Sci. 2021, 22, 533. [Google Scholar] [CrossRef]
- Tiffner, A.; Schober, R.; Höglinger, C.; Bonhenry, D.; Pandey, S.; Lunz, V.; Sallinger, M.; Frischauf, I.; Fahrner, M.; Lindinger, S.; et al. CRAC Channel Opening Is Determined by a Series of Orai1 Gating Checkpoints in the Transmembrane and Cytosolic Regions. J. Biol. Chem. 2021, 296, 100224. [Google Scholar] [CrossRef]
- Tiffner, A.; Maltan, L.; Fahrner, M.; Sallinger, M.; Weiss, S.; Grabmayr, H.; Hoglinger, C.; Derler, I. Transmembrane Domain 3 (TM3) Governs Orai1 and Orai3 Pore Opening in an Isoform-Specific Manner. Front. Cell Dev. Biol. 2021, 9, 635705. [Google Scholar] [CrossRef]
- Yeung, P.S.; Yamashita, M.; Ing, C.E.; Pomes, R.; Freymann, D.M.; Prakriya, M. Mapping the Functional Anatomy of Orai1 Transmembrane Domains for CRAC Channel Gating. Proc. Natl. Acad. Sci. USA 2018, 115, E5193–E5202. [Google Scholar] [CrossRef]
- Fahrner, M.; Pandey, S.K.; Muik, M.; Traxler, L.; Butorac, C.; Stadlbauer, M.; Zayats, V.; Krizova, A.; Plenk, P.; Frischauf, I.; et al. Communication between N Terminus and Loop2 Tunes Orai Activation. J. Biol. Chem. 2018, 293, 1271–1285. [Google Scholar] [CrossRef]
- Palty, R.; Isacoff, E.Y. Cooperative Binding of Stromal Interaction Molecule 1 (STIM1) to the N and C Termini of Calcium Release-Activated Calcium Modulator 1 (Orai1). J. Biol. Chem. 2016, 291, 334–341. [Google Scholar] [CrossRef]
- Butorac, C.; Muik, M.; Derler, I.; Stadlbauer, M.; Lunz, V.; Krizova, A.; Lindinger, S.; Schober, R.; Frischauf, I.; Bhardwaj, R.; et al. A Novel STIM1-Orai1 Gating Interface Essential for CRAC Channel Activation. Cell Calcium 2019, 79, 57–67. [Google Scholar] [CrossRef] [PubMed]
- Derler, I.; Butorac, C.; Krizova, A.; Stadlbauer, M.; Muik, M.; Fahrner, M.; Frischauf, I.; Romanin, C. Authentic CRAC Channel Activity Requires STIM1 and the Conserved Portion of the Orai N Terminus. J. Biol. Chem. 2018, 293, 1259–1270. [Google Scholar] [CrossRef] [PubMed]
- Krizova, A.; Maltan, L.; Derler, I. Critical Parameters Maintaining Authentic CRAC Channel Hallmarks. Eur. Biophys. J. 2019, 48, 425–445. [Google Scholar] [CrossRef] [PubMed]
- McNally, B.A.; Somasundaram, A.; Jairaman, A.; Yamashita, M.; Prakriya, M. The C- and N-Terminal STIM1 Binding Sites on Orai1 Are Required for Both Trapping and Gating CRAC Channels. J. Physiol. 2013, 591, 2833–2850. [Google Scholar] [CrossRef] [PubMed]
- Wagner, H.J.; Engesser, R.; Ermes, K.; Geraths, C.; Timmer, J.; Weber, W. Synthetic Biology-Inspired Design of Signal-Amplifying Materials Systems. Mater. Today 2019, 22, 25–34. [Google Scholar] [CrossRef]
- Mohammad, N.B.; Lam, C.C.K.; Truong, K. Synthetic Biology Approaches in Immunology. Biochemistry 2019, 58, 1484–1491. [Google Scholar] [CrossRef] [PubMed]
- Bibi, A.; Ahmed, A. Synthetic Biology: Approaches, Opportunities, Applications and Challenges. Abasyn J. Life Sci. 2020, 3, 25–40. [Google Scholar] [CrossRef]
- Stanton, B.Z.; Chory, E.J.; Crabtree, G.R. Chemically Induced Proximity in Biology and Medicine. Science 2018, 359, eaao5902. [Google Scholar] [CrossRef]
- Fink, T.; Lonzarić, J.; Praznik, A.; Plaper, T.; Merljak, E.; Leben, K.; Jerala, N.; Lebar, T.; Strmšek, Ž.; Lapenta, F.; et al. Design of Fast Proteolysis-Based Signaling and Logic Circuits in Mammalian Cells. Nat. Chem. Biol. 2019, 15, 115–122. [Google Scholar] [CrossRef]
- Lan, T.-H.; He, L.; Huang, Y.; Zhou, Y. Optogenetics for Transcriptional Programming and Genetic Engineering. Trends Genet. 2022, 38, 1253–1270. [Google Scholar] [CrossRef]
- Ho, S.N.; Biggar, S.R.; Spencer, D.M.; Schreiber, S.L.; Crabtree, G.R. Dimeric Ligands Define a Role for Transcriptional Activation Domains in Reinitiation. Nature 1996, 382, 822–826. [Google Scholar] [CrossRef] [PubMed]
- Voß, S.; Klewer, L.; Wu, Y.-W. Chemically Induced Dimerization: Reversible and Spatiotemporal Control of Protein Function in Cells. Curr. Opin. Chem. Biol. 2015, 28, 194–201. [Google Scholar] [CrossRef] [PubMed]
- Spencer, D.M.; Wandless, T.J.; Schreiber, S.L.; Crabtree, G.R. Controlling Signal Transduction with Synthetic Ligands. Science 1993, 262, 1019–1024. [Google Scholar] [CrossRef] [PubMed]
- Fegan, A.; White, B.; Carlson, J.C.T.; Wagner, C.R. Chemically Controlled Protein Assembly: Techniques and Applications. Chem. Rev. 2010, 110, 3315–3336. [Google Scholar] [CrossRef] [PubMed]
- Dumont, F.J. FK506, An Immunosuppressant Targeting Calcineurin Function. Curr. Med. Chem. 2000, 7, 731–748. [Google Scholar] [CrossRef]
- Clipstone, N.A.; Crabtree, G.R. Identification of Calcineurin as a Key Signalling Enzyme in T-Lymphocyte Activation. Nature 1992, 357, 695–697. [Google Scholar] [CrossRef]
- Zheng, X.-F.; Fiorentino, D.; Chen, J.; Crabtree, G.R.; Schreiber, S.L. TOR Kinase Domains Are Required for Two Distinct Functions, Only One of Which Is Inhibited by Rapamycin. Cell 1995, 82, 121–130. [Google Scholar] [CrossRef]
- Michnick, S.W.; Rosen, M.K.; Wandless, T.J.; Karplus, M.; Schreiber, S.L. Solution Structure of FKBP, a Rotamase Enzyme and Receptor for FK506 and Rapamycin. Science 1991, 252, 836–839. [Google Scholar] [CrossRef]
- Holsinger, L.J.; Spencer, D.M.; Austin, D.J.; Schreiber, S.L.; Crabtree, G.R. Signal Transduction in T Lymphocytes Using a Conditional Allele of Sos. Proc. Natl. Acad. Sci. USA 1995, 92, 9810–9814. [Google Scholar] [CrossRef]
- Ye, X.; Rivera, V.M.; Zoltick, P.; Cerasoli, F.; Schnell, M.A.; Gao, G.; Hughes, J.V.; Gilman, M.; Wilson, J.M. Regulated Delivery of Therapeutic Proteins After in Vivo Somatic Cell Gene Transfer. Science 1999, 283, 88–91. [Google Scholar] [CrossRef]
- Rivera, V.M.; Clackson, T.; Natesan, S.; Pollock, R.; Amara, J.F.; Keenan, T.; Magari, S.R.; Phillips, T.; Courage, N.L.; Cerasoli, F.; et al. A Humanized System for Pharmacologic Control of Gene Expression. Nat. Med. 1996, 2, 1028–1032. [Google Scholar] [CrossRef] [PubMed]
- Rivera, V.M.; Gao, G.; Grant, R.L.; Schnell, M.A.; Zoltick, P.W.; Rozamus, L.W.; Clackson, T.; Wilson, J.M. Long-Term Pharmacologically Regulated Expression of Erythropoietin in Primates following AAV-Mediated Gene Transfer. Blood 2005, 105, 1424–1430. [Google Scholar] [CrossRef]
- Pajvani, U.B.; Trujillo, M.E.; Combs, T.P.; Iyengar, P.; Jelicks, L.; Roth, K.A.; Kitsis, R.N.; Scherer, P.E. Fat Apoptosis through Targeted Activation of Caspase 8: A New Mouse Model of Inducible and Reversible Lipoatrophy. Nat. Med. 2005, 11, 797–803. [Google Scholar] [CrossRef]
- Baker, D.J.; Childs, B.G.; Durik, M.; Wijers, M.E.; Sieben, C.J.; Zhong, J.A.; Saltness, R.; Jeganathan, K.B.; Verzosa, G.C.; Pezeshki, A.; et al. Naturally Occurring P16Ink4a-Positive Cells Shorten Healthy Lifespan. Nature 2016, 530, 184–189. [Google Scholar] [CrossRef] [PubMed]
- Baker, D.J.; Wijshake, T.; Tchkonia, T.; LeBrasseur, N.K.; Childs, B.G.; van de Sluis, B.; Kirkland, J.L.; van Deursen, J.M. Clearance of P16Ink4a-Positive Senescent Cells Delays Ageing-Associated Disorders. Nature 2011, 479, 232–236. [Google Scholar] [CrossRef]
- Fan, L.; Freeman, K.W.; Khan, T.; Pham, E.; Spencer, D.M. Improved Artificial Death Switches Based on Caspases and FADD. Hum. Gene Ther. 1999, 10, 2273–2285. [Google Scholar] [CrossRef]
- MacCorkle, R.A.; Freeman, K.W.; Spencer, D.M. Synthetic Activation of Caspases: Artificial Death Switches. Proc. Natl. Acad. Sci. USA 1998, 95, 3655–3660. [Google Scholar] [CrossRef] [PubMed]
- Spencer, D.M.; Belshaw, P.J.; Chen, L.; Ho, S.N.; Randazzo, F.; Crabtree, G.R.; Schreiber, S.L. Functional Analysis of Fas Signaling in Vivo Using Synthetic Inducers of Dimerization. Curr. Biol. 1996, 6, 839–847. [Google Scholar] [CrossRef]
- Zhou, X.; Di Stasi, A.; Tey, S.-K.; Krance, R.A.; Martinez, C.; Leung, K.S.; Durett, A.G.; Wu, M.-F.; Liu, H.; Leen, A.M.; et al. Long-Term Outcome after Haploidentical Stem Cell Transplant and Infusion of T Cells Expressing the Inducible Caspase 9 Safety Transgene. Blood 2014, 123, 3895–3905. [Google Scholar] [CrossRef]
- Zhou, X.; Dotti, G.; Krance, R.A.; Martinez, C.A.; Naik, S.; Kamble, R.T.; Durett, A.G.; Dakhova, O.; Savoldo, B.; Di Stasi, A.; et al. Inducible Caspase-9 Suicide Gene Controls Adverse Effects from Alloreplete T Cells after Haploidentical Stem Cell Transplantation. Blood 2015, 125, 4103–4113. [Google Scholar] [CrossRef]
- Clackson, T.; Yang, W.; Rozamus, L.W.; Hatada, M.; Amara, J.F.; Rollins, C.T.; Stevenson, L.F.; Magari, S.R.; Wood, S.A.; Courage, N.L.; et al. Redesigning an FKBP–Ligand Interface to Generate Chemical Dimerizers with Novel Specificity. Proc. Natl. Acad. Sci. USA 1998, 95, 10437–10442. [Google Scholar] [CrossRef] [PubMed]
- Di Stasi, A.; Tey, S.-K.; Dotti, G.; Fujita, Y.; Kennedy-Nasser, A.; Martinez, C.; Straathof, K.; Liu, E.; Durett, A.G.; Grilley, B.; et al. Inducible Apoptosis as a Safety Switch for Adoptive Cell Therapy. N. Engl. J. Med. 2011, 365, 1673–1683. [Google Scholar] [CrossRef] [PubMed]
- Dueber, J.E.; Yeh, B.J.; Chak, K.; Lim, W.A. Reprogramming Control of an Allosteric Signaling Switch Through Modular Recombination. Science 2003, 301, 1904–1908. [Google Scholar] [CrossRef] [PubMed]
- Gordley, R.M.; Williams, R.E.; Bashor, C.J.; Toettcher, J.E.; Yan, S.; Lim, W.A. Engineering Dynamical Control of Cell Fate Switching Using Synthetic Phospho-Regulons. Proc. Natl. Acad. Sci. USA 2016, 113, 13528–13533. [Google Scholar] [CrossRef] [PubMed]
- Good, M.C.; Zalatan, J.G.; Lim, W.A. Scaffold Proteins: Hubs for Controlling the Flow of Cellular Information. Science 2011, 332, 680–686. [Google Scholar] [CrossRef] [PubMed]
- Nandagopal, N.; Elowitz, M.B. Synthetic Biology: Integrated Gene Circuits. Science 2011, 333, 1244–1248. [Google Scholar] [CrossRef] [PubMed]
- Lonzaric, J.; Fink, T.; Jerala, R. Design and Applications of Synthetic Information Processing Circuits in Mammalian Cells; Royal Society of Chemistry: Cambridge, UK, 2017; pp. 1–34. [Google Scholar]
- Kitada, T.; DiAndreth, B.; Teague, B.; Weiss, R. Programming Gene and Engineered-Cell Therapies with Synthetic Biology. Science 2018, 359, eaad1067. [Google Scholar] [CrossRef]
- Haellman, V.; Fussenegger, M. Synthetic Biology—Engineering Cell-Based Biomedical Devices. Curr. Opin. Biomed. Eng. 2017, 4, 50–56. [Google Scholar] [CrossRef]
- Jazbec, V.; Jerala, R.; Benčina, M. Proteolytically Activated CRAC Effectors through Designed Intramolecular Inhibition. ACS Synth. Biol. 2022, 11, 2756–2765. [Google Scholar] [CrossRef]
- Yi, L.; Gebhard, M.C.; Li, Q.; Taft, J.M.; Georgiou, G.; Iverson, B.L. Engineering of TEV Protease Variants by Yeast ER Sequestration Screening (YESS) of Combinatorial Libraries. Proc. Natl. Acad. Sci. USA 2013, 110, 7229–7234. [Google Scholar] [CrossRef]
- Zheng, N.; Pérez, J.d.J.; Zhang, Z.; Domínguez, E.; Garcia, J.A.; Xie, Q. Specific and Efficient Cleavage of Fusion Proteins by Recombinant Plum Pox Virus NIa Protease. Protein Expr. Purif. 2008, 57, 153–162. [Google Scholar] [CrossRef]
- Seo, J.-K.; Choi, H.-S.; Kim, K.-H. Engineering of Soybean Mosaic Virus as a Versatile Tool for Studying Protein–Protein Interactions in Soybean. Sci. Rep. 2016, 6, 22436. [Google Scholar] [CrossRef]
- Fernandez-Rodriguez, J.; Voigt, C.A. Post-Translational Control of Genetic Circuits Using Potyvirus Proteases. Nucleic Acids Res. 2016, 44, 6493–6502. [Google Scholar] [CrossRef]
- Gradišar, H.; Jerala, R.D. Novo Design of Orthogonal Peptide Pairs Forming Parallel Coiled-coil Heterodimers. J. Pept. Sci. 2011, 17, 100–106. [Google Scholar] [CrossRef]
- Woolfson, D.N. The Design of Coiled-Coil Structures and Assemblies. Adv. Protein Chem. 2005, 70, 79–112. [Google Scholar] [CrossRef]
- GRIGORYAN, G.; KEATING, A. Structural Specificity in Coiled-Coil Interactions. Curr. Opin. Struct. Biol. 2008, 18, 477–483. [Google Scholar] [CrossRef]
- Reinke, A.W.; Grant, R.A.; Keating, A.E. A Synthetic Coiled-Coil Interactome Provides Heterospecific Modules for Molecular Engineering. J. Am. Chem. Soc. 2010, 132, 6025–6031. [Google Scholar] [CrossRef]
- Lupas, A. Coiled Coils: New Structures and New Functions. Trends Biochem. Sci. 1996, 21, 375–382. [Google Scholar] [CrossRef] [PubMed]
- Chambers, P.; Pringle, C.R.; Easton, A.J. Heptad Repeat Sequences Are Located Adjacent to Hydrophobic Regions in Several Types of Virus Fusion Glycoproteins. J. General. Virol. 1990, 71, 3075–3080. [Google Scholar] [CrossRef] [PubMed]
- CRICK, F.H.C. Is α-Keratin a Coiled Coil? Nature 1952, 170, 882–883. [Google Scholar] [CrossRef] [PubMed]
- Truebestein, L.; Leonard, T.A. Coiled-coils: The Long and Short of It. BioEssays 2016, 38, 903–916. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.; Shen, Y.; Campbell, R.E. Engineering Photosensory Modules of Non-Opsin-Based Optogenetic Actuators. Int. J. Mol. Sci. 2020, 21, 6522. [Google Scholar] [CrossRef]
- Sancar, A. Structure and Function of DNA Photolyase and Cryptochrome Blue-Light Photoreceptors. Chem. Rev. 2003, 103, 2203–2238. [Google Scholar] [CrossRef]
- Cashmore, A.R.; Jarillo, J.A.; Wu, Y.-J.; Liu, D. Cryptochromes: Blue Light Receptors for Plants and Animals. Science 1999, 284, 760–765. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.; Shalitin, D. Cryptochrome Structure and Signal Transduction. Annu. Rev. Plant Biol. 2003, 54, 469–496. [Google Scholar] [CrossRef]
- Liu, H.; Liu, B.; Zhao, C.; Pepper, M.; Lin, C. The Action Mechanisms of Plant Cryptochromes. Trends Plant Sci. 2011, 16, 684–691. [Google Scholar] [CrossRef] [PubMed]
- Kennedy, M.J.; Hughes, R.M.; Peteya, L.A.; Schwartz, J.W.; Ehlers, M.D.; Tucker, C.L. Rapid Blue-Light–Mediated Induction of Protein Interactions in Living Cells. Nat. Methods 2010, 7, 973–975. [Google Scholar] [CrossRef]
- Che, D.L.; Duan, L.; Zhang, K.; Cui, B. The Dual Characteristics of Light-Induced Cryptochrome 2, Homo-Oligomerization and Heterodimerization, for Optogenetic Manipulation in Mammalian Cells. ACS Synth. Biol. 2015, 4, 1124–1135. [Google Scholar] [CrossRef]
- Zemelman, B.V.; Nesnas, N.; Lee, G.A.; Miesenböck, G. Photochemical Gating of Heterologous Ion Channels: Remote Control over Genetically Designated Populations of Neurons. Proc. Natl. Acad. Sci. USA 2003, 100, 1352–1357. [Google Scholar] [CrossRef]
- Levskaya, A.; Weiner, O.D.; Lim, W.A.; Voigt, C.A. Spatiotemporal Control of Cell Signalling Using a Light-Switchable Protein Interaction. Nature 2009, 461, 997–1001. [Google Scholar] [CrossRef]
- Maltan, L.; Najjar, H.; Tiffner, A.; Derler, I. Deciphering Molecular Mechanisms and Intervening in Physiological and Pathophysiological Processes of Ca2+ Signaling Mechanisms Using Optogenetic Tools. Cells 2021, 10, 3340. [Google Scholar] [CrossRef]
- Kyung, T.; Lee, S.; Kim, J.E.; Cho, T.; Park, H.; Jeong, Y.-M.; Kim, D.; Shin, A.; Kim, S.; Baek, J.; et al. Optogenetic Control of Endogenous Ca2+ Channels in Vivo. Nat. Biotechnol. 2015, 33, 1092–1096. [Google Scholar] [CrossRef]
- Bohineust, A.; Garcia, Z.; Corre, B.; Lemaître, F.; Bousso, P. Optogenetic Manipulation of Calcium Signals in Single T Cells in Vivo. Nat. Commun. 2020, 11, 1143. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Kyung, T.; Chung, J.-H.; Kim, N.; Keum, S.; Lee, J.; Park, H.; Kim, H.M.; Lee, S.; Shin, H.-S.; et al. Non-Invasive Optical Control of Endogenous Ca2+ Channels in Awake Mice. Nat. Commun. 2020, 11, 210. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, N.T.; Ma, G.; Lin, E.; D’Souza, B.; Jing, J.; He, L.; Huang, Y.; Zhou, Y. CRAC Channel-Based Optogenetics. Cell Calcium 2018, 75, 79–88. [Google Scholar] [CrossRef] [PubMed]
- Krauss, U.; Minh, B.Q.; Losi, A.; Gärtner, W.; Eggert, T.; von Haeseler, A.; Jaeger, K.-E. Distribution and Phylogeny of Light-Oxygen-Voltage-Blue-Light-Signaling Proteins in the Three Kingdoms of Life. J. Bacteriol. 2009, 191, 7234–7242. [Google Scholar] [CrossRef]
- Harper, S.M.; Neil, L.C.; Gardner, K.H. Structural Basis of a Phototropin Light Switch. Science 2003, 301, 1541–1544. [Google Scholar] [CrossRef] [PubMed]
- Renicke, C.; Schuster, D.; Usherenko, S.; Essen, L.-O.; Taxis, C. A LOV2 Domain-Based Optogenetic Tool to Control Protein Degradation and Cellular Function. Chem. Biol. 2013, 20, 619–626. [Google Scholar] [CrossRef]
- Okajima, K.; Aihara, Y.; Takayama, Y.; Nakajima, M.; Kashojiya, S.; Hikima, T.; Oroguchi, T.; Kobayashi, A.; Sekiguchi, Y.; Yamamoto, M.; et al. Light-Induced Conformational Changes of LOV1 (Light Oxygen Voltage-Sensing Domain 1) and LOV2 Relative to the Kinase Domain and Regulation of Kinase Activity in Chlamydomonas Phototropin. J. Biol. Chem. 2014, 289, 413–422. [Google Scholar] [CrossRef]
- Halavaty, A.S.; Moffat, K. N- and C-Terminal Flanking Regions Modulate Light-Induced Signal Transduction in the LOV2 Domain of the Blue Light Sensor Phototropin 1 from Avena Sativa. Biochemistry 2007, 46, 14001–14009. [Google Scholar] [CrossRef] [PubMed]
- Motta-Mena, L.B.; Reade, A.; Mallory, M.J.; Glantz, S.; Weiner, O.D.; Lynch, K.W.; Gardner, K.H. An Optogenetic Gene Expression System with Rapid Activation and Deactivation Kinetics. Nat. Chem. Biol. 2014, 10, 196–202. [Google Scholar] [CrossRef] [PubMed]
- Pudasaini, A.; El-Arab, K.K.; Zoltowski, B.D. LOV-Based Optogenetic Devices: Light-Driven Modules to Impart Photoregulated Control of Cellular Signaling. Front. Mol. Biosci. 2015, 2, 146196. [Google Scholar] [CrossRef] [PubMed]
- Ma, G.; Wen, S.; Huang, Y.; Zhou, Y. The STIM-Orai Pathway: Light-Operated Ca2+ Entry through Engineered CRAC Channels. Adv. Exp. Med. Biol. 2017, 993, 117–138. [Google Scholar] [CrossRef] [PubMed]
- Wonnacott, S.; Bermudez, I.; Millar, N.S.; Tzartos, S.J. Nicotinic Acetylcholine Receptors. Br. J. Pharmacol. 2018, 175, 1785–1788. [Google Scholar] [CrossRef] [PubMed]
- Guntas, G.; Hallett, R.A.; Zimmerman, S.P.; Williams, T.; Yumerefendi, H.; Bear, J.E.; Kuhlman, B. Engineering an Improved Light-Induced Dimer (ILID) for Controlling the Localization and Activity of Signaling Proteins. Proc. Natl. Acad. Sci. USA 2015, 112, 112–117. [Google Scholar] [CrossRef]
- He, L.; Zhang, Y.; Ma, G.; Tan, P.; Li, Z.; Zang, S.; Wu, X.; Jing, J.; Fang, S.; Zhou, L.; et al. Near-Infrared Photoactivatable Control of Ca2+ Signaling and Optogenetic Immunomodulation. Elife 2015, 4, e10024. [Google Scholar] [CrossRef]
- He, L.; Jing, J.; Zhu, L.; Tan, P.; Ma, G.; Zhang, Q.; Nguyen, N.T.; Wang, J.; Zhou, Y.; Huang, Y. Optical Control of Membrane Tethering and Interorganellar Communication at Nanoscales. Chem. Sci. 2017, 8, 5275–5281. [Google Scholar] [CrossRef] [PubMed]
- Ishii, T.; Sato, K.; Kakumoto, T.; Miura, S.; Touhara, K.; Takeuchi, S.; Nakata, T. Light Generation of Intracellular Ca(2+) Signals by a Genetically Encoded Protein BACCS. Nat. Commun. 2015, 6, 8021. [Google Scholar] [CrossRef]
- He, L.; Wang, L.; Zeng, H.; Tan, P.; Ma, G.; Zheng, S.; Li, Y.; Sun, L.; Dou, F.; Siwko, S.; et al. Engineering of a Bona Fide Light-Operated Calcium Channel. Nat. Commun. 2021, 12, 164. [Google Scholar] [CrossRef]
- Srikanth, S.; Jung, H.J.; Ribalet, B.; Gwack, Y. The Intracellular Loop of Orai1 Plays a Central Role in Fast Inactivation of Ca2+ Release-Activated Ca2+ Channels. J. Biol. Chem. 2010, 285, 5066–5075. [Google Scholar] [CrossRef]
- Hsu, P.D.; Lander, E.S.; Zhang, F. Development and Applications of CRISPR-Cas9 for Genome Engineering. Cell 2014, 157, 1262–1278. [Google Scholar] [CrossRef] [PubMed]
- Ran, F.A.; Hsu, P.D.; Wright, J.; Agarwala, V.; Scott, D.A.; Zhang, F. Genome Engineering Using the CRISPR-Cas9 System. Nat. Protoc. 2013, 8, 2281–2308. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, L.A.; Larson, M.H.; Morsut, L.; Liu, Z.; Brar, G.A.; Torres, S.E.; Stern-Ginossar, N.; Brandman, O.; Whitehead, E.H.; Doudna, J.A.; et al. CRISPR-Mediated Modular RNA-Guided Regulation of Transcription in Eukaryotes. Cell 2013, 154, 442. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, N.T.; He, L.; Martinez-Moczygemba, M.; Huang, Y.; Zhou, Y. Rewiring Calcium Signaling for Precise Transcriptional Reprogramming. ACS Synth. Biol. 2018, 7, 814–821. [Google Scholar] [CrossRef]
- Chen, Y.; Lu, L.; Ye-Lehmann, S. Focus: Genome Editing: Genetic Code Expansion and Optoproteomics. Yale J. Biol. Med. 2017, 90, 599. [Google Scholar]
- Lang, K.; Chin, J.W. Cellular Incorporation of Unnatural Amino Acids and Bioorthogonal Labeling of Proteins. Chem. Rev. 2014, 114, 4764–4806. [Google Scholar] [CrossRef]
- Liu, C.C.; Schultz, P.G. Adding New Chemistries to the Genetic Code. Annu. Rev. Biochem. 2010, 79, 413–444. [Google Scholar] [CrossRef]
- Klippenstein, V.; Mony, L.; Paoletti, P. Probing Ion Channel Structure and Function Using Light-Sensitive Amino Acids. Trends Biochem. Sci. 2018, 43, 436–451. [Google Scholar] [CrossRef]
- Coin, I. Application of Non-Canonical Crosslinking Amino Acids to Study Protein–Protein Interactions in Live Cells. Curr. Opin. Chem. Biol. 2018, 46, 156–163. [Google Scholar] [CrossRef] [PubMed]
- Lang, K.; Davis, L.; Chin, J.W. Genetic Encoding of Unnatural Amino Acids for Labeling Proteins. Methods Mol. Biol. 2015, 1266, 217–228. [Google Scholar] [CrossRef] [PubMed]
- Lancia, J.K.; Nwokoye, A.; Dugan, A.; Joiner, C.; Pricer, R.; Mapp, A.K. Sequence Context and Crosslinking Mechanism Affect the Efficiency of in Vivo Capture of a Protein-Protein Interaction. Biopolymers 2014, 101, 391–397. [Google Scholar] [CrossRef]
- Nodling, A.R.; Spear, L.A.; Williams, T.L.; Luk, L.Y.P.; Tsai, Y.H. Using Genetically Incorporated Unnatural Amino Acids to Control Protein Functions in Mammalian Cells. Essays Biochem. 2019, 63, 237–266. [Google Scholar] [CrossRef]
- Edwards, W.F.; Young, D.D.; Deiters, A. Light-Activated Cre Recombinase as a Tool for the Spatial and Temporal Control of Gene Function in Mammalian Cells. ACS Chem. Biol. 2009, 4, 441–445. [Google Scholar] [CrossRef]
- Kang, J.Y.; Kawaguchi, D.; Coin, I.; Xiang, Z.; O’Leary, D.D.M.; Slesinger, P.A.; Wang, L. In Vivo Expression of a Light-Activatable Potassium Channel Using Unnatural Amino Acids. Neuron 2013, 80, 358–370. [Google Scholar] [CrossRef]
- Ren, W.; Ji, A.; Ai, H.W. Light Activation of Protein Splicing with a Photocaged Fast Intein. J. Am. Chem. Soc. 2015, 137, 2155–2158. [Google Scholar] [CrossRef]
- Nguyen, D.P.; Mahesh, M.; Elsässer, S.J.; Hancock, S.M.; Uttamapinant, C.; Chin, J.W. Genetic Encoding of Photocaged Cysteine Allows Photoactivation of TEV Protease in Live Mammalian Cells. J. Am. Chem. Soc. 2014, 136, 2240–2243. [Google Scholar] [CrossRef] [PubMed]
- Nikić-Spiegel, I. Expanding the Genetic Code for Neuronal Studies. ChemBioChem 2020, 21, 3169–3179. [Google Scholar] [CrossRef]
- Kaiser, A.; Coin, I. Capturing Peptide–GPCR Interactions and Their Dynamics. Molecules 2020, 25, 4724. [Google Scholar] [CrossRef] [PubMed]
- Maltan, L.; Weiß, S.; Najjar, H.; Leopold, M.; Lindinger, S.; Höglinger, C.; Höbarth, L.; Sallinger, M.; Grabmayr, H.; Berlansky, S.; et al. Photocrosslinking-Induced CRAC Channel-like Orai1 Activation Independent of STIM1. Nat. Commun. 2023, 14, 1286. [Google Scholar] [CrossRef] [PubMed]
- Hoppmann, C.; Lacey, V.K.; Louie, G.V.; Wei, J.; Noel, J.P.; Wang, L. Genetically Encoding Photoswitchable Click Amino Acids in Escherichia Coli and Mammalian Cells. Angew. Chem. Int. Ed. 2014, 53, 3932–3936. [Google Scholar] [CrossRef]
- Hoppmann, C.; Schmieder, P.; Heinrich, N.; Beyermann, M. Photoswitchable Click Amino Acids: Light Control of Conformation and Bioactivity. Chembiochem 2011, 12, 2555–2559. [Google Scholar] [CrossRef] [PubMed]
- Hoppmann, C.; Kühne, R.; Beyermann, M. Intramolecular Bridges Formed by Photoswitchable Click Amino Acids. Beilstein J. Org. Chem. 2012, 8, 884–889. [Google Scholar] [CrossRef]
- Hoppmann, C.; Maslennikov, I.; Choe, S.; Wang, L. In Situ Formation of an Azo Bridge on Proteins Controllable by Visible Light. J. Am. Chem. Soc. 2015, 137, 11218–11221. [Google Scholar] [CrossRef]
- Klippenstein, V.; Hoppmann, C.; Ye, S.; Wang, L.; Paoletti, P. Optocontrol of Glutamate Receptor Activity by Single Side-Chain Photoisomerization. Elife 2017, 6, e25808. [Google Scholar] [CrossRef]
- Puljung, M.C. ANAP: A Versatile, Fluorescent Probe of Ion Channel Gating and Regulation. Methods Enzymol. 2021, 654, 49–84. [Google Scholar] [CrossRef] [PubMed]
- Brauchi, S.E.; Steinberg, X.P. Studying Ion Channel Conformation Dynamics by Encoding Coumarin as Unnatural Amino Acid. Methods Enzymol. 2021, 653, 239–266. [Google Scholar] [CrossRef]
- Coin, I.; Katritch, V.; Sun, T.; Xiang, Z.; Siu, F.Y.; Beyermann, M.; Stevens, R.C.; Wang, L. Genetically Encoded Chemical Probes in Cells Reveal the Binding Path of Urocortin-I to CRF Class B GPCR. Cell 2013, 155, 1258. [Google Scholar] [CrossRef] [PubMed]
- Coin, I.; Perrin, M.H.; Vale, W.W.; Wang, L. Photo-Cross-Linkers Incorporated into G-Protein-Coupled Receptors in Mammalian Cells: A Ligand Comparison. Angew. Chem. Int. Ed. Engl. 2011, 50, 8077–8081. [Google Scholar] [CrossRef]
- Grunbeck, A.; Sakmar, T.P. Probing G Protein-Coupled Receptor-Ligand Interactions with Targeted Photoactivatable Cross-Linkers. Biochemistry 2013, 52, 8625–8632. [Google Scholar] [CrossRef]
- Murray, C.I.; Westhoff, M.; Eldstrom, J.; Thompson, E.; Emes, R.; Fedida, D. Unnatural Amino Acid Photo-Crosslinking of the IKs Channel Complex Demonstrates a KCNE1:KCNQ1 Stoichiometry of up to 4:4. Elife 2016, 5, e11815. [Google Scholar] [CrossRef]
- Westhoff, M.; Murray, C.I.; Eldstrom, J.; Fedida, D. Photo-Cross-Linking of IKs Demonstrates State-Dependent Interactions between KCNE1 and KCNQ1. Biophys. J. 2017, 113, 415–425. [Google Scholar] [CrossRef]
- Zhu, S.; Riou, M.; Yao, C.A.; Carvalho, S.; Rodriguez, P.C.; Bensaude, O.; Paoletti, P.; Ye, S. Genetically Encoding a Light Switch in an Ionotropic Glutamate Receptor Reveals Subunit-Specific Interfaces. Proc. Natl. Acad. Sci. USA 2014, 111, 6081–6086. [Google Scholar] [CrossRef]
- Serfling, R.; Seidel, L.; Bock, A.; Lohse, M.J.; Annibale, P.; Coin, I. Quantitative Single-Residue Bioorthogonal Labeling of G Protein-Coupled Receptors in Live Cells. ACS Chem. Biol. 2019, 14, 1141–1149. [Google Scholar] [CrossRef]
- Steinberg, X.; Kasimova, M.A.; Cabezas-Bratesco, D.; Galpin, J.D.; Ladron-de-Guevara, E.; Villa, F.; Carnevale, V.; Islas, L.; Ahern, C.A.; Brauchi, S.E. Conformational Dynamics in TRPV1 Channels Reported by an Encoded Coumarin Amino Acid. Elife 2017, 6, e28626. [Google Scholar] [CrossRef]
- Brauchi, S.; Orio, P. Voltage Sensing in Thermo-TRP Channels. Adv. Exp. Med. Biol. 2011, 704, 517–530. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Han, Y.; Chen, X.; Aierken, A.; Wen, H.; Zheng, W.; Wang, H.; Lu, X.; Zhao, Z.; Ma, C.; et al. Molecular Mechanisms Underlying Menthol Binding and Activation of TRPM8 Ion Channel. Nat. Commun. 2020, 11, 3790. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Lu, X.; Wang, Y.; Xu, L.; Chen, X.; Yang, F.; Lai, R. A Paradigm of Thermal Adaptation in Penguins and Elephants by Tuning Cold Activation in TRPM8. Proc. Natl. Acad. Sci. USA 2020, 117, 8633–8638. [Google Scholar] [CrossRef] [PubMed]
- Zagotta, W.N.; Gordon, M.T.; Senning, E.N.; Munari, M.A.; Gordon, S.E. Measuring Distances between TRPV1 and the Plasma Membrane Using a Noncanonical Amino Acid and Transition Metal Ion FRET. J. General. Physiol. 2016, 147, 201–216. [Google Scholar] [CrossRef] [PubMed]
- Sun, M.-Y.; Zhang, X.; Yu, P.-C.; Liu, D.; Yang, Y.; Cui, W.-W.; Yang, X.-N.; Lei, Y.-T.; Li, X.-H.; Wang, W.-H.; et al. Vanilloid Agonist-Mediated Activation of TRPV1 Channels Requires Coordinated Movement of the S1–S4 Bundle Rather than a Quiescent State. Sci. Bull. 2022, 67, 1062–1076. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Hu, M.; Ai, X.; Zhang, Z.; Xing, B. Near-Infrared Manipulation of Membrane Ion Channels via Upconversion Optogenetics. Adv. Biosyst. 2019, 3, 1800233. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.; Yao, Y.; Zhang, W.; Fang, Q.; Zhang, L.; Zhang, Y.; Xu, Y. Applications of Upconversion Nanoparticles in Cellular Optogenetics. Acta Biomater. 2021, 135, 1–12. [Google Scholar] [CrossRef]
- Nguyen, N.T.; Ma, G.; Zhou, Y.; Jing, J. Optogenetic Approaches to Control Ca2+-Modulated Physiological Processes. Curr. Opin. Physiol. 2020, 17, 187–196. [Google Scholar] [CrossRef]
- Sun, Y.; Feng, W.; Yang, P.; Huang, C.; Li, F. The Biosafety of Lanthanide Upconversion Nanomaterials. Chem. Soc. Rev. 2015, 44, 1509–1525. [Google Scholar] [CrossRef]
- Gnach, A.; Lipinski, T.; Bednarkiewicz, A.; Rybka, J.; Capobianco, J.A. Upconverting Nanoparticles: Assessing the Toxicity. Chem. Soc. Rev. 2015, 44, 1561–1584. [Google Scholar] [CrossRef]
- Wu, Y.I.; Frey, D.; Lungu, O.I.; Jaehrig, A.; Schlichting, I.; Kuhlman, B.; Hahn, K.M. A Genetically Encoded Photoactivatable Rac Controls the Motility of Living Cells. Nature 2009, 461, 104–108. [Google Scholar] [CrossRef]
- Ostrowski, A.D.; Chan, E.M.; Gargas, D.J.; Katz, E.M.; Han, G.; Schuck, P.J.; Milliron, D.J.; Cohen, B.E. Controlled Synthesis and Single-Particle Imaging of Bright, Sub-10 Nm Lanthanide-Doped Upconverting Nanocrystals. ACS Nano 2012, 6, 2686–2692. [Google Scholar] [CrossRef]
- Hososhima, S.; Yuasa, H.; Ishizuka, T.; Hoque, M.R.; Yamashita, T.; Yamanaka, A.; Sugano, E.; Tomita, H.; Yawo, H. Near-Infrared (NIR) up-Conversion Optogenetics. Sci. Rep. 2015, 5, 16533. [Google Scholar] [CrossRef]
- Shah, S.; Liu, J.-J.; Pasquale, N.; Lai, J.; McGowan, H.; Pang, Z.P.; Lee, K.-B. Hybrid Upconversion Nanomaterials for Optogenetic Neuronal Control. Nanoscale 2015, 7, 16571–16577. [Google Scholar] [CrossRef] [PubMed]
- Ding, H.; Lu, L.; Shi, Z.; Wang, D.; Li, L.; Li, X.; Ren, Y.; Liu, C.; Cheng, D.; Kim, H.; et al. Microscale Optoelectronic Infrared-to-Visible Upconversion Devices and Their Use as Injectable Light Sources. Proc. Natl. Acad. Sci. USA 2018, 115, 6632–6637. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Jayakumar, M.K.G.; Zheng, X.; Shikha, S.; Zhang, Y.; Bansal, A.; Poon, D.J.J.; Chu, P.L.; Yeo, E.L.L.; Chua, M.L.K.; et al. Upconversion Superballs for Programmable Photoactivation of Therapeutics. Nat. Commun. 2019, 10, 4586. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Weitemier, A.Z.; Zeng, X.; He, L.; Wang, X.; Tao, Y.; Huang, A.J.Y.; Hashimotodani, Y.; Kano, M.; Iwasaki, H.; et al. Near-Infrared Deep Brain Stimulation via Upconversion Nanoparticle–Mediated Optogenetics. Science 2018, 359, 679–684. [Google Scholar] [CrossRef]
- Lin, X.; Wang, Y.; Chen, X.; Yang, R.; Wang, Z.; Feng, J.; Wang, H.; Lai, K.W.C.; He, J.; Wang, F.; et al. Multiplexed Optogenetic Stimulation of Neurons with Spectrum-Selective Upconversion Nanoparticles. Adv. Healthc. Mater. 2017, 6, 1700446. [Google Scholar] [CrossRef] [PubMed]
- Ai, X.; Lyu, L.; Zhang, Y.; Tang, Y.; Mu, J.; Liu, F.; Zhou, Y.; Zuo, Z.; Liu, G.; Xing, B. Remote Regulation of Membrane Channel Activity by Site-Specific Localization of Lanthanide-Doped Upconversion Nanocrystals. Angew. Chem. Int. Ed. 2017, 56, 3031–3035. [Google Scholar] [CrossRef]
- Sood, R.; English, M.A.; Jones, M.; Mullikin, J.; Wang, D.-M.; Anderson, M.; Wu, D.; Chandrasekharappa, S.C.; Yu, J.; Zhang, J.; et al. Methods for Reverse Genetic Screening in Zebrafish by Resequencing and TILLING. Methods 2006, 39, 220–227. [Google Scholar] [CrossRef]
- Lepage, S.E.; Bruce, A.E.E. Zebrafish Epiboly: Mechanics and Mechanisms. Int. J. Dev. Biol. 2010, 54, 1213–1228. [Google Scholar] [CrossRef]
- Ma, G.; Liu, J.; Ke, Y.; Liu, X.; Li, M.; Wang, F.; Han, G.; Huang, Y.; Wang, Y.; Zhou, Y. Optogenetic Control of Voltage-Gated Calcium Channels. Angew. Chem. Int. Ed. Engl. 2018, 57, 7019. [Google Scholar] [CrossRef]
- Broichhagen, J.; Frank, J.A.; Trauner, D. A Roadmap to Success in Photopharmacology. Acc. Chem. Res. 2015, 48, 1947–1960. [Google Scholar] [CrossRef] [PubMed]
- Lerch, M.M.; Hansen, M.J.; van Dam, G.M.; Szymanski, W.; Feringa, B.L. Emerging Targets in Photopharmacology. Angew. Chem. Int. Ed. Engl. 2016, 55, 10978–10999. [Google Scholar] [CrossRef] [PubMed]
- Szymański, W.; Beierle, J.M.; Kistemaker, H.A.V.; Velema, W.A.; Feringa, B.L. Reversible Photocontrol of Biological Systems by the Incorporation of Molecular Photoswitches. Chem. Rev. 2013, 113, 6114–6178. [Google Scholar] [CrossRef]
- Velema, W.A.; Szymanski, W.; Feringa, B.L. Photopharmacology: Beyond Proof of Principle. J. Am. Chem. Soc. 2014, 136, 2178–2191. [Google Scholar] [CrossRef]
- Yang, X.; Ma, G.; Zheng, S.; Qin, X.; Li, X.; Du, L.; Wang, Y.; Zhou, Y.; Li, M. Optical Control of CRAC Channels Using Photoswitchable Azopyrazoles. J. Am. Chem. Soc. 2020, 142, 9460–9470. [Google Scholar] [CrossRef] [PubMed]
- Broichhagen, J.; Schönberger, M.; Cork, S.C.; Frank, J.A.; Marchetti, P.; Bugliani, M.; Shapiro, A.M.J.; Trapp, S.; Rutter, G.A.; Hodson, D.J.; et al. Optical Control of Insulin Release Using a Photoswitchable Sulfonylurea. Nat. Commun. 2014, 5, 5116. [Google Scholar] [CrossRef] [PubMed]
- Schönberger, M.; Althaus, M.; Fronius, M.; Clauss, W.; Trauner, D. Controlling Epithelial Sodium Channels with Light Using Photoswitchable Amilorides. Nat. Chem. 2014, 6, 712–719. [Google Scholar] [CrossRef] [PubMed]
- Hauwert, N.J.; Mocking, T.A.M.; Da Costa Pereira, D.; Kooistra, A.J.; Wijnen, L.M.; Vreeker, G.C.M.; Verweij, E.W.E.; De Boer, A.H.; Smit, M.J.; De Graaf, C.; et al. Synthesis and Characterization of a Bidirectional Photoswitchable Antagonist Toolbox for Real-Time GPCR Photopharmacology. J. Am. Chem. Soc. 2018, 140, 4232–4243. [Google Scholar] [CrossRef] [PubMed]
- Hauwert, N.J.; Mocking, T.A.M.; Da Costa Pereira, D.; Lion, K.; Huppelschoten, Y.; Vischer, H.F.; De Esch, I.J.P.; Wijtmans, M.; Leurs, R. A Photoswitchable Agonist for the Histamine H3 Receptor, a Prototypic Family A G-Protein-Coupled Receptor. Angew. Chem. Int. Ed. 2019, 58, 4531–4535. [Google Scholar] [CrossRef]
- Donthamsetti, P.; Konrad, D.B.; Hetzler, B.; Fu, Z.; Trauner, D.; Isacoff, E.Y. Selective Photoswitchable Allosteric Agonist of a G Protein-Coupled Receptor. J. Am. Chem. Soc. 2021, 143, 8951–8956. [Google Scholar] [CrossRef]
- Ashmole, I.; Duffy, S.M.; Leyland, M.L.; Morrison, V.S.; Begg, M.; Bradding, P. CRACM/Orai Ion Channel Expression and Function in Human Lung Mast Cells. J. Allergy Clin. Immunol. 2012, 129, 1628–1635.e2. [Google Scholar] [CrossRef]
- Derler, I.; Schindl, R.; Fritsch, R.; Heftberger, P.; Riedl, M.C.; Begg, M.; House, D.; Romanin, C. The Action of Selective CRAC Channel Blockers Is Affected by the Orai Pore Geometry. Cell Calcium 2013, 53, 139–151. [Google Scholar] [CrossRef] [PubMed]
- van Kruchten, R.; Braun, A.; Feijge, M.A.H.; Kuijpers, M.J.E.; Rivera-Galdos, R.; Kraft, P.; Stoll, G.; Kleinschnitz, C.; Bevers, E.M.; Nieswandt, B.; et al. Antithrombotic Potential of Blockers of Store-Operated Calcium Channels in Platelets. Arterioscler. Thromb. Vasc. Biol. 2012, 32, 1717–1723. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Xin, P.; Yoast, R.E.; Emrich, S.M.; Johnson, M.T.; Pathak, T.; Benson, J.C.; Azimi, I.; Gill, D.L.; Monteith, G.R.; et al. Distinct Pharmacological Profiles of ORAI1, ORAI2, and ORAI3 Channels. Cell Calcium 2020, 91, 102281. [Google Scholar] [CrossRef]
- Waldherr, L.; Tiffner, A.; Mishra, D.; Sallinger, M.; Schober, R.; Frischauf, I.; Schmidt, T.; Handl, V.; Sagmeister, P.; Köckinger, M.; et al. Blockage of Store-Operated Ca2+ Influx by Synta66 Is Mediated by Direct Inhibition of the Ca2+ Selective Orai1 Pore. Cancers 2020, 12, 2876. [Google Scholar] [CrossRef]
- Udasin, R.; Sil, A.; Zomot, E.; Achildiev Cohen, H.; Haj, J.; Engelmayer, N.; Lev, S.; Binshtok, A.M.; Shaked, Y.; Kienzler, M.A.; et al. Photopharmacological Modulation of Native CRAC Channels Using Azoboronate Photoswitches. Proc. Natl. Acad. Sci. USA 2022, 119, e2118160119. [Google Scholar] [CrossRef] [PubMed]
- Han, D.; Xu, Z.; Zhuang, Y.; Ye, Z.; Qian, Q. Current Progress in CAR-T Cell Therapy for Hematological Malignancies. J. Cancer 2021, 12, 326–334. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Liu, J.; Zhong, J.F.; Zhang, X. Engineering CAR-T Cells. Biomark. Res. 2017, 5, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Lindner, S.E.; Johnson, S.M.; Brown, C.E.; Wang, L.D. Chimeric Antigen Receptor Signaling: Functional Consequences and Design Implications. Sci. Adv. 2020, 6, eaaz3223. [Google Scholar] [CrossRef]
- Abrantes, R.; Duarte, H.O.; Gomes, C.; Wälchli, S.; Reis, C.A. CAR-Ts: New Perspectives in Cancer Therapy. FEBS Lett. 2022, 596, 403–416. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, N.T.; Huang, K.; Zeng, H.; Jing, J.; Wang, R.; Fang, S.; Chen, J.; Liu, X.; Huang, Z.; You, M.J.; et al. Nano-Optogenetic Engineering of CAR T Cells for Precision Immunotherapy with Enhanced Safety. Nat. Nanotechnol. 2021, 16, 1424–1434. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Yu, Z.; Cai, Y.; Du, R.; Cai, L. Engineering of an Enhanced Synthetic Notch Receptor by Reducing Ligand-Independent Activation. Commun. Biol. 2020, 3, 116. [Google Scholar] [CrossRef] [PubMed]
- Ruffo, E.; Butchy, A.A.; Tivon, Y.; So, V.; Kvorjak, M.; Parikh, A.; Adams, E.L.; Miskov-Zivanov, N.; Finn, O.J.; Deiters, A.; et al. Post-Translational Covalent Assembly of CAR and SynNotch Receptors for Programmable Antigen Targeting. Nat. Commun. 2023, 14, 2463. [Google Scholar] [CrossRef]
- Yamamura, H.; Yamamura, A.; Ko, E.A.; Pohl, N.M.; Smith, K.A.; Zeifman, A.; Powell, F.L.; Thistlethwaite, P.A.; Yuan, J.X.-J. Activation of Notch Signaling by Short-Term Treatment with Jagged-1 Enhances Store-Operated Ca 2+ Entry in Human Pulmonary Arterial Smooth Muscle Cells. Am. J. Physiol.-Cell Physiol. 2014, 306, C871–C878. [Google Scholar] [CrossRef]
- Song, S.; Babicheva, A.; Zhao, T.; Ayon, R.J.; Rodriguez, M.; Rahimi, S.; Balistrieri, F.; Harrington, A.; Shyy, J.Y.-J.; Thistlethwaite, P.A.; et al. Notch Enhances Ca2+ Entry by Activating Calcium-Sensing Receptors and Inhibiting Voltage-Gated K + Channels. Am. J. Physiol.-Cell Physiol. 2020, 318, C954–C968. [Google Scholar] [CrossRef]
- Zhang, S.; Gumpper, R.H.; Huang, X.-P.; Liu, Y.; Krumm, B.E.; Cao, C.; Fay, J.F.; Roth, B.L. Molecular Basis for Selective Activation of DREADD-Based Chemogenetics. Nature 2022, 612, 354–362. [Google Scholar] [CrossRef]
- Chaki, S.; Alkanfari, I.; Roy, S.; Amponnawarat, A.; Hui, Y.; Oskeritzian, C.A.; Ali, H. Inhibition of Orai Channel Function Regulates Mas-Related G Protein-Coupled Receptor-Mediated Responses in Mast Cells. Front. Immunol. 2022, 12, 803335. [Google Scholar] [CrossRef]
- Hutchings, C.J. Mini-Review: Antibody Therapeutics Targeting G Protein-Coupled Receptors and Ion Channels. Antib. Ther. 2020, 3, 257–264. [Google Scholar] [CrossRef]
- Muyldermans, S. Nanobodies: Natural Single-Domain Antibodies. Annu. Rev. Biochem. 2013, 82, 775–797. [Google Scholar] [CrossRef]
- Lucchi, R.; Bentanachs, J.; Oller-Salvia, B. The Masking Game: Design of Activatable Antibodies and Mimetics for Selective Therapeutics and Cell Control. ACS Cent. Sci. 2021, 7, 724–738. [Google Scholar] [CrossRef]
- Gil, A.A.; Carrasco-López, C.; Zhu, L.; Zhao, E.M.; Ravindran, P.T.; Wilson, M.Z.; Goglia, A.G.; Avalos, J.L.; Toettcher, J.E. Optogenetic Control of Protein Binding Using Light-Switchable Nanobodies. Nat. Commun. 2020, 11, 4044. [Google Scholar] [CrossRef] [PubMed]
- Carrasco-López, C.; Zhao, E.M.; Gil, A.A.; Alam, N.; Toettcher, J.E.; Avalos, J.L. Development of Light-Responsive Protein Binding in the Monobody Non-Immunoglobulin Scaffold. Nat. Commun. 2020, 11, 4045. [Google Scholar] [CrossRef] [PubMed]
- Yu, D.; Lee, H.; Hong, J.; Jung, H.; Jo, Y.; Oh, B.-H.; Park, B.O.; Heo, W. Do Optogenetic Activation of Intracellular Antibodies for Direct Modulation of Endogenous Proteins. Nat. Methods 2019, 16, 1095–1100. [Google Scholar] [CrossRef]
- Lee, Y.-T.; He, L.; Zhou, Y. Expanding the Chemogenetic Toolbox by Circular Permutation. J. Mol. Biol. 2020, 432, 3127–3136. [Google Scholar] [CrossRef] [PubMed]
- Jedlitzke, B.; Mootz, H.D. A Light-Activatable Photocaged Variant of the Ultra-High Affinity ALFA-Tag Nanobody. ChemBioChem 2022, 23, e202200079. [Google Scholar] [CrossRef] [PubMed]
- Hoppmann, C.; Wang, L. Genetically Encoding Photoswitchable Click Amino Acids for General Optical Control of Conformation and Function of Proteins. Methods Enzymol. 2019, 624, 249–264. [Google Scholar] [CrossRef] [PubMed]
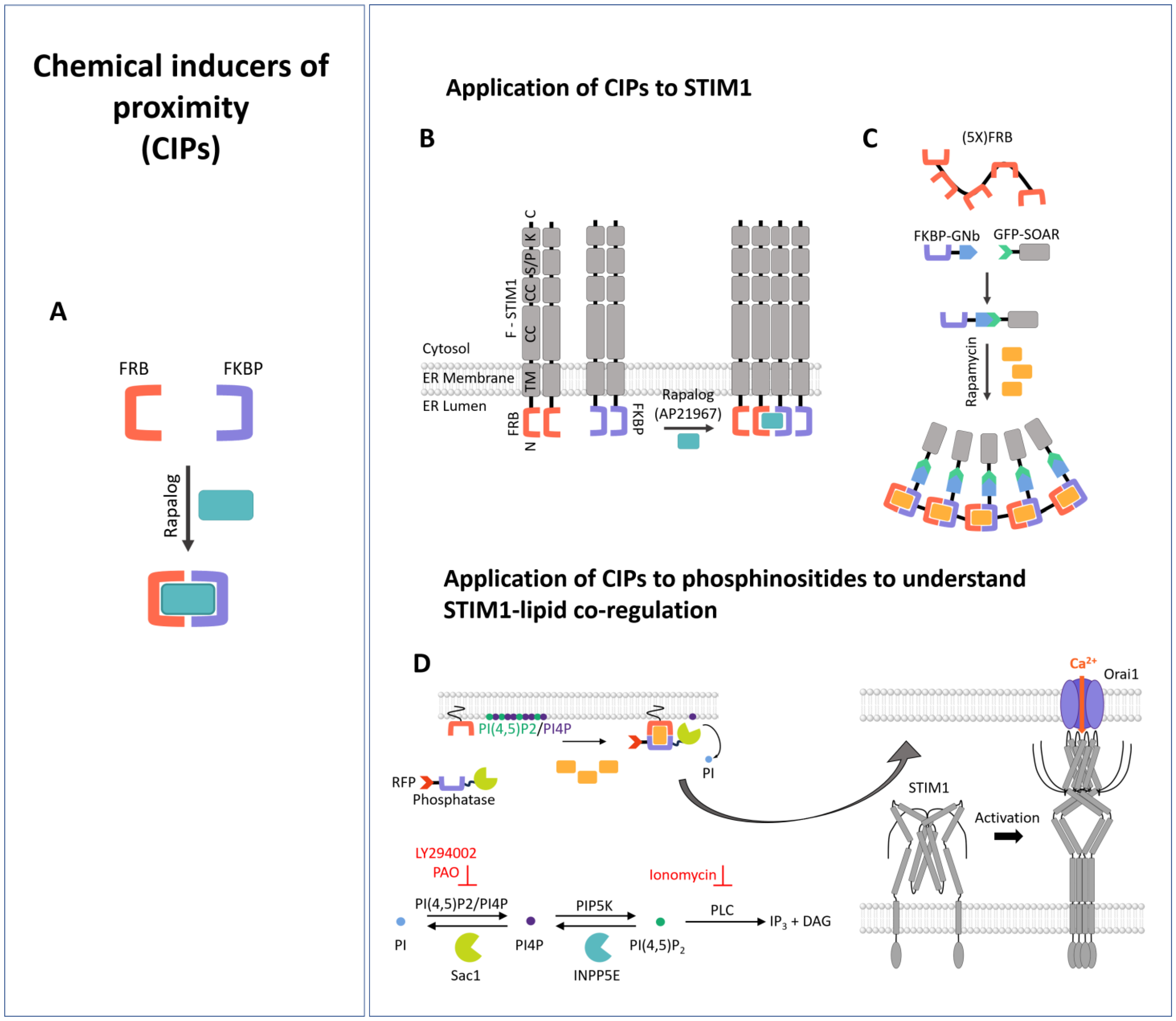
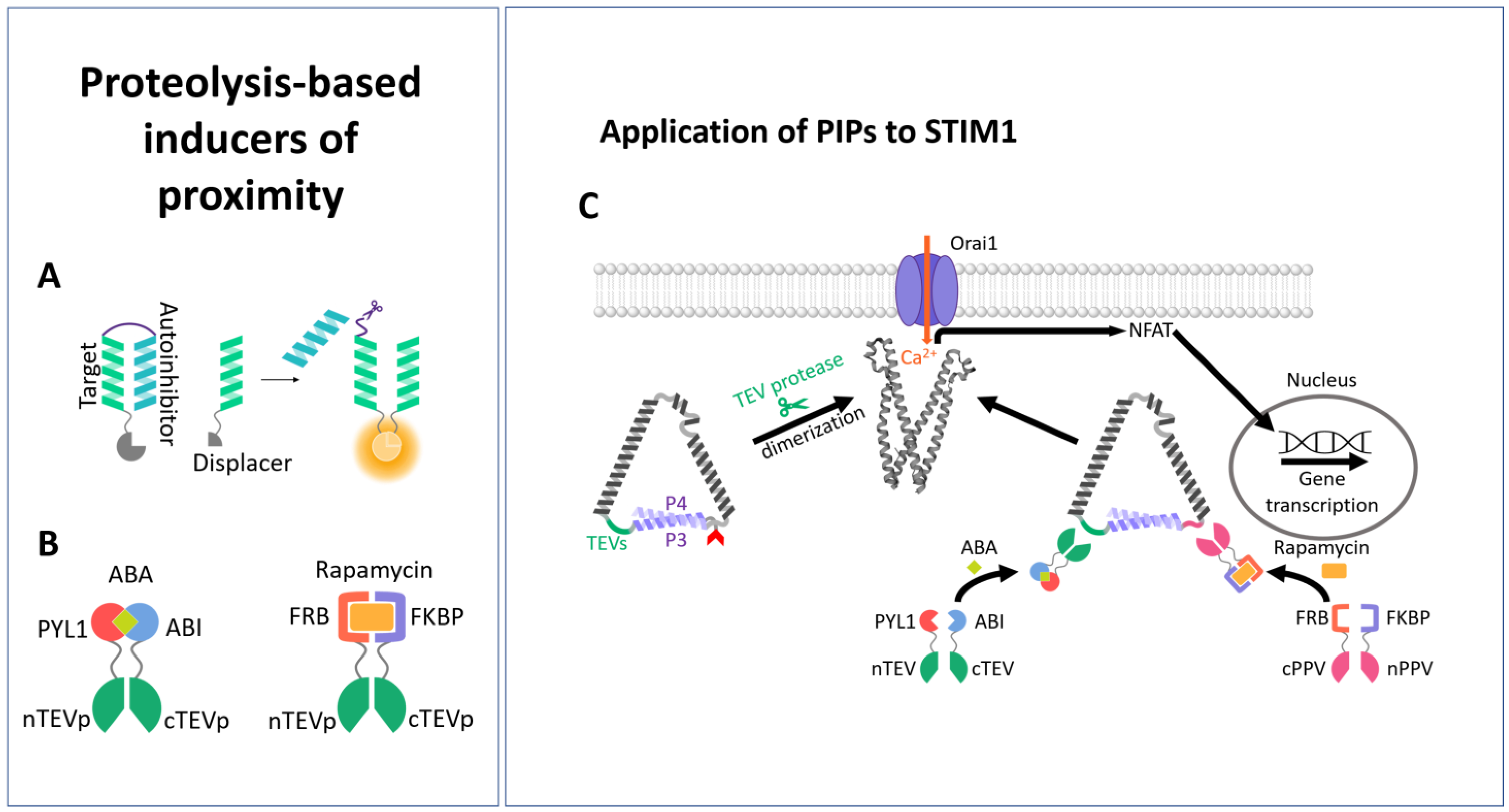


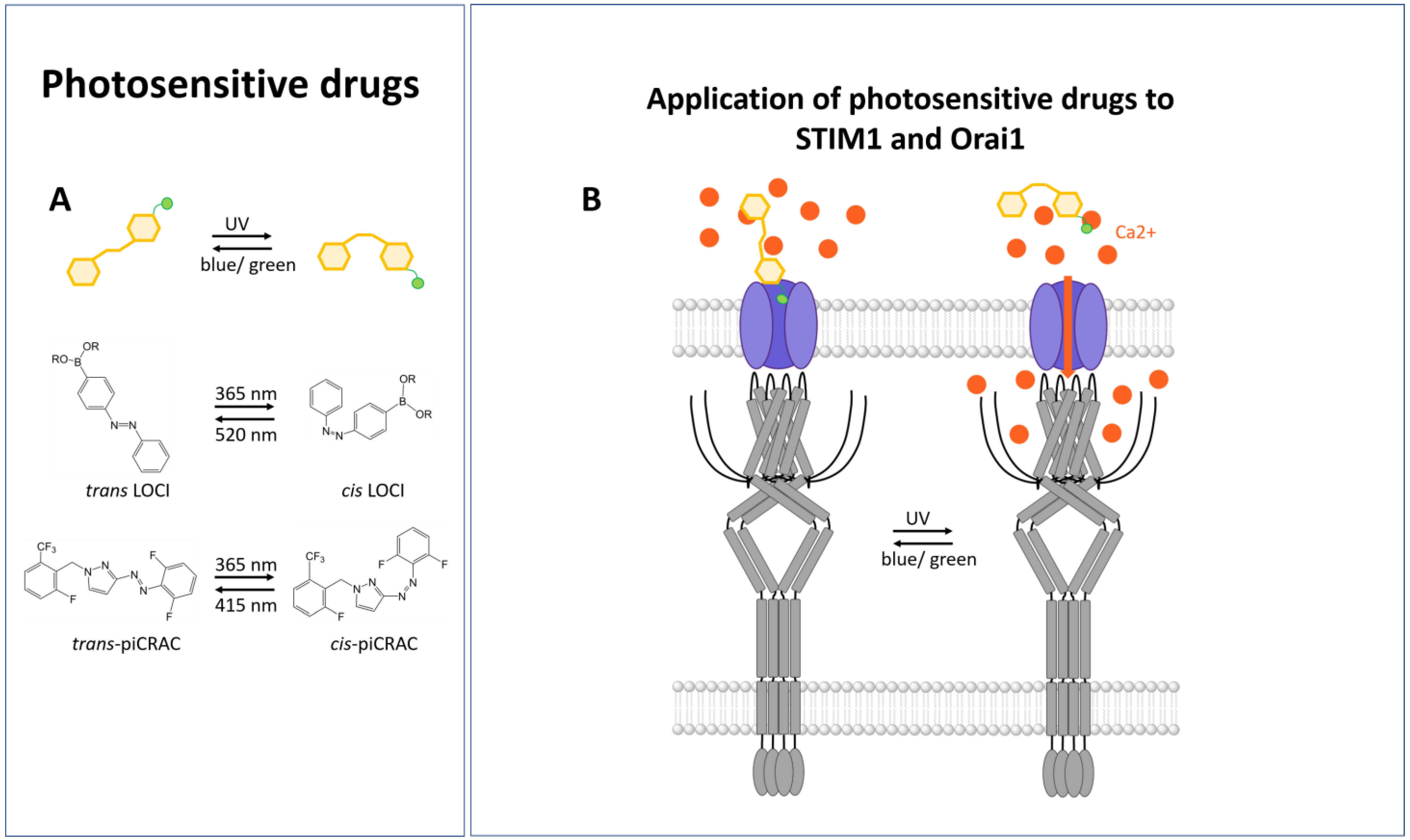
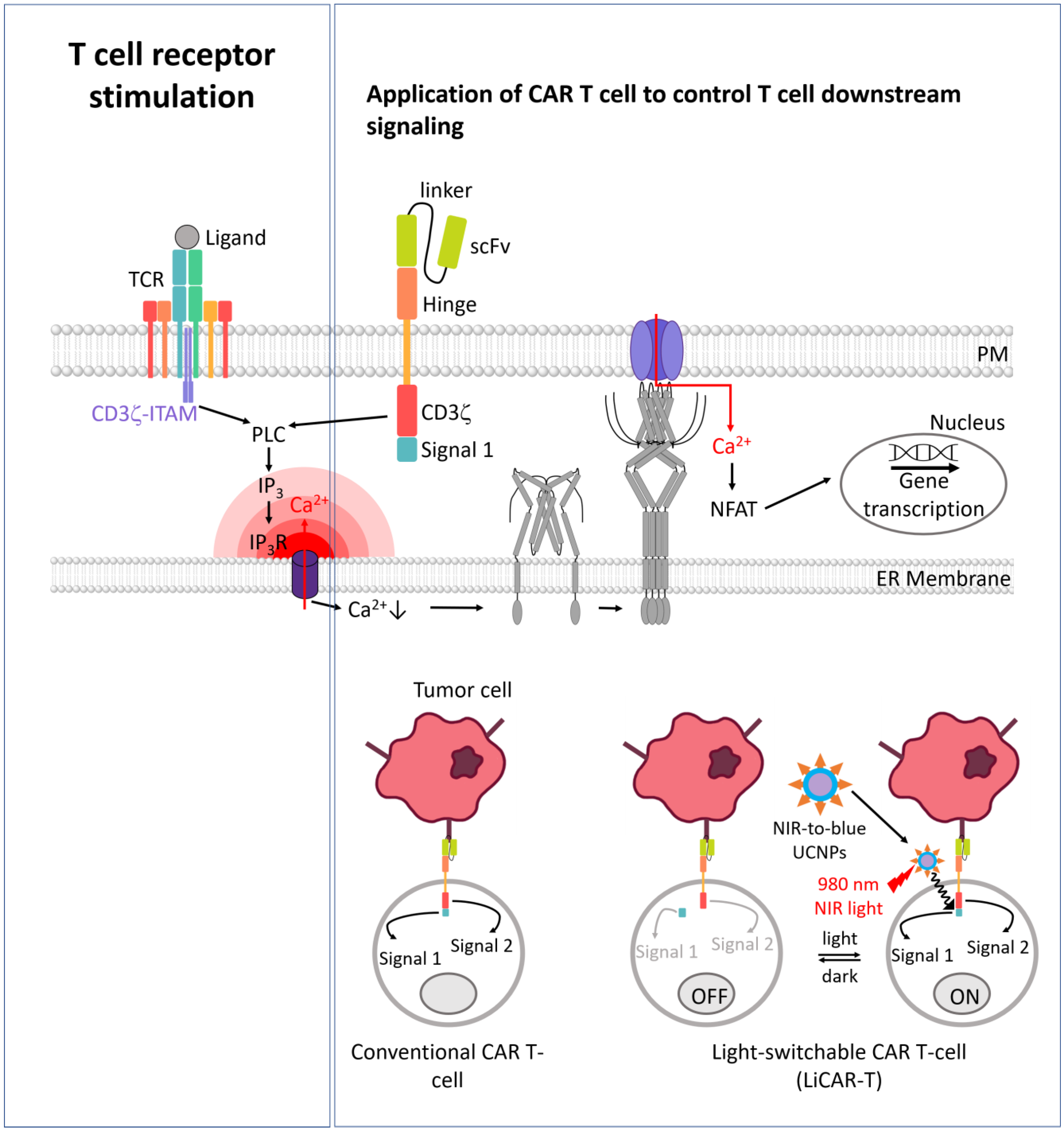
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bacsa, B.; Hopl, V.; Derler, I. Synthetic Biology Meets Ca2+ Release-Activated Ca2+ Channel-Dependent Immunomodulation. Cells 2024, 13, 468. https://doi.org/10.3390/cells13060468
Bacsa B, Hopl V, Derler I. Synthetic Biology Meets Ca2+ Release-Activated Ca2+ Channel-Dependent Immunomodulation. Cells. 2024; 13(6):468. https://doi.org/10.3390/cells13060468
Chicago/Turabian StyleBacsa, Bernadett, Valentina Hopl, and Isabella Derler. 2024. "Synthetic Biology Meets Ca2+ Release-Activated Ca2+ Channel-Dependent Immunomodulation" Cells 13, no. 6: 468. https://doi.org/10.3390/cells13060468
APA StyleBacsa, B., Hopl, V., & Derler, I. (2024). Synthetic Biology Meets Ca2+ Release-Activated Ca2+ Channel-Dependent Immunomodulation. Cells, 13(6), 468. https://doi.org/10.3390/cells13060468






