Upregulation of the Renin–Angiotensin System Is Associated with Patient Survival and the Tumour Microenvironment in Glioblastoma
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics
2.2. TCGA In Silico Analysis
2.3. Cell Lines and Reagents
2.4. RNA Sequencing Analysis
2.5. Statistical Analysis
3. Results
3.1. Higher Gene Expression of RAS Components Associates with Poorer Survival Outcomes for Glioblastoma
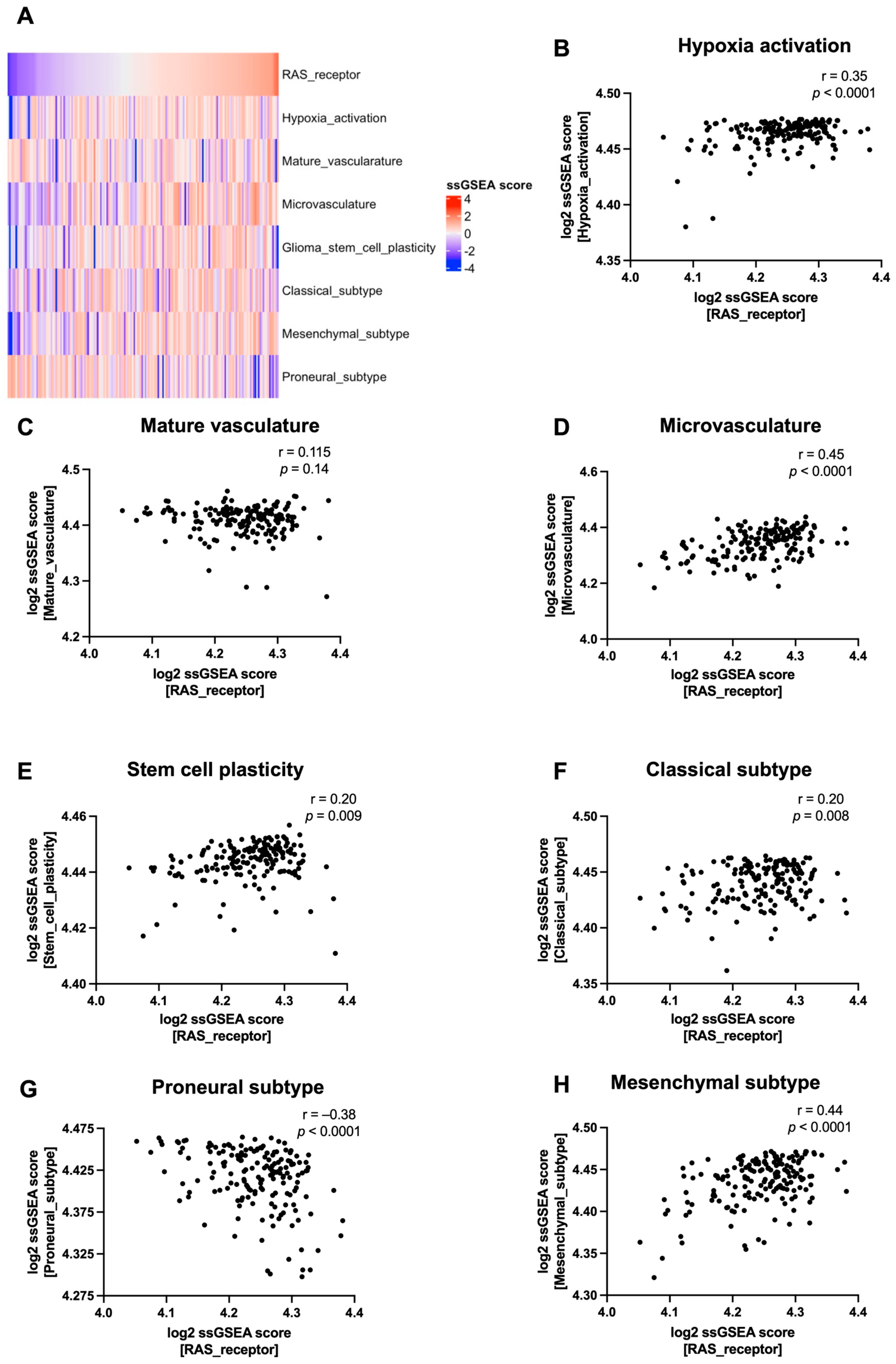
3.2. RAS Receptor Expression Correlates with Tumour Microenvironment Features in Glioblastoma
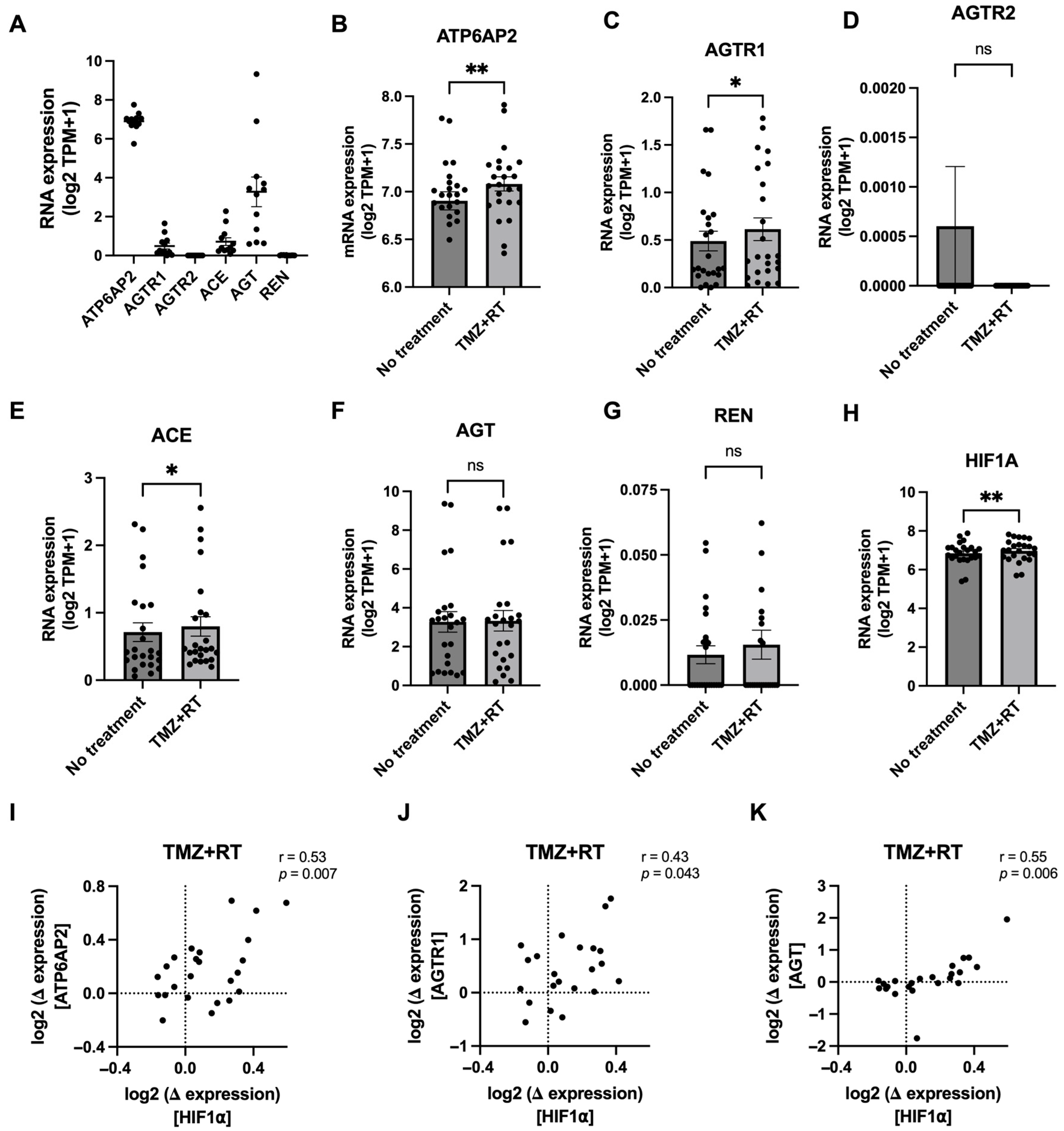
3.3. Glioblastoma Chemoradiation Increases Gene Expression of RAS Components in Patient-Derived Cell Lines
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Brown, N.F.; Ottaviani, D.; Tazare, J.; Gregson, J.; Kitchen, N.; Brandner, S.; Fersht, N.; Mulholland, P. Survival Outcomes and Prognostic Factors in Glioblastoma. Cancers 2022, 14, 3161. [Google Scholar] [CrossRef] [PubMed]
- Tamimi, A.F.; Juweid, M. Epidemiology and Outcome of Glioblastoma. In Glioblastoma; Codon Publications: Singapore, 2017; pp. 143–153. ISBN 9780994438126. [Google Scholar]
- Mohammed, S.; Dinesan, M.; Ajayakumar, T. Survival and quality of life analysis in glioblastoma multiforme with adjuvant chemoradiotherapy: A retrospective study. Rep. Pract. Oncol. Radiother. 2022, 27, 1026–1036. [Google Scholar] [CrossRef] [PubMed]
- Weller, M.; van den Bent, M.; Preusser, M.; Le Rhun, E.; Tonn, J.C.; Minniti, G.; Bendszus, M.; Balana, C.; Chinot, O.; Dirven, L.; et al. EANO guidelines on the diagnosis and treatment of diffuse gliomas of adulthood. Nat. Rev. Clin. Oncol. 2021, 18, 170–186. [Google Scholar] [CrossRef] [PubMed]
- Ropolo, M.; Daga, A.; Griffero, F.; Foresta, M.; Casartelli, G.; Zunino, A.; Poggi, A.; Cappedi, E.; Zona, G.; Spaziante, R.; et al. Comparative analysis of DNA repair in stem and nonstem glioma cell cultures. Mol. Cancer Res. 2009, 7, 383–392. [Google Scholar] [CrossRef] [PubMed]
- Lozinski, M.; Bowden, N.A.; Graves, M.C.; Fay, M.; Day, B.W.; Stringer, B.W.; Tooney, P.A. Transcriptomic Profiling of DNA Damage Response in Patient-Derived Glioblastoma Cells before and after Radiation and Temozolomide Treatment. Cells 2022, 11, 1215. [Google Scholar] [CrossRef] [PubMed]
- DeCordova, S.; Shastri, A.; Tsolaki, A.G.; Yasmin, H.; Klein, L.; Singh, S.K.; Kishore, U. Molecular Heterogeneity and Immunosuppressive Microenvironment in Glioblastoma. Front. Immunol. 2020, 11, 1402. [Google Scholar] [CrossRef] [PubMed]
- Clara, C.A.; Marie, S.K.N.; de Almeida, J.R.W.; Wakamatsu, A.; Oba-Shinjo, S.M.; Uno, M.; Neville, M.; Rosemberg, S. Angiogenesis and expression of PDGF-C, VEGF, CD105 and HIF-1α in human glioblastoma. Neuropathology 2014, 34, 343–352. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, E.M.; Bandopadhyay, G.; Coyle, B.; Grabowska, A. A HIF-independent, CD133-mediated mechanism of cisplatin resistance in glioblastoma cells. Cell. Oncol. 2018, 41, 319–328. [Google Scholar] [CrossRef]
- Ge, X.; Pan, M.H.; Wang, L.; Li, W.; Jiang, C.; He, J.; Abouzid, K.; Liu, L.Z.; Shi, Z.; Jiang, B.H. Hypoxia-mediated mitochondria apoptosis inhibition induces temozolomide treatment resistance through miR-26a/Bad/Bax axis. Cell Death Dis. 2018, 9, 1128. [Google Scholar] [CrossRef]
- Sparks, M.A.; Crowley, S.D.; Gurley, S.B.; Mirotsou, M.; Coffman, T.M. Classical renin–angiotensin system in kidney physiology. Compr. Physiol. 2014, 4, 1201–1228. [Google Scholar] [CrossRef]
- Campbell, D.J. Critical review of prorenin and (pro)renin receptor research. Hypertension 2008, 51, 1259–1264. [Google Scholar] [CrossRef]
- Klickstein, L.B.; Kaempfer, C.E.; Wintroub, B.U. The granulocyte-angiotensin system. Angiotensin I-converting activity of cathepsin G. J. Biol. Chem. 1982, 257, 15042–15046. [Google Scholar] [CrossRef] [PubMed]
- Jackson, L.; Eldahshan, W.; Fagan, S.C.; Ergul, A. Within the brain: The renin angiotensin system. Int. J. Mol. Sci. 2018, 19, 876. [Google Scholar] [CrossRef]
- Grobe, J.L.; Xu, D.; Sigmund, C.D. An intracellular renin–angiotensin system in neurons: Fact, hypothesis, or fantasy. Physiology 2008, 23, 187–193. [Google Scholar] [CrossRef]
- Almutlaq, M.; Alamro, A.A.; Alamri, H.S.; Alghamdi, A.A.; Barhoumi, T. The Effect of Local Renin Angiotensin System in the Common Types of Cancer. Front. Endocrinol. 2021, 12, 6361. [Google Scholar] [CrossRef] [PubMed]
- Wegman-Ostrosky, T.; Soto-Reyes, E.; Vidal-Millán, S.; Sánchez-Corona, J. The renin–angiotensin system meets the hallmarks of cancer. J. Renin. Angiotensin. Aldosterone. Syst. 2015, 16, 227–233. [Google Scholar] [CrossRef] [PubMed]
- Arrieta, O.; Pineda-Olvera, B.; Guevara-Salazar, P.; Hernández-Pedro, N.; Morales-Espinosa, D.; Cerón-Lizarraga, T.L.; González-De La Rosa, C.H.; Rembao, D.; Segura-Pacheco, B.; Sotelo, J. Expression of AT1 and AT2 angiotensin receptors in astrocytomas is associated with poor prognosis. Br. J. Cancer 2008, 99, 160–166. [Google Scholar] [CrossRef]
- Singh, A.; Srivastava, N.; Yadav, A.; Ateeq, B. Targeting AGTR1/NF-κB/CXCR4 axis by miR-155 attenuates oncogenesis in glioblastoma. Neoplasia 2020, 22, 497–510. [Google Scholar] [CrossRef]
- Bradshaw, A.R.; Wickremesekera, A.C.; Brasch, H.D.; Chibnall, A.M.; Davis, P.F.; Tan, S.T.; Itinteang, T. Glioblastoma Multiforme Cancer Stem Cells Express Components of the Renin–Angiotensin System. Front. Surg. 2016, 3, 51. [Google Scholar] [CrossRef][Green Version]
- Kouchi, M.; Shibayama, Y.; Ogawa, D.; Miyake, K.; Nishiyama, A.; Tamiya, T. (Pro)renin receptor is crucial for glioma development via the Wnt/b-catenin signaling pathway. J. Neurosurg. 2017, 127, 819–828. [Google Scholar] [CrossRef]
- Panza, S.; Malivindi, R.; Caruso, A.; Russo, U.; Giordano, F.; Győrffy, B.; Gelsomino, L.; De Amicis, F.; Barone, I.; Conforti, F.L.; et al. Novel insights into the antagonistic effects of losartan against angiotensin ii/agtr1 signaling in glioblastoma cells. Cancers 2021, 13, 4555. [Google Scholar] [CrossRef] [PubMed]
- Perryman, R.; Renziehausen, A.; Shaye, H.; Kostagianni, A.D.; Tsiailanis, A.D.; Thorne, T.; Chatziathanasiadou, M.V.; Sivolapenko, G.B.; El Mubarak, M.A.; Han, G.W.; et al. Inhibition of the angiotensin II type 2 receptor AT2R is a novel therapeutic strategy for glioblastoma. Proc. Natl. Acad. Sci. USA 2022, 119, e2116289119. [Google Scholar] [CrossRef]
- Urup, T.; Michaelsen, S.R.; Olsen, L.R.; Toft, A.; Christensen, I.J.; Grunnet, K.; Winther, O.; Broholm, H.; Kosteljanetz, M.; Issazadeh-Navikas, S.; et al. Angiotensinogen and HLA class II predict bevacizumab response in recurrent glioblastoma patients. Mol. Oncol. 2016, 10, 1160–1168. [Google Scholar] [CrossRef] [PubMed]
- Urup, T.; Gillberg, L.; Kaastrup, K.; Maya Jeje Schuang, L.; Michaelsen, S.R.; Andrée Larsen, V.; Christensen, I.J.; Broholm, H.; Lassen, U.; Grønbæk, K.; et al. Angiotensinogen promoter methylation predicts bevacizumab treatment response of patients with recurrent glioblastoma. Mol. Oncol. 2020, 14, 964–973. [Google Scholar] [CrossRef] [PubMed]
- Juillerat-Jeanneret, L.; Celerier, J.; Chapuis Bernasconi, C.; Nguyen, G.; Wostl, W.; Maerki, H.P.; Janzer, R.C.; Corvol, P.; Gasc, J.M. Renin and angiotensinogen expression and functions in growth and apoptosis of human glioblastoma. Br. J. Cancer 2004, 90, 1059–1068. [Google Scholar] [CrossRef]
- Cerami, E.; Gao, J.; Dogrusoz, U.; Gross, B.E.; Sumer, S.O.; Aksoy, B.A.; Jacobsen, A.; Byrne, C.J.; Heuer, M.L.; Larsson, E.; et al. The cBio Cancer Genomics Portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012, 2, 401–404. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Aksoy, B.A.; Dogrusoz, U.; Dresdner, G.; Gross, B.; Sumer, S.O.; Sun, Y.; Jacobsen, A.; Sinha, R.; Larsson, E.; et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 2013, 6, pl1. [Google Scholar] [CrossRef] [PubMed]
- Reich, M.; Liefeld, T.; Gould, J.; Lerner, J.; Tamayo, P.; Mesirov, J.P. GenePattern 2.0 [2]. Nat. Genet. 2006, 38, 500–501. [Google Scholar] [CrossRef] [PubMed]
- Barbie, D.A.; Tamayo, P.; Boehm, J.S.; Kim, S.Y.; Moody, S.E.; Dunn, I.F.; Schinzel, A.C.; Sandy, P.; Meylan, E.; Scholl, C.; et al. Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature 2009, 462, 108–112. [Google Scholar] [CrossRef]
- Jin, X.; Kim, L.J.Y.; Wu, Q.; Wallace, L.C.; Prager, B.C.; Sanvoranart, T.; Gimple, R.C.; Wang, X.; Mack, S.C.; Miller, T.E.; et al. Targeting glioma stem cells through combined BMI1 and EZH2 inhibition. Nat. Med. 2017, 23, 1352–1361. [Google Scholar] [CrossRef]
- Zheng, H.; Ying, H.; Yan, H.; Kimmelman, A.C.; Hiller, D.J.; Chen, A.J.; Perry, S.R.; Tonon, G.; Chu, G.C.; Ding, Z.; et al. p53 and Pten control neural and glioma stem/progenitor cell renewal and differentiation. Nature 2008, 455, 1129–1133. [Google Scholar] [CrossRef] [PubMed]
- Verhaak, R.G.W.; Hoadley, K.A.; Purdom, E.; Wang, V.; Qi, Y.; Wilkerson, M.D.; Miller, C.R.; Ding, L.; Golub, T.; Mesirov, J.P.; et al. Integrated Genomic Analysis Identifies Clinically Relevant Subtypes of Glioblastoma Characterized by Abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell 2010, 17, 98–110. [Google Scholar] [CrossRef] [PubMed]
- Stringer, B.W.; Day, B.W.; D’Souza, R.C.J.; Jamieson, P.R.; Ensbey, K.S.; Bruce, Z.C.; Lim, Y.C.; Goasdoué, K.; Offenhäuser, C.; Akgül, S.; et al. A reference collection of patient-derived cell line and xenograft models of proneural, classical and mesenchymal glioblastoma. Sci. Rep. 2019, 9, 4902. [Google Scholar] [CrossRef] [PubMed]
- Lozinski, M.; Bowden, N.A.; Graves, M.C.; Fay, M.; Day, B.W.; Stringer, B.W.; Tooney, P.A. ATR inhibition using gartisertib enhances cell death and synergises with temozolomide and radiation in patient-derived glioblastoma cell lines. Oncotarget 2024, 15, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Weinstein, J.N.; Collisson, E.A.; Mills, G.B.; Shaw, K.R.M.; Ozenberger, B.A.; Ellrott, K.; Sander, C.; Stuart, J.M.; Chang, K.; Creighton, C.J.; et al. The cancer genome atlas pan-cancer analysis project. Nat. Genet. 2013, 45, 1113–1120. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, B.R.; Stodola, T.J.; Wagner, J.R.; Didier, D.N.; Exner, E.C.; Lombard, J.H.; Greene, A.S. Mechanisms of Mas1 Receptor-Mediated Signaling in the Vascular Endothelium. Arterioscler. Thromb. Vasc. Biol. 2017, 37, 433–445. [Google Scholar] [CrossRef] [PubMed]
- Turner, A.J. ACE2 Cell Biology, Regulation, and Physiological Functions. In The Protective Arm of the Renin Angiotensin System (RAS): Functional Aspects and Therapeutic Implications; Elsevier: Amsterdam, The Netherlands, 2015; pp. 185–189. ISBN 9780128014851. [Google Scholar]
- Lo, S.K.; Li, I.T.; Tsou, T.S.; See, L. Non-significant in univariate but significant in multivariate analysis: A discussion with examples. Chang. Yi Xue Za Zhi 1995, 18, 95–101. [Google Scholar]
- Lee, J.W.; Ko, J.; Ju, C.; Eltzschig, H.K. Hypoxia signaling in human diseases and therapeutic targets. Exp. Mol. Med. 2019, 51, 1–13. [Google Scholar] [CrossRef]
- O’rawe, M.; Kilmister, E.J.; Mantamadiotis, T.; Kaye, A.H.; Tan, S.T.; Wickremesekera, A.C. The renin–angiotensin system in the tumor microenvironment of glioblastoma. Cancers 2021, 13, 4004. [Google Scholar] [CrossRef]
- Li, W.; Peng, H.; Seth, D.M.; Feng, Y. The prorenin and (Pro)renin receptor: New players in the brain renin–angiotensin system? Int. J. Hypertens. 2012, 2012, 290635. [Google Scholar] [CrossRef]
- Matsusaka, T.; Niimura, F.; Shimizu, A.; Pastan, I.; Saito, A.; Kobori, H.; Nishiyama, A.; Ichikawa, I. Liver angiotensinogen is the primary source of renal angiotensin II. J. Am. Soc. Nephrol. 2012, 23, 1181–1189. [Google Scholar] [CrossRef] [PubMed]
- Morimoto, S.; Cassell, M.D.; Sigmund, C.D. Neuron-specific expression of human angiotensinogen in brain causes increased salt appetite. Physiol. Genomics 2002, 2002, 113–120. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Velásquez, C.; Mansouri, S.; Gutiérrez, O.; Mamatjan, Y.; Mollinedo, P.; Karimi, S.; Singh, O.; Terán, N.; Martino, J.; Zadeh, G.; et al. Hypoxia can induce migration of glioblastoma cells through a methylation-dependent control of ODZ1 gene expression. Front. Oncol. 2019, 9, 488046. [Google Scholar] [CrossRef] [PubMed]
- Yahata, Y.; Shirakata, Y.; Tokumaru, S.; Yang, L.; Dai, X.; Tohyama, M.; Tsuda, T.; Sayama, K.; Iwai, M.; Horiuchi, M.; et al. A novel function of angiotensin II in skin wound healing: Induction of fibroblast and keratinocyte migration by angiotensin II via heparin-binding epidermal growth factor (EGF)-like growth factor-mediated EGF receptor transactivation. J. Biol. Chem. 2006, 281, 13209–13216. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Zhang, J.W.; Hu, L.; Song, Y.C.; Zhou, L.; Fan, Y.; Zhu, H.Y.; Wang, Y.; Li, Q.P. Activation of the AT1R/HIF-1 α/ACE Axis Mediates Angiotensin II-Induced VEGF Synthesis in Mesenchymal Stem Cells. Biomed Res. Int. 2014, 2014, 627380. [Google Scholar] [CrossRef]
- Pagé, E.L.; Robitaille, G.A.; Pouysségur, J.; Richard, D.E. Induction of hypoxia-inducible factor-1α by transcriptional and translational mechanisms. J. Biol. Chem. 2002, 277, 48403–48409. [Google Scholar] [CrossRef] [PubMed]
- Colman, H.; Zhang, L.; Sulman, E.P.; McDonald, J.M.; Shooshtari, N.L.; Rivera, A.; Popoff, S.; Nutt, C.L.; Louis, D.N.; Cairncross, J.G.; et al. A multigene predictor of outcome in glioblastoma. Neuro. Oncol. 2010, 12, 49–57. [Google Scholar] [CrossRef] [PubMed]
- Latour, M.; Her, N.G.; Kesari, S.; Nurmemmedov, E. WNT Signaling as a Therapeutic Target for Glioblastoma. Int. J. Mol. Sci. 2021, 22, 8428. [Google Scholar] [CrossRef] [PubMed]
- Perng, P.; Lim, M. Immunosuppressive mechanisms of malignant gliomas: Parallels at non-CNS sites. Front. Oncol. 2015, 5, 153. [Google Scholar] [CrossRef]
- Sottoriva, A.; Spiteri, I.; Piccirillo, S.G.M.; Touloumis, A.; Collins, V.P.; Marioni, J.C.; Curtis, C.; Watts, C.; Tavaré, S. Intratumor heterogeneity in human glioblastoma reflects cancer evolutionary dynamics. Proc. Natl. Acad. Sci. USA 2013, 110, 4009–4014. [Google Scholar] [CrossRef]
- Fujimori, T.; Shibayama, Y.; Kanda, T.; Suzuki, K.; Ogawa, D.; Ishikawa, R.; Kadota, K.; Matsunaga, T.; Tamiya, T.; Nishiyama, A.; et al. Effects of a monoclonal antibody against (pro)renin receptor on gliomagenesis. Sci. Rep. 2023, 13, 808. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.L.; Chou, C.H.; Li, Y.F.; Huang, L.C.; Kao, Y.; Hueng, D.Y.; Tsai, C.K. Antiproliferative and apoptotic effects of telmisartan in human glioma cells. Cancer Cell Int. 2023, 23, 111. [Google Scholar] [CrossRef] [PubMed]
- Datta, M.; Chatterjee, S.; Perez, E.M.; Gritsch, S.; Roberge, S.; Duquette, M.; Chen, I.X.; Naxerova, K.; Kumar, A.S.; Ghosh, M.; et al. Losartan controls immune checkpoint blocker-induced edema and improves survival in glioblastoma mouse models. Proc. Natl. Acad. Sci. USA 2023, 120, e2219199120. [Google Scholar] [CrossRef] [PubMed]
- Ho, J.K.; Moriarty, F.; Manly, J.J.; Larson, E.B.; Evans, D.A.; Rajan, K.B.; Hudak, E.M.; Hassan, L.; Liu, E.; Sato, N.; et al. Blood-Brain Barrier Crossing Renin–Angiotensin Drugs and Cognition in the Elderly: A Meta-Analysis. Hypertension 2021, 78, 629–643. [Google Scholar] [CrossRef]
- O’Rawe, M.; Wickremesekera, A.C.; Pandey, R.; Young, D.; Sim, D.; FitzJohn, T.; Burgess, C.; Kaye, A.H.; Tan, S.T. Treatment of glioblastoma with re-purposed renin–angiotensin system modulators: Results of a phase I clinical trial. J. Clin. Neurosci. 2022, 95, 48–54. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lozinski, M.; Lumbers, E.R.; Bowden, N.A.; Martin, J.H.; Fay, M.F.; Pringle, K.G.; Tooney, P.A. Upregulation of the Renin–Angiotensin System Is Associated with Patient Survival and the Tumour Microenvironment in Glioblastoma. Cells 2024, 13, 634. https://doi.org/10.3390/cells13070634
Lozinski M, Lumbers ER, Bowden NA, Martin JH, Fay MF, Pringle KG, Tooney PA. Upregulation of the Renin–Angiotensin System Is Associated with Patient Survival and the Tumour Microenvironment in Glioblastoma. Cells. 2024; 13(7):634. https://doi.org/10.3390/cells13070634
Chicago/Turabian StyleLozinski, Mathew, Eugenie R. Lumbers, Nikola A. Bowden, Jennifer H. Martin, Michael F. Fay, Kirsty G. Pringle, and Paul A. Tooney. 2024. "Upregulation of the Renin–Angiotensin System Is Associated with Patient Survival and the Tumour Microenvironment in Glioblastoma" Cells 13, no. 7: 634. https://doi.org/10.3390/cells13070634
APA StyleLozinski, M., Lumbers, E. R., Bowden, N. A., Martin, J. H., Fay, M. F., Pringle, K. G., & Tooney, P. A. (2024). Upregulation of the Renin–Angiotensin System Is Associated with Patient Survival and the Tumour Microenvironment in Glioblastoma. Cells, 13(7), 634. https://doi.org/10.3390/cells13070634





