Abstract
Astrocytes and ependymal cells have been reported to be able to switch from a mature cell identity towards that of a neural stem/progenitor cell. Astrocytes are widely scattered in the brain where they exert multiple functions and are routinely targeted for in vitro and in vivo reprogramming. Ependymal cells serve more specialized functions, lining the ventricles and the central canal, and are multiciliated, epithelial-like cells that, in the spinal cord, act as bi-potent progenitors in response to injury. Here, we isolate or generate ependymal cells and post-mitotic astrocytes, respectively, from the lateral ventricles of the mouse brain and we investigate their capacity to reverse towards a progenitor-like identity in culture. Inhibition of the GSK3 and TGFβ pathways facilitates the switch of mature astrocytes to Sox2-expressing, mitotic cells that generate oligodendrocytes. Although this medium allows for the expansion of quiescent NSCs, isolated from live rats by “milking of the brain”, it does not fully reverse astrocytes towards the bona fide NSC identity; this is a failure correlated with a concomitant lack of neurogenic activity. Ependymal cells could be induced to enter mitosis either via exposure to neuraminidase-dependent stress or by culturing them in the presence of FGF2 and EGF. Overall, our data confirm that astrocytes and ependymal cells retain a high capacity to reverse to a progenitor identity and set up a simple and highly controlled platform for the elucidation of the molecular mechanisms that regulate this reversal.
1. Introduction
Astrocytes constitute the largest glial population in the mammalian brain and they are scattered throughout the parenchyma [1]. In mice, they are the progeny of radial glial cells, generated either via direct final divisions during early postnatal life [2,3] or via embryonic, intermediate, and astroglial progenitors [4,5,6]. Historically, they were considered a relatively homogeneous population of supporting cells, even though the presence of subpopulations of different morphologies (protoplasmic versus fibrotic or even cortical layer-specific types) [5,7] and of differential marker expressions (for example, regarding GFAP or S100β) has been described [8,9]. Recently, the astrocytic heterogeneity has been confirmed at the single-cell transcription level [10,11,12,13] and it has revealed that gene expression varies significantly between astrocytes located at different areas of the brain and, surprisingly, showed common expression patterns between astrocytes and neurons of the same area [14].
There are, at least, three major indications suggesting that astrocytes retain a high plasticity capacity in vivo. Firstly, in response to injury, degeneration, or stress, they show different levels of activation, manifested with changes in shape, marker expression, and mitotic activity [15]. The astrocytes closest to the injury become highly intercalated and form the gliotic scar, a structure that encircles the area of the lesion and limits the expansion of secondary degenerative processes [16], with ambiguous effects on tissue repair and axon regeneration [17]. Secondly, the neural stem cells (NSCs) that survive in the postnatal brain, clustered within specialized microenvironments called niches, are of astroglial morphology [18]. In other words, the small pool of multipotent, self-renewing cells of the mature brain is a GFAP-expressing cell population, their existence being confirmed only by the generation of a progeny of different lineages (neurons, oligodendrocytes, and astrocytes). For example, the NSCs of the subependymal zone (SEZ) niche cannot be clearly distinguished from the other astrocytes of the niche with any definitive marker [19,20]; albeit, one morphologic difference is that they extend one monociliated process in between ependymal cells to reach the cerebrospinal fluid [21] and one longer process to contact blood vessels [18,22]. Thirdly, experimental work has shown that in response to an injury, such as a mechanical cortical lesion, a fraction of astrocytes exhibit stem/progenitor properties when isolated and cultured [23,24]. It is now established that Notch signalling is crucial for this spontaneous reprogramming [25] and accumulating reports have provided evidence that astrocytes can be converted (via targeted reprogramming) to neurons [26,27,28,29].
Ependymal cells are a glial population of specialized morphology (multiciliated, epithelial-like, and cuboid cells) that form the walls of the ventricular system of the brain and of the spinal cord [30]. Those lining the central canal have been shown to retain high plasticity potential. In response to injury, they can generate astrocytes and oligodendrocytes while they exhibit neurogenic potential in vitro [31,32]. In contrast, ependymal cells of the brain lack such potential, even though their contribution to the formation of the niche microenvironment of the SEZ is well established [33,34,35].
Based on the above, astrocytes and ependymal cells can be considered cell populations positioned closer to the neural stem/progenitor identity in a hypothetical “stem cell” to “fully mature cell” continuum, especially when compared to neurons or oligodendrocytes. The abundance of astrocytes, in combination with their still not fully known functional repertoire, as well as the functional importance of ependymal cells, in combination with their limited numbers and their inability to regenerate, make these two cell populations ideal targets for investigation. Here, we show that both these pools can be induced to reverse, to different degrees, towards a progenitor state with simple changes in their culture conditions. This confirms the continuum hypothesis and paves the way for a more targeted investigation of key molecular pathways that control the plasticity capacity of postnatal brain cells.
2. Materials and Methods
2.1. Animals
Adult male and female B6CBAC wild-type mice (1–3 months old) were used for the isolation of postnatal brain neural stem cells and ependymal cells. Adult female Wistar rats (2–4 months old) were used for the isolation of SEZ-derived cells via brain milking. Animals were kept in the animal facilities of the University of Patras, in standard laboratory polyacrylic cages (3–5 mice/cage), under a relative humidity of 50–60%, a controlled temperature (22 ± 1 °C), and a steady light–dark cycle (12 h/12 h), with free access to water and food. Their breeding and maintenance were in accordance with the European Communities Council Directive Guidelines (86/609/EEC) for the care and use of laboratory animals, as implemented in Greece by the Presidential Decree 56/2013 and approved and scrutinized by the Prefectural Animal Care and Use Committee (Protocol number: 5675/39/18-01-2021 and reference number of establishment: EL 13BIO04) and the Animal Welfare and Ethical Review Committee of the University of Patras.
2.2. Brain Milking
The isolation of neural stem and progenitor cells from the brains of live rats was conducted with the “brain milking” protocol described in detail in [36]. Briefly, rats were stereotaxically injected (coordinates: anterioposterior = 0.3 mm, lateral = +1.2 mm, depth = 3.5 mm) with 2 μL of a release cocktail, consisting of Clostridium perfringens neuraminidase (500 mU, Merck, Sigma-Aldrich, Sain Louis, MO, USA, Cat: N2876), integrin-β1-blocking antibody (1 μg, BD Pharmingen, San Diego, CA, USA, Cat: 555002), and FGF-2 (0.5 μg, Peprotech, London, UK, Cat: 100-18B). After three days, we performed four CSF liquid biopsies (“draining” of the brain) from the cisterna magna of the anaesthetized animals, acquiring approximately 120 μL of CSF each time, with an interval of 40 min between the collections. Biopsies were immediately mixed with an ice-cold neurosphere medium.
2.3. Reagents for Cell Cultures
Dulbecco’s modified Eagle’s mediumcontaining—high levels of glucose and pyruvate (DMEM, 11995-065), Neurobasal medium (21103049), DMEM-F12 (10565018), and Poly-D-Lysine (PDL, A3890401)—as well as B27 (17504-044), B27 without retinoic acid (12587010), and N2 (17502-048) culture supplements, were obtained from Thermo Fisher Scientific (Waltham, MA, USA). Fibroblast growth factor-2 (FGF-2, 100-18B) and epidermal growth factor (EGF, 315-09) were purchased from Peprotech (Londin, UK). Accutase was obtained from PAN-Biotech (P10-21100, Aidenbach, Germany). The glycogen synthase kinase-3 (GSK-3), CHIR99021 (SML1046), the TGFβ kinase/activin receptor-like kinase (ALK 5) inhibitor, A 83-01 (SML0788), and neuraminidase (N2876) were purchased from Merck, Sigma-Aldrich (Sain Louis, MO, USA).
2.4. Neurosphere Cultures
To culture postnatal brain neural stem and progenitor cells in the form of neurospheres, the whole lateral walls of the lateral ventricles (where the SEZs are located) of adult mice were dissected under a stereoscope, dissociated with accutase (37 °C, 20 min), and were resuspended in a standard NSC proliferation medium containing a high-glucose DMEM medium, supplemented with 2% B27 and 1% N2, as well as with FGF-2 and EGF (final concentration for both 20 ng/mL). In these conditions, NSCs are grown in the form of 3D, freely floating aggregates with a self-renewing capacity, called neurospheres, and are passaged every 5–7 days.
2.5. Astrocyte Generation and Culture
Astrocytes were generated by dissociating neurospheres (passages 5–10) with accutase and plating 60,000 cells/well on PDL-coated coverslips in wells of 24-well plates or 300,000 cells on PDL-coated wells of 6-well plates. Cells were cultured in DMEM, complemented with 10% heat-inactivated FBS, for 7 days, changing the medium every two days. To achieve the de-differentiation of astrocytes, the cultures’ medium was changed to either (i) a NSC-stemness medium, made of Neurobasal (49%) and DMEM-F12 (49%) media, complemented with B27-RA (1%), N2 (0,5%), FGF-2 (20 ng/mL), GSK-3 inhibitor CHIR99021 (3 μM), and ALK4/5/7 or TGF-beta/smad inhibitor A83-01 (0.5 μM), or (ii) a lineage medium, made of Neurobasal (49%) and DMEM-F12 (49%) media, complemented with B27-RA (1%), N2 (0,5%), and FGF-2 (20 ng/mL). Cells were kept in culture for 7 days, changing the medium every 2 days. To test the final differentiation potential of cultured astrocytes, the medium was changed to a growth-factor-free, serum-free medium made of DMEM, B27 (2%), and N2 (1%) for 5 days, changing the medium every 2 days.
2.6. Ependymal Cell Culture Generation and Neuraminidase Assays
Ependymal cells were obtained by dissecting the periventricular areas, around the lateral ventricles of adult mice, under a stereoscope. The tissue was dissociated with accutase (37 °C, 20 min) and cells obtained from 3 mice were pooled together and resuspended in a total of 450 μL of DMEM+ 10% heat-inactivated FBS medium. Cells were plated on 9 PDL-coated coverslips in wells of 48-well plates in 50 μL droplets and were left to adhere at room temperature for 20 min. Cells were then kept in culture with 200 μL of medium for up to 5 days without changing the media, only supplementing them to retain volume. Neuraminidase (stock solution: 20 U/mL in water) was added directly to the wells in order to create the desired final concentration.
2.7. Immunocytochemistry and Antibodies
Cells were fixed with 2% paraformaldehyde (PFA) (15 min at room temperature/RT) and were processed for immunofluorescence staining using standard protocols. Briefly, cells were incubated with blocking buffer (3%BSA, 0.1% Triton x-100 from Merck, Sigma-Aldrich, Sain Louis, MO, USA, in PBS) for 1 h at RT and primary antibody incubation (in blocking buffer) was performed overnight at 4 °C. The next day, samples were incubated with the appropriate secondary antibodies for 2 h at RT and were mounted with mowiol. The following antibodies were used (name, species raised in, dilution, provider, and catalogue number):
DCX (rabbit, 1/800, Abcam, Cambridge, UK, ab18723), EGFR (mouse 1/200, Abcam, ab30), GFAP (goat, 1/700, Abcam, ab53554), ID3 (mouse, 1/200, Santa Cruz Biotechnology, Dallas, TX, USA, sc-56712), Ki67 (rabbit, 1/500, Abcam, ab16667), NESTIN (chicken, 1/200, Abcam, 130417), OLIG2 (rabbit, 1/300, Millipore, Livingston, UK, AB9610), PCNT (mouse, 1/500, BD Biosciences, Wokingham, UK, 611815), Phosphorylated Histone-3 (rabbit, 1/500, Abcam, ab80612), s100β (rabbit, 1/500, Abcam, ab52642 and mouse Sigma, 1/200, S2532), SOX2 (goat, 1/200, Santa Cruz, sc-17320), TUBULIN βIII (mouse, 1/500, Abcam, ab7751), TUBULIN acetylated (mouse, 1/1500, Sigma-Aldrich, T6793), and β-catenin (mouse, 1/200, Santa Cruz Biotechnology and rabbit, 1/500, Abcam, ab16051).
Appropriate secondary antibodies conjugated with fluorescence dyes were purchased from Thermo Fisher Scientific (molecular probes, Eugene, OR, USA) and Biotium (Fremont, CA, USA) and raised in donkey or goat IgGs with fluorophores of 568 nm, 488 nm, or 647 nm.
2.8. Immunohistochemistry
Original images have been produced and archived from tissue used and processed as described in previous publications. For the post-brain milking tissue shown in Figure S2, see [37].
2.9. Quantitative Reverse Transcription Polymerase Chain Reaction
Total RNA was extracted using the QIAGEN miRNeasy Mini Kit (Germantown, MD, USA, Cat: 217004) and cDNA synthesis was conducted using the High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems, San Francisco, CA, USA, Cat: 4374966), following the manufacturer’s protocols. A quantitative reverse transcription polymerase chain reaction (qRT-PCR) was performed as per the manufacturer’s instructions using the Kapa Sybr Fast Universal qPCR Kit (Sigma-Aldrich, Cat: KK4600) and results were generated with the MxPro QPCR 4.10 software. The primers (Sigma-Aldrich) used in the experiments were as follows:
- (1)
- Rpl19 (housekeeping gene)
fp CCGACGAAAGGGTATGCTCA
rp GGGCAACAGACAAAGGCTTG
- (2)
- Sox9
fp GTACCCGCATCTGCACAAC
rp CTCCTCCACGAAGGGTCTCT
- (3)
- Glast
fp GTTCCCTGGGGAGCTTCT
rp TTACTATCTAGGGCCGCCATT
- (4)
- Aldh1l1
fp CTCGGTTTGCTGATGGGGACG
rp GCTTGAATCCTCCAAAAGGTGCGG
- (5)
- Glut-1
fp GAAGTGAAAGAGCGGGTGAG
rp CTGTTGACCAGCGCAAAG
- (6)
- Tbr1
fp CAAGGGAGCATCAAACAACA
rp GTCCTCTGTGCCATCCTCAT
- (7)
- Pitx3
fp ACCCTCCGCTTCCAGAAC
rp GAGGCCTTCTCCGAGTCAC
- (8)
- Gbx2
fp GCTGCTCGCTTTCTCTGC
rp GCTGTAATCCACATCGCTCTC
2.10. Imaging, Cell Counts, and Statistical Analysis
Images were taken using a Leica SP8 confocal microscope. For cell counts, at least 15 random optical fields per coverslip (10 mm diameter) were acquired with the x63 objective lens. Statistical analyses were performed using the IBM SPSS 29 statistical software and Microsoft Office Excel and the graphs were constructed in the GraphPad Prism 5.0 software. For statistical comparisons, we performed Student’s t-tests (for 2 groups) or one-way ANOVA, followed by post hoc tests. Probability values lower than p = 0.05 were considered statistically significant.
3. Results
3.1. De-Differentiation of Postnatal Brain Astrocytes via Cell Culture Media Modifications
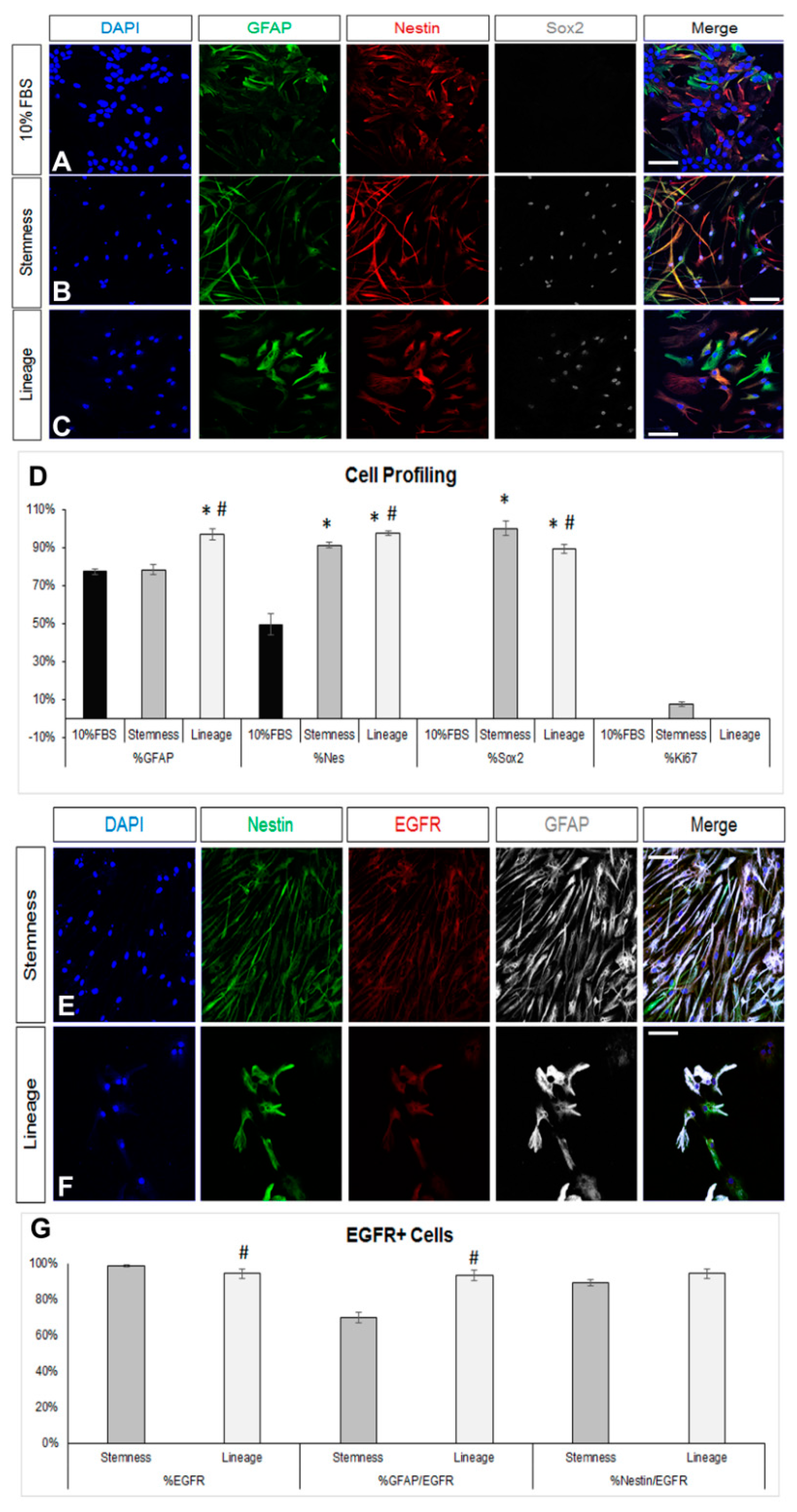
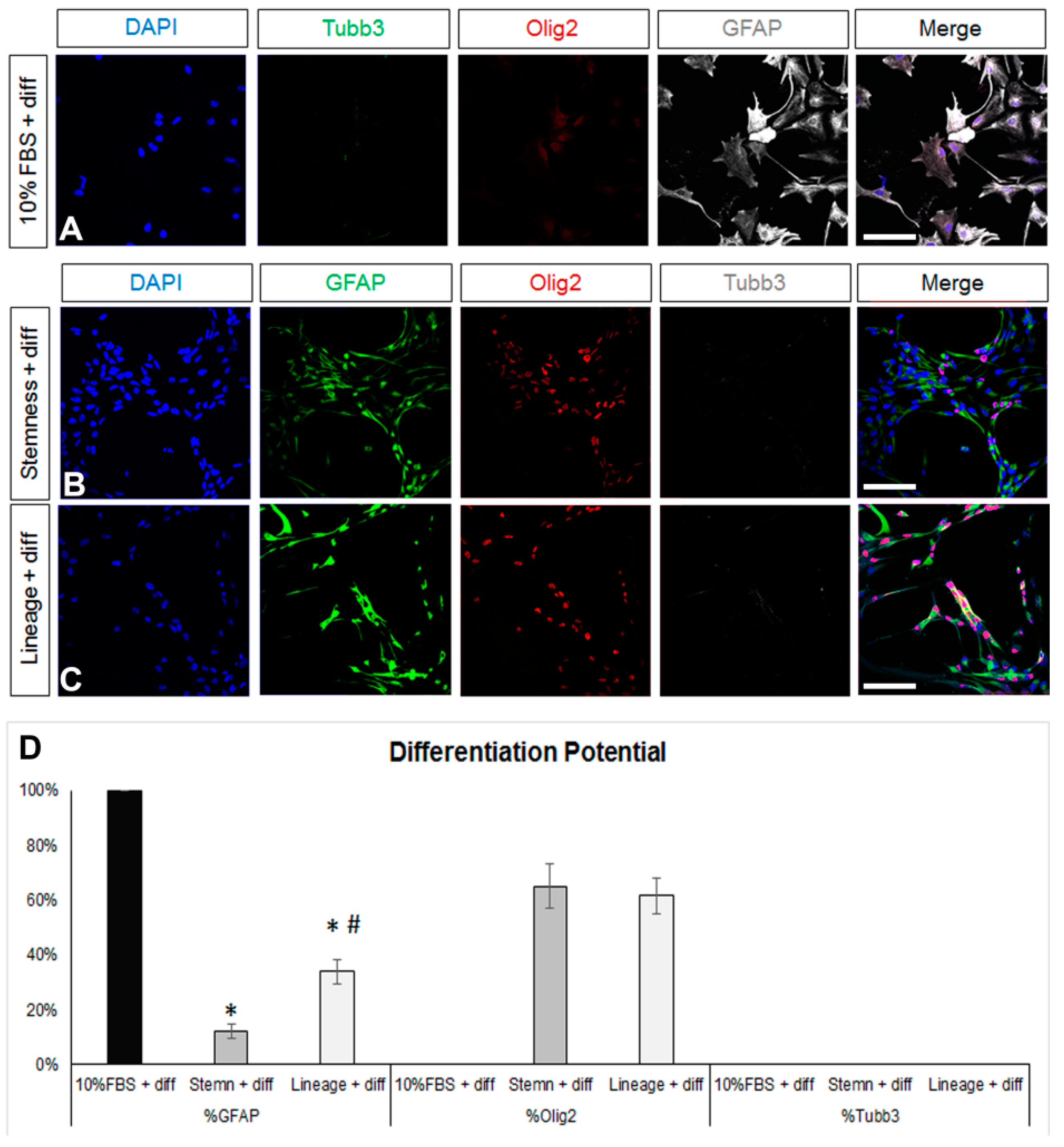
We first obtained postnatal brain neural stem and progenitor cells (NSPCs) by dissecting and dissociating the SEZ stem cell niches of postnatal mice and growing the cells in a typical NSPC medium, which includes FGF-2 and EGF. NSPCs were maintained in the form of free-floating colonies, called neurospheres, which were passaged between five and eight times before use. Mouse astrocytes were generated by differentiating NSPCs for 7 days in DMEM complemented with 10% FBS. At the end of the 7 days, we ended up with cultures consisting of approximately 80% GFAP immunopositive cells (Figure 1A,D). Almost 50% of the cells were immunopositive for nestin (a marker of neural progenitor identity with expression carried on in the glial lineage) [38,39], the majority being double-positive for GFAP and nestin, while there were no Sox2 [40] or Ki67+ immunopositive cells, indicating that the cells had lost their progenitor identity and were post-mitotic, respectively (Figure 1D). Cells in these cultures exhibited the typical morphology of protoplasmic astrocytes (Figure 2A and Supplementary Figure S1A) and their molecular characterization, using RT-qPCR, revealed expression of the astrocytic markers Sox9, GLAST, Aldh1l1, and i(Supplementary Figure S1D). Based on recent experimental work showing that astrocytes, at different anatomical positions, express genes that mark local populations of neurons [14], we looked for the expression of genes, such as Tbr1, Pitx3, and Gbx2, that act to determine and specify cortical, nigral, and thalamic neurons, respectively, and only Pitx3 was found to be expressed (data not shown). We subsequently switched culture conditions by growing the cells in supplemented DMEM (serum-free, growth-factor-free), which induces the final differentiation of cells, without instructing specific lineage [41] and we fixed cells after 5 days. We found a homogeneously astrocytic (GFAP+) population devoid of Olig2+ or βIII-tubulin+ (Tubb3) cells (marking cells of oligodendroglial and neuronal lineage, respectively) (Figure 2A,D; black bars). Overall, we concluded that by culturing NSPCs in 10%FBS for one week, we were able to generate cells that exhibited key properties and markers of terminally differentiated astrocytes.
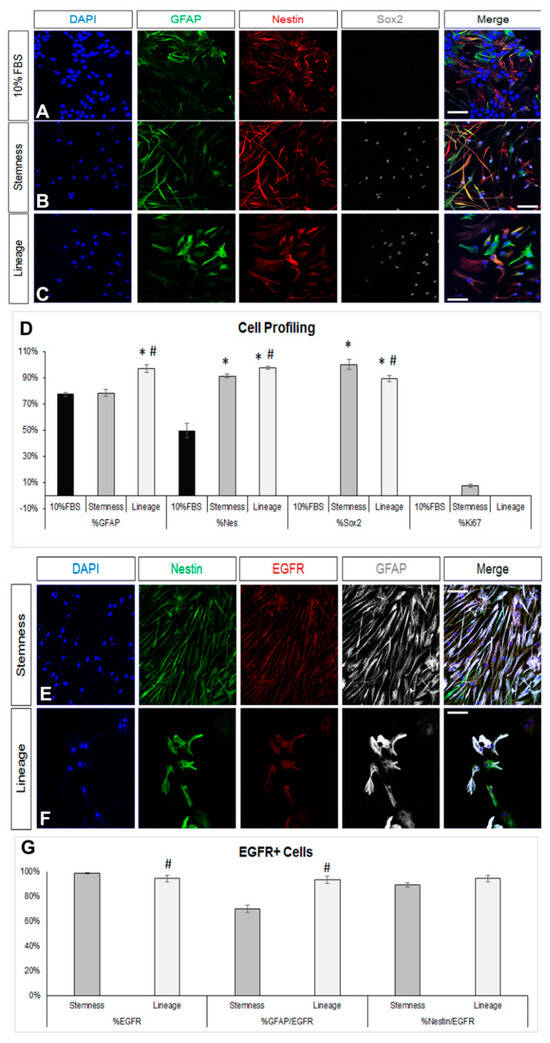
Figure 1.
De-differentiation of astrocytes via modification of the culture medium. (A–C) Images of cells immunostained for GFAP (in green), nestin (in red), and Sox2 (in grey) immediately after culture in DMEM+ 10%FBS for 7 days (A) or after a further 7-day culture in two different de-differentiation media (B,C). (D) Graph showing the marker profile quantification of cells grown in the different media. (E,F) Comparative images of astrocytes immunostained for nestin (in green), EGFR (in red), and GFAP (in grey) after culture in the two de-differentiation media. (G) Graph showing the profile of EGFR+ cells after culture in the two de-differentiation media. [Scale bars: 20 μm; (D) *: p < 0.05 for comparisons to the 10%FBS medium and #: p < 0.05 for comparisons to the stemness medium; one-way ANOVA followed by post hoc analysis; (G) #: p < 0.05 for comparisons to the stemness medium; Student’s t-test].
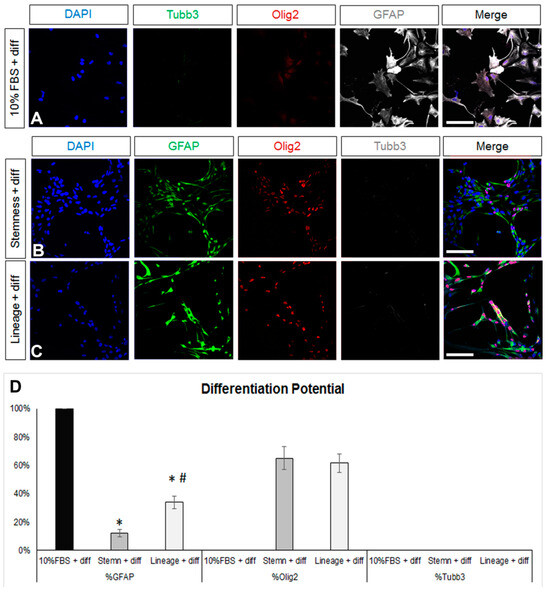
Figure 2.
Investigation of the differentiation potential of mature and de-differentiated astrocytes. (A) Images of astrocytes generated after a 7-day culture in DMEM+ 10%FBS and then being differentiated for another 5 days in supplemented serum-free, growth-factor-free DMEM immunostained for β3-tubulin (Tubb3, to mark neurons, in green), Olig2 (to mark oligodendroglial lineage cells, in red), and GFAP (in grey). (B,C) Images of astrocytes de-differentiated for 7 days in two different media and then differentiated for another 5 days in supplemented serum-free, growth-factor-free DMEM immunostained for GFAP (in green), Olig2 (in red), and Tubb3 (in grey). (D) Graph showing the marker profile quantification of cells grown in the different media after 5 days of differentiation. [Scale bars: 20 μm; (D) *: p < 0.05 for comparisons to the 10%FBS medium and #: p < 0.05 for comparisons to the stemness medium; one-way ANOVA followed by post hoc analysis].
To assess the de-differentiation capacity of the post-10% FBS astrocytes, we cultured them, for 7 more days, in two different media: either a NSC-lineage medium, that included only FGF-2 and promoted lineage progress, or a medium that has been previously shown to enhance “stemness” and to expand the bona fide NSC pool [42], that included inhibitors of GSK-3β and of TGF-β. Culture in the NSC-lineage medium resulted in cells with diverse morphology and an almost homogeneous co-expression of GFAP and a panel of neural progenitor identity markers, such as nestin, Sox2, and the EGF receptor (EGFR) [43] (Figure 1B,D,E,G; dark grey bars and Supplementary Figure S1B). Culture in the NSC-stemness medium led to cells acquiring a homogeneous, fibrous morphology (Supplementary Figure S1C), as well as to a strikingly uniform expression of Sox2 and a strong increase in the presence of nestin and EGFR immunopositive cells. The most notable differences to the effects of the NSC-lineage medium were that (i) nestin+ cells were significantly fewer while EGFR+ cells were significantly more; (ii) the percentage of GFAP+ cells did not change compared to the starting 10% FBS population; and (iii) a low number of Ki67+ cells emerged (Figure 1C,D,F,G). We also examined the presence of ID3, a marker of quiescent NSCs, and it was absent from cells cultured in either media (Supplementary Figure S1E,F).
Any reversal towards a de-differentiated, progenitor-like identity should be accompanied by the re-emergence of multipotency; thus, after the 7-day de-differentiation step, (NSC-lineage or -stemness media) cells were cultured in supplemented (serum and growth-factor free) DMEM for another 5 days. As expected, astrocytes were present after the differentiation of both the NSC-lineage and the NSC-stemness cell populations, albeit at significantly lower percentages when compared to the differentiated post-10% FBS cells and with NSC-stemness cells exhibiting the lowest astrogliogenic potential (Figure 2B,D). Importantly, both cell populations re-acquired a (similar) oligodendrogenic potential but failed to generate neurons.
Overall, our data revealed that the most efficient de-differentiation of a population of post-mitotic, Sox2 immunonegative, and uniformly astrogliogenic cells was achieved using the NSC-stemness medium that transformed them into cells with low astrogliogenic and high oligodendrogenic potential, with uniform re-expression of Sox2 and with re-emerging mitotic activity. Nevertheless, these significantly de-differentiated cells did not show neurogenic activity and did not include any quiescent NSCs.
3.2. Quiescence Can Be Promoted in Postnatal Brain, SEZ-Derived, Astroglia-Like NSPCs in Culture
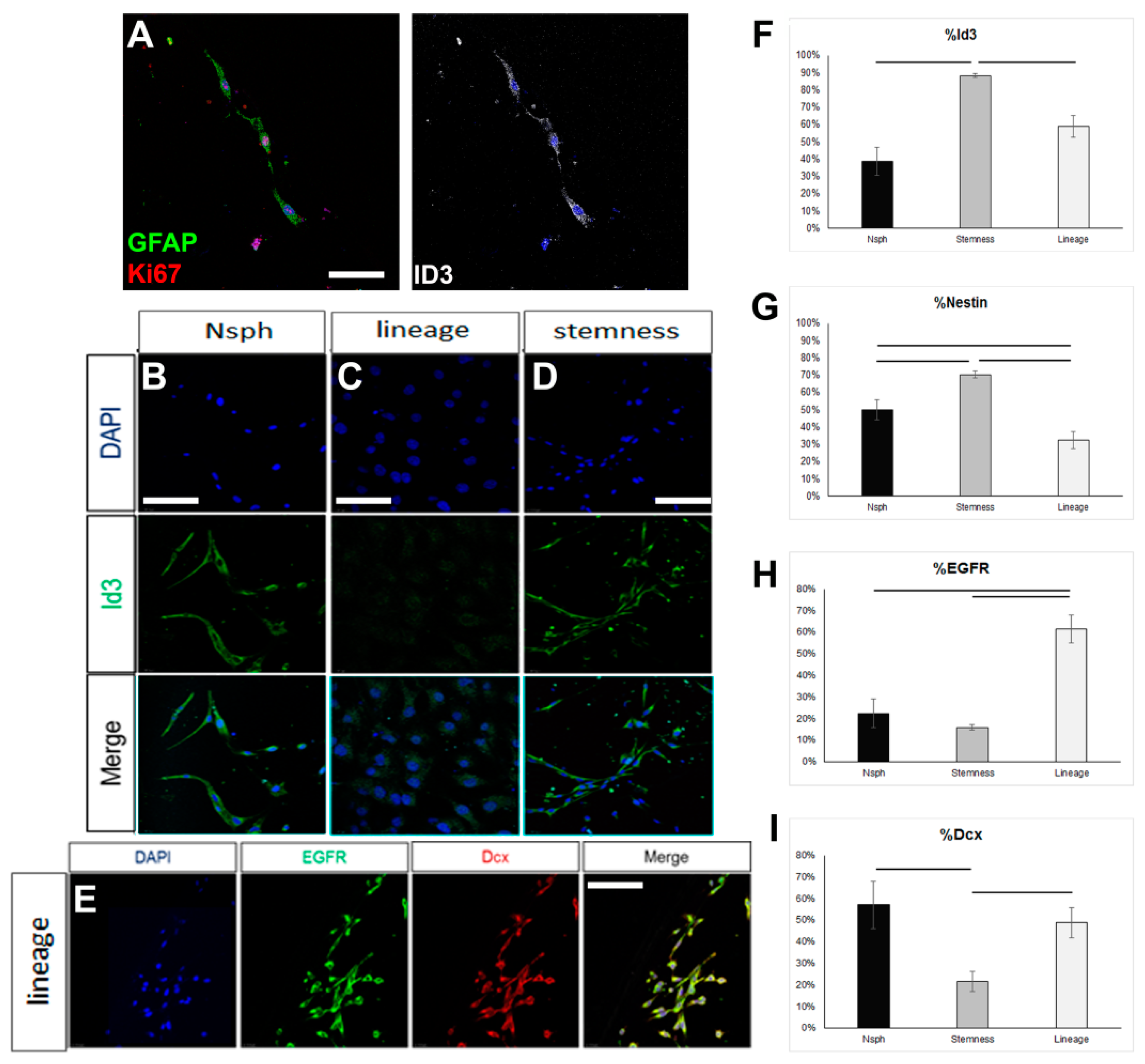
In order to assess if the failure to reverse the astrocytes up to the level of ID3 expression, which would be compatible with an endogenous quiescent NSC identity, could be due to the inability of the NSC-stemness medium to foster a bona fide NSC identity, we tested this medium on rat NSPCs obtained by brain milking. This method allows the direct isolation of NSPCs from the brains of live adult rats, without the use of aggressive, tissue-dissociation protocols, thus allowing the cells to retain in higher fidelity their endogenous properties [36,37]. We confirmed the presence of cells phenotypically compatible with endogenous, quiescent NSCs (GFAP+, ID3+, EGFR-) in the samples obtained after milking (Figure 3A). When these cells were cultured for 7 days in the typical neurosphere-growing medium (in the presence of EGF and FGF2), expression of ID3 was observed in approximately 40% of the cells, EGFR started to be expressed, and a significant fraction of cells expressed Dcx, a marker of neuronal commitment (Figure 3B–I). When milking-derived cells were cultured in the NSC-lineage medium, the expression of ID3 remained at similar levels, with the percentage of nestin+ progenitor cells becoming significantly reduced and the percentage of EGFR+ cells becoming significantly increased. Neuronal commitment remained at similar levels. On the other hand, culture in the NSC-stemness medium resulted in a significant increase in the percentage of ID3+ and of nestin+ cells, as well as a significant reduction in neuronal commitment (Figure 3A–I). Mitotic activity remained at similar levels irrespective of the culture medium, with approximately 50% of the cells expressing Ki67 (data not shown). Overall, our data confirmed that the behaviour of postnatal brain NSCs, which are of the astroglial phenotype, can be significantly manipulated in vitro, with the NSC-stemness medium specifically expanding the ID3+ pool (uncommitted NSCs) and restricting the pool of EGFR+ and Dcx+ cells.
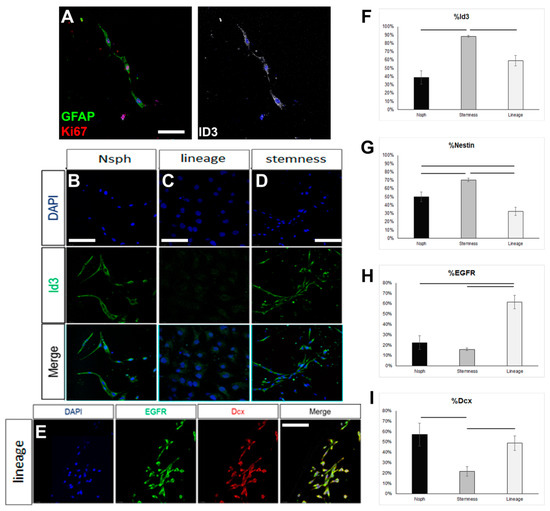
Figure 3.
Investigation of the properties of rat NSCs cultured in different media. (A) Image of mitotic astrocytes (co-expressing GFAP, in green, and Ki67, in red), as well as ID3, in cell samples obtained via the milking of the brains of live rats and cultured for 7 days. (B–D) Images of ID3+ cells (in green) in samples obtained via the milking of the brains of live rats and cultured for 7 days in different media (Neurosphere/Nsph medium; NSC-lineage and NSC-stemness media). (E) Image of milking-derived cells immunostained for EGFR (in green) and Doublecortin (Dcx, in red) after culture for 7 days in the NSC-lineage progression medium. (F–I) Graphs showing the marker profile quantifications of milking-derived cells grown in the different media for 7 days. [Scale bars: 15 μm in (A) and 20 μm in (B–E); horizontal lines indicate difference (p < 0.05); one-way ANOVA followed by post hoc analysis].
3.3. Postnatal Brain-Derived Ependymal Cells Regain Mitotic Activity in Culture, under Stress, and in the Presence of Growth Factors
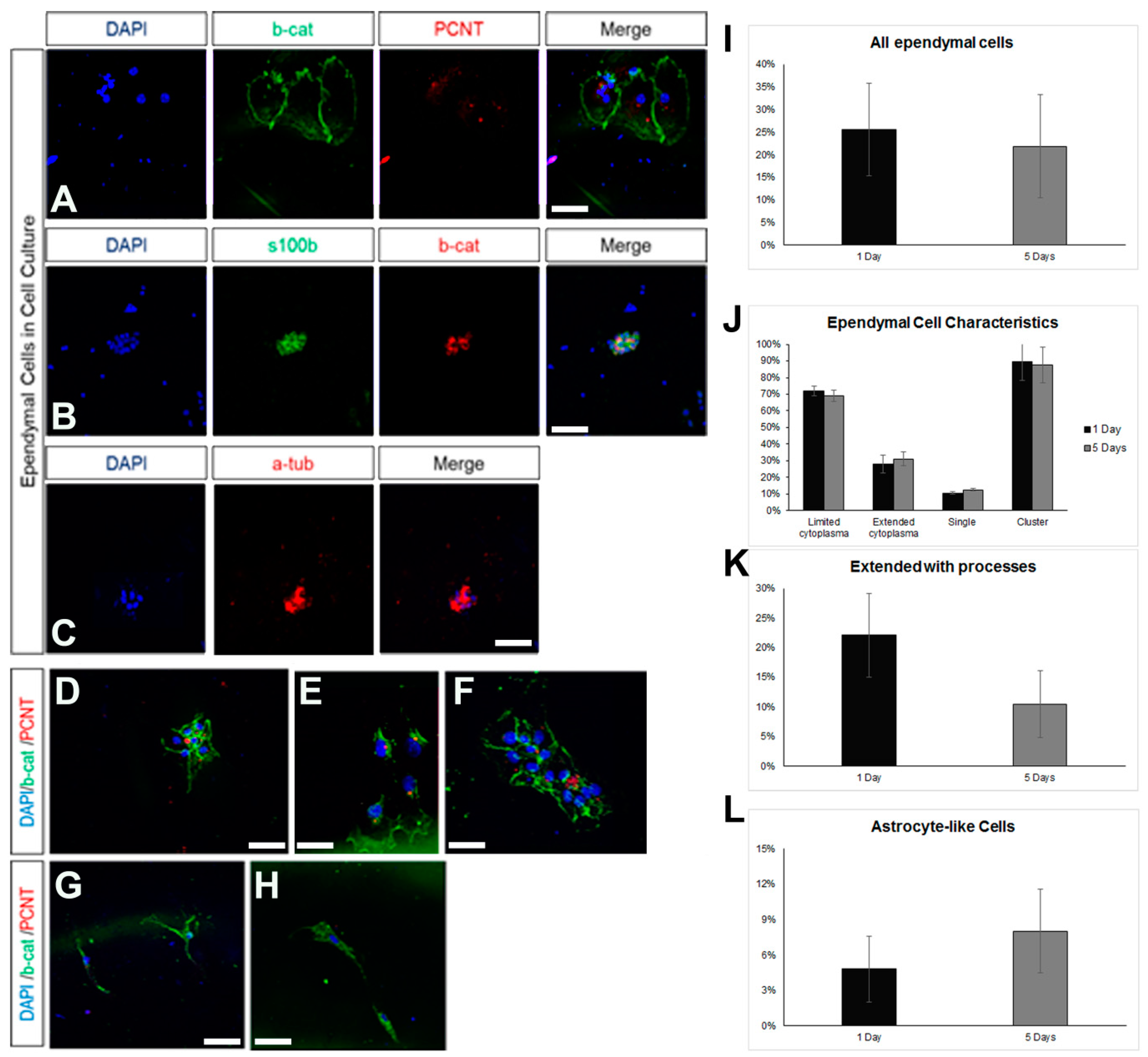
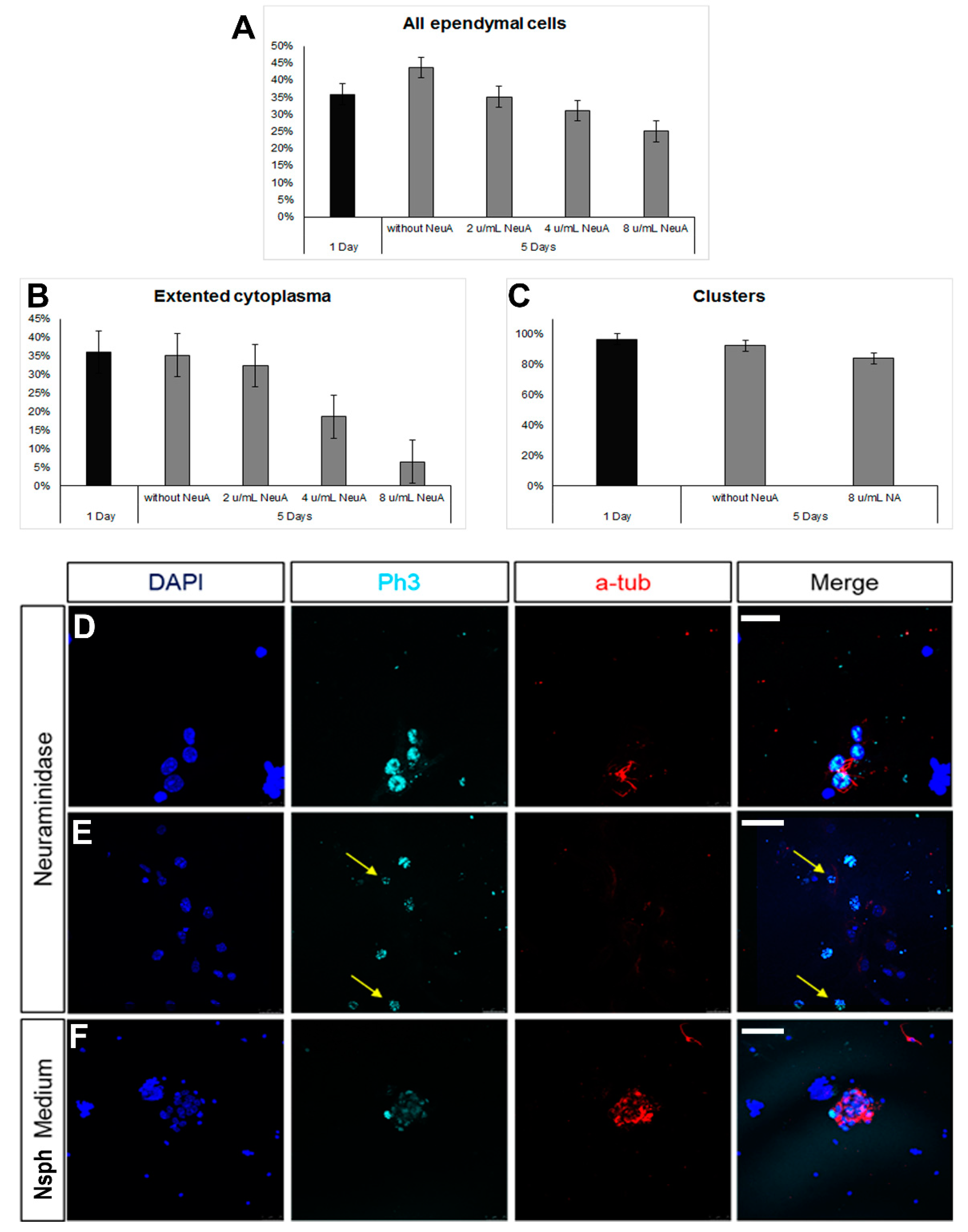
Ependymal cells are ciliated cells that form the walls of the ventricular system. Limited ependymal loss, specifically in the SEZ area, can be restored by NSCs of the niche [44]; however, larger ependyma denudations result in gliosis and hydrocephalus [30]. Thus, the consensus is that the brain’s ependymal cells lack mitotic and regenerative capacity. However, the ependymal cells that line the spinal cord’s central canal retain the capacity to proliferate [32], indicating that the ependymal fate is not dominantly restrictive for proliferation. Therefore, we investigated if postnatal mouse brain ependymal cells can exhibit mitotic activity in vitro. Mixed cell cultures, which included approximately 25% healthy ependymal cells, could be prepared only by dissecting and dissociating periventricular areas of the lateral ventricles that were subsequently cultured in DMEM complemented with 10% FBS. Ependymal cells were identified by the combined expression of at least two of the following markers: S100β (a marker of astrocytes and ependymal cells) [37], β-catenin [22], pericentrin (PCNT, a protein that marks basal bodies, for which we did not quantify the staining but followed an on/off approach), and acetylated α-tubulin (to mark directly the cilia) (Figure 4A–C). The cells were kept in culture for 5 days, with the percentage of ependymal cells resting stable (Figure 4D). The majority of ependymal cells remained in, or formed, clusters (Figure 4D) and we could identify three major subtypes: (a) ependyma with large, typically cuboid cytoplasm; (b) ependymal cells of small, again, typically cuboid cytoplasm; (c) ependymal cells without α-tub+ cilia, with an elongated shape and extending processes (Figure 4E,F). The presence of the last ependymal pool decreased over time in the culture. Finally, we could also detect GFAP+ astrocytes, with a subpopulation co-expressing β-catenin but no other ependymal markers. The presence of these cells increased over time (Figure 4G,I). As with the astroglial cells cultured in 10% FBS, we did not detect any mitotic cells throughout the 5 days.
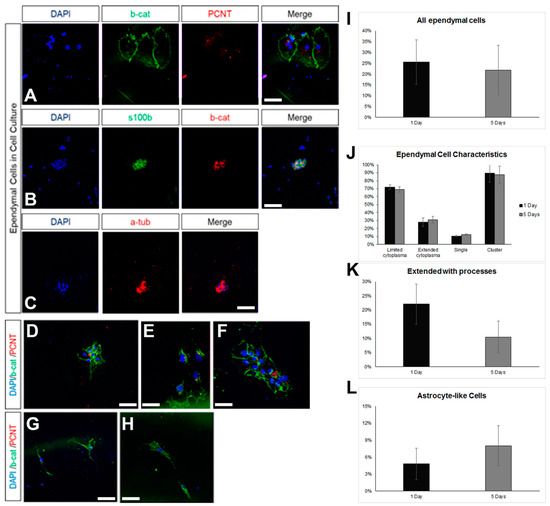
Figure 4.
Characterization of ependymal cells in primary cell cultures. (A) Indicative image of ependymal cells (co-expressing beta-catenin, in green, and PCNT, in red) in cell samples obtained via the milking of the brains of live rats and cultured for 7 days. (B) Indicative image of ependymal cells (co-expressing s100beta, in green, and beta-catenin, in red). (C) Indicative image of ependymal cells expressing acetylated tubulin (in red). (D–H) Indicative images of ependymal cells (D–F) of different sizes and morphologies, as well as of astrocyte-like cells (G–H), expressing beta-catenin (in green) but not PCNT (in red). (I–L) Graphs showing the quantification of different types of cells after different time points in culture. [Scale bars: 15 μm].
The milking of the brain protocol involves the compromise of the integrity of the ependymal cell layer at the level of the lateral ventricles, induced by the administration of neuraminidase [36,37]. Even though the areas of ependymal damage are later characterized by the formation of astroglial scars, in some cases, we also observed that some long-term, surviving ependymal cells were expressing markers of mitosis (Supplementary Figure S2). To assess this phenomenon further, we used the ependymal cell cultures and exposed the cells to different doses of neuraminidase. We observed a reduction in the number of ependymal cells with the increasing dose of neuraminidase; although, this did not reach statistical significance (Figure 5A). Even though the presence of clusters remained unaltered (Figure 5B) at the higher dose, the morphology of ependymal cells was affected, with cells losing cell volume (Figure 5C). Notably, the presence of high doses of neuraminidase led to the appearance of mitotic ependymal cells, identified by the expression of phospho-histone 3, a protein expressed only during the M phase of the cell cycle. Mitotic activation was detected both in ciliated ependymal cells (Figure 5D) and in non-ciliated, elongated ependymal cells (Figure 5E). Since we found that ependymal cells, kept in differentiation conditions, could be induced to re-enter the cell cycle after ependyma-specific stress, we also assessed the behaviour of ependymal cells when cultured in the strongly pro-mitotic NSPC medium and we confirmed that the presence of FGF-2 and EGF led to the emergence of mitotic ependymal cells (Figure 5F).
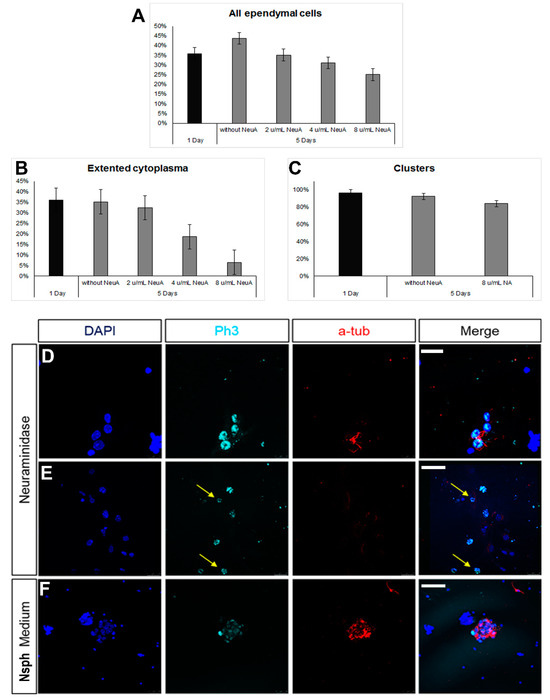
Figure 5.
Emergence of mitotic ependymal cells in primary cultures. (A–C) Graphs showing numbers and morphological features of ependymal cells after exposure to different doses of neuraminidase. (D–F) Images of cells expressing phospho-histone3 (in cyan, to mark cells in the M phase). Some of these cells were ependymal cells (expressing acetylated tubulin, in red) and some were not (examples shown with yellow arrows). [Scale bars: 10 μm in (D), 20 μm in (E,F)].
4. Discussion
Astrocytes are the most abundant and the most multifaceted cell type of the CNS [45]. They play a role in regulating metabolic processes, the extracellular microenvironment, synaptic activity, and the response of the tissue to cell stress and degeneration. In addition, NSCs assume an astrocytic phenotype in order to survive in the postnatal brain [46] while astrocytes are the cells of choice in any attempt to induce glia-to-neuron conversion [47]. Therefore, the detailed investigation of the molecular processes that operate during the transition of a “mature” astrocyte towards a more progenitor-like identity is of high importance in any effort to harness the endogenous regenerative potential of the brain.
Here, we generated astrocytes by inducing NSCs isolated from postnatal mice to differentiate towards this fate by culturing them in 10% FBS. The astrocytes that were produced expressed a range of characteristic astroglial markers (GFAP, S100β, Glast, Aldh1, and Sox9), were not mitotic, were immunonegative for Sox2, and, upon exposure to further differentiation conditions, generated only astrocytes. Based on the above, this protocol allowed the generation, in high numbers and in standardized conditions, of a homogeneous population of mature astrocytes. It relieves the process from the introduction of confounding factors resulting from (i) the different levels of cell stress and damage occurring during the acute isolation of astrocytes from the brain [48], (ii) or during the application of complicated, typically involving serum, culture protocols [49], and (iii) the spatial heterogeneity of the brain’s astrocytes. It should be noted, however, that NSCs populating the SEZ are not of common embryonic origin; although, embryonic NSCs of ventral origin dominate in the formation of the niche [50]. The expression of Pitx3 in the astrocytes we generated suggests a positional identity link with dopaminergic neurons of the substantia nigra in the midbrain [51] or with dopaminergic interneurons in the olfactory bulb, both being areas that receive SEZ-derived neurons [19,52]. A detailed investigation of the positional identity of these astrocytes and of any cells they might generate after reprogramming has to be performed in the future, taking also into account the effects of poorly defined serum factors included in the initial steps (e.g., pro-gliotic BMPs) [53]. This is why we then sought to explore the plasticity of these mature astrocytes, in terms of their de-differentiation capacity, in simple and clear ways by replacing the pro-astroglial medium with two different serum-free media. One that has been shown to favour the uncommitted NSC identity (referred to as NSC-stemness medium) [42], which includes a GSK3 and a TGF-β inhibitor, or another that did not contain the inhibitors but with the addition of FGF-2 in order to support cell survival and to promote lineage (referred to as NSC-lineage medium). The key results of this experiment were the massive re-appearance of the expression of Sox2, which was significantly higher in the NSC-stemness medium, and the re-emergence of mitotic cells, only in the NSC-stemness medium. We, therefore, were able to reverse astrocytes towards a progenitor identity but to different degrees. The highest reversal was achieved by inhibiting the GSK3 and TGFβ pathways, resulting in astrocytes that not only re-expressed the key progenitor factor Sox2 but also re-entered mitosis. To further assess the degree of de-differentiation we also investigated the expression of ID3, a marker of quiescent NSCs [43,54], and we found a ubiquitous lack of expression, in both conditions. This could be due either to a real limitation of the capacity of astrocytes to fully de-differentiate all the way back to a bona fide NSC or to a limitation of the medium to support the quiescent NSC (qNSC) identity [42,55].
To test for the latter, we isolated NSCs from the brains of live rats using the brain-milking method, with which cells are released in the CSF and subsequently collected via liquid biopsies, without the use of aggressive cell-dissociation protocols [36,37]. NSCs isolated with this method retain high levels of quiescence as they generate slowly growing colonies and exhibit limited self-renewal capacity, similar to endogenous SEZ NSCs [56,57]. When milking-derived cells were cultured in the NSC-stemness medium, we found a significant expansion of ID3+ cells, as compared to cells cultured in the NSC-lineage and the neurosphere medium, at the respective expense of the expression of EGFR, which characterises activated NSCs and their progeny [58,59]. Therefore, we demonstrated that the NSC-stemness medium not only can sustain the qNSC identity [42] but can also foster the expansion of these cells in vitro.
Lastly, we switched the culture medium of de-differentiated astrocytes to a typical differentiation-inducing, serum-free, growth-factor-free medium that does not bias towards specific identities or cell subtypes. Our results showed the emergence of oligodendrogenic potential, with astrogliogenesis remaining significantly lower in the astrocytes de-differentiated with the NSC-stemness medium. The observed lack of neurogenic activity, even in the most de-differentiated astrocytes, might be linked to the failure to generate ID3+ quiescent NSC since quiescence is necessary to unlock the full potential of stem cells [31,60,61]. Additional experimental work is necessary to clearly establish if the re-emergence of neurogenesis will require a deeper reversal of astrocytes towards the qNSC state and what the essential factors to facilitate this in culture are [55]. It should be, also, noted that neurogenesis might require longer time frames and is a much more complicated process, influenced by the developmental origin [62] and the exact location in the niche of the parent NSCs [18,19,58]. Nevertheless, the appearance of neuroblasts, initially expressing doublecortin and subsequently βIII tubulin, is typically observed within the first three days of differentiation [63].
Ependymal cells of the spinal cord are another glial type that has been shown to exhibit astrogliogenic and oligodendrogenic properties in vivo in response to injury [32]. Similar properties have not been reported for the brain’s ependyma, which cannot be regenerated after damage; instead, it is replaced by gliotic tissue [37,64] and ependymal loss can lead to hydrocephalus. Nevertheless, an immunohistochemical analysis of the lateral ventricles’ ependyma, at three months post-milking, revealed the presence of ependymal cells expressing the proliferation marker PCNA. Therefore, we set up ependymal cell cultures and we confirmed the presence of multiciliated, cuboid ependymal cells of different sizes. When we mimicked the stress that ependymal cells experience in vivo after the injection of neuraminidase in the lateral ventricles, by exposing ependymal cells isolated from the same area to neuraminidase, we detected the appearance of mitotic ependymal cells. Furthermore, we detected mitotic ependymal cells in the high presence of FGF-2 and EGF, both being NSC-supporting factors [43] that are present in the SEZ niche [21,65].
5. Conclusions
Overall, our data reinforce the hypothesis that astrocytes and ependymal cells retain the intrinsic potential to reverse toward a progenitor identity. They also show that different levels of de-differentiation can be achieved in vitro by manipulating the culture conditions; thus, they can be used in order to dissect the molecular signatures that govern the flow from the qNSC state to that of fully mature cells (such as neurons and oligodendrocytes) or of cell types with enhanced plasticity potential, such as astrocytes and ependymal cells. The aim of this work was to provide a well-characterized and simple platform for the investigation of the reprogramming potential of the brain’s astroglial and ependymal cells. It is intended to serve as a starting point that can be modified in order to investigate the activity of signalling pathways (e.g., the role of BMPs or of Notch that are known to play a role in the maintenance of the astroglial and neuronal identities [25,53,66,67] using serum-free, more tightly defined, conditions. In the absence of large NSC pools in the human brain, especially the ageing one [68], the capacity to reprogram in vivo astrocytes or ependymal cells remains a valid therapeutic target [14,26,47]. The employment of such cell assays, especially with the use of human neural progenitors of embryonic or iPSC origin, will accelerate the effort to devise reprogramming strategies using highly controllable small molecules or signalling factors, rather than the induced expression of transcription factors [69].
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/cells13080668/s1, Figure S1: Morphologic and marker profile comparison of typical and de-differentiated astrocytes; Figure S2: Appearance of proliferating ependymal cells after brain milking.
Author Contributions
D.K., M.K. and D.D.: Methodology, Investigation, Formal Analysis, Data Curation, Writing—Original Draft Preparation, Review and Editing. I.K.: Conceptualization, Methodology, Investigation, Formal Analysis, Data Curation, Writing—Original Draft Preparation, Review and Editing, Funding Acquisition, Supervision, Project Administration. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
The study was conducted in accordance with the European Communities Council Directive Guidelines (86/609/EEC) for the care and use of laboratory animals, as implemented in Greece by the Presidential Decree 56/2013 and as approved and scrutinized by the Animal Care and Use Committee of the Western Greece Regional General Administration (Protocol number: 5675/39/18-01-2021).
Informed Consent Statement
Not applicable.
Data Availability Statement
All raw data supporting the conclusions of this article will be made available by the authors on request.
Acknowledgments
This work was supported by a Erasmus+ traineeship to DK and the authors are grateful to Benedikt Berninger (Johannes Gutenberg University, Mainz, Germany) for his support. The contribution of Filippos Katsaitis in the scientific and artistic task of preparation of the graphical abstract, using the BioRender basic plan application, are also gratefully acknowledged.
Conflicts of Interest
The authors have no conflicts of interest to declare.
References
- Rowitch, D.H.; Kriegstein, A.R. Developmental Genetics of Vertebrate Glial–Cell Specification. Nature 2010, 468, 214–222. [Google Scholar] [CrossRef] [PubMed]
- Masahira, N.; Takebayashi, H.; Ono, K.; Watanabe, K.; Ding, L.; Furusho, M.; Ogawa, Y.; Nabeshima, Y.; Alvarez-Buylla, A.; Shimizu, K.; et al. Olig2-Positive Progenitors in the Embryonic Spinal Cord Give Rise not Only to Motoneurons and Oligodendrocytes, but also to a Subset of Astrocytes and Ependymal Cells. Dev. Biol. 2006, 293, 358–369. [Google Scholar] [CrossRef]
- Clavreul, S.; Abdeladim, L.; Hernández-Garzón, E.; Niculescu, D.; Durand, J.; Ieng, S.-H.; Barry, R.; Bonvento, G.; Beaurepaire, E.; Livet, J.; et al. Cortical Astrocytes Develop in a Plastic Manner at Both Clonal and Cellular Levels. Nat. Commun. 2019, 10, 4884. [Google Scholar] [CrossRef]
- Ge, W.-P.; Miyawaki, A.; Gage, F.H.; Jan, Y.N.; Jan, L.Y. Local Generation of Glia Is a Major Astrocyte Source in Postnatal Cortex. Nature 2012, 484, 376–380. [Google Scholar] [CrossRef]
- Molofsky, A.V.; Deneen, B. Astrocyte Development: A Guide for the Perplexed. Glia 2015, 63, 1320–1329. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Liu, G.; Yang, L.; Li, Z.; Zhang, Z.; Xu, Z.; Cai, Y.; Du, H.; Su, Z.; Wang, Z.; et al. Decoding Cortical Glial Cell Development. Neurosci. Bull. 2021, 37, 440–460. [Google Scholar] [CrossRef]
- Lanjakornsiripan, D.; Pior, B.-J.; Kawaguchi, D.; Furutachi, S.; Tahara, T.; Katsuyama, Y.; Suzuki, Y.; Fukazawa, Y.; Gotoh, Y. Layer-Specific Morphological and Molecular Differences in Neocortical Astrocytes and Their Dependence on Neuronal Layers. Nat. Commun. 2018, 9, 1623. [Google Scholar] [CrossRef]
- Bushong, E.A.; Martone, M.E.; Ellisman, M.H. Maturation of Astrocyte Morphology and the Establishment of Astrocyte Domains during Postnatal Hippocampal Development. Int. J. Dev. Neurosci. 2004, 22, 73–86. [Google Scholar] [CrossRef] [PubMed]
- Ogata, K.; Kosaka, T. Structural and Quantitative Analysis of Astrocytes in the Mouse Hippocampus. Neuroscience 2002, 113, 221–233. [Google Scholar] [CrossRef]
- Bayraktar, O.A.; Bartels, T.; Holmqvist, S.; Kleshchevnikov, V.; Martirosyan, A.; Polioudakis, D.; Ben Haim, L.; Young, A.M.H.; Batiuk, M.Y.; Prakash, K.; et al. Astrocyte Layers in the Mammalian Cerebral Cortex Revealed by a Single-Cell in Situ Transcriptomic Map. Nat. Neurosci. 2020, 23, 500–509. [Google Scholar] [CrossRef]
- Batiuk, M.Y.; Martirosyan, A.; Wahis, J.; de Vin, F.; Marneffe, C.; Kusserow, C.; Koeppen, J.; Viana, J.F.; Oliveira, J.F.; Voet, T.; et al. Identification of Region-Specific Astrocyte Subtypes at Single Cell Resolution. Nat. Commun. 2020, 11, 1220. [Google Scholar] [CrossRef]
- Yao, Z.; van Velthoven, C.T.J.; Kunst, M.; Zhang, M.; McMillen, D.; Lee, C.; Jung, W.; Goldy, J.; Abdelhak, A.; Aitken, M.; et al. A High-Resolution Transcriptomic and Spatial Atlas of Cell Types in the Whole Mouse Brain. Nature 2023, 624, 317–332. [Google Scholar] [CrossRef]
- Zhang, M.; Pan, X.; Jung, W.; Halpern, A.R.; Eichhorn, S.W.; Lei, Z.; Cohen, L.; Smith, K.A.; Tasic, B.; Yao, Z.; et al. Molecularly Defined and Spatially Resolved Cell Atlas of the Whole Mouse Brain. Nature 2023, 624, 343–354. [Google Scholar] [CrossRef]
- Herrero-Navarro, Á.; Puche-Aroca, L.; Moreno-Juan, V.; Sempere-Ferràndez, A.; Espinosa, A.; Susín, R.; Torres-Masjoan, L.; Leyva-Díaz, E.; Karow, M.; Figueres-Oñate, M.; et al. Astrocytes and Neurons Share Region-Specific Transcriptional Signatures That Confer Regional Identity to Neuronal Reprogramming. Sci. Adv. 2021, 7, eabe8978. [Google Scholar] [CrossRef]
- Escartin, C.; Galea, E.; Lakatos, A.; O’Callaghan, J.P.; Petzold, G.C.; Serrano-Pozo, A.; Steinhäuser, C.; Volterra, A.; Carmignoto, G.; Agarwal, A.; et al. Reactive Astrocyte Nomenclature, Definitions, and Future Directions. Nat. Neurosci. 2021, 24, 312–325. [Google Scholar] [CrossRef] [PubMed]
- Wanner, I.B.; Anderson, M.A.; Song, B.; Levine, J.; Fernandez, A.; Gray-Thompson, Z.; Ao, Y.; Sofroniew, M.V. Glial Scar Borders are Formed by Newly Proliferated, Elongated Astrocytes that Interact to Corral Inflammatory and Fibrotic Cells via STAT3-Dependent Mechanisms after Spinal Cord Injury. J. Neurosci. 2013, 33, 12870–12886. [Google Scholar] [CrossRef] [PubMed]
- Adams, K.L.; Gallo, V. The Diversity and Disparity of the Glial Scar. Nat. Neurosci. 2018, 21, 9–15. [Google Scholar] [CrossRef] [PubMed]
- Obernier, K.; Alvarez-Buylla, A. Neural Stem Cells: Origin, Heterogeneity and Regulation in the Adult Mammalian Brain. Development 2019, 146, dev156059. [Google Scholar] [CrossRef] [PubMed]
- Mizrak, D.; Levitin, H.M.; Delgado, A.C.; Crotet, V.; Yuan, J.; Chaker, Z.; Silva-Vargas, V.; Sims, P.A.; Doetsch, F. Single-Cell Analysis of Regional Differences in Adult V-SVZ Neural Stem Cell Lineages. Cell Rep. 2019, 26, 394–406.e5. [Google Scholar] [CrossRef]
- Velloso, F.J.; Shankar, S.; Parpura, V.; Rakic, P.; Levison, S.W. Neural Stem Cells in Adult Mammals are not Astrocytes. ASN Neuro 2022, 14, 17590914221134739. [Google Scholar] [CrossRef]
- Doetsch, F.; Petreanu, L.; Caille, I.; Garcia-Verdugo, J.-M.; Alvarez-Buylla, A. EGF Converts Transit-Amplifying Neurogenic Precursors in the Adult Brain into Multipotent Stem Cells. Neuron 2002, 36, 1021–1034. [Google Scholar] [CrossRef]
- Mirzadeh, Z.; Merkle, F.T.; Soriano-Navarro, M.; Garcia-Verdugo, J.M.; Alvarez-Buylla, A. Neural Stem Cells Confer Unique Pinwheel Architecture to the Ventricular Surface in Neurogenic Regions of the Adult Brain. Cell Stem Cell 2008, 3, 265–278. [Google Scholar] [CrossRef]
- Sirko, S.; Behrendt, G.; Johansson, P.A.; Tripathi, P.; Costa, M.R.; Bek, S.; Heinrich, C.; Tiedt, S.; Colak, D.; Dichgans, M.; et al. Reactive Glia in the Injured Brain Acquire Stem Cell Properties in Response to Sonic Hedgehog. Cell Stem Cell 2013, 12, 426–439. [Google Scholar] [CrossRef]
- Sirko, S.; Neitz, A.; Mittmann, T.; Horvat-Bröcker, A.; von Holst, A.; Eysel, U.T.; Faissner, A. Focal Laser-Lesions Activate an Endogenous Population of Neural Stem/Progenitor Cells in the Adult Visual Cortex. Brain 2009, 132, 2252–2264. [Google Scholar] [CrossRef]
- Magnusson, J.P.; Zamboni, M.; Santopolo, G.; Mold, J.E.; Barrientos-Somarribas, M.; Talavera-Lopez, C.; Andersson, B.; Frisén, J. Activation of a Neural Stem Cell Transcriptional Program in Parenchymal Astrocytes. Elife 2020, 9, e59733. [Google Scholar] [CrossRef]
- Berninger, B.; Costa, M.R.; Koch, U.; Schroeder, T.; Sutor, B.; Grothe, B.; Götz, M. Functional Properties of Neurons Derived from In Vitro Reprogrammed Postnatal Astroglia. J. Neurosci. 2007, 27, 8654–8664. [Google Scholar] [CrossRef]
- Heinrich, C.; Gascón, S.; Masserdotti, G.; Lepier, A.; Sanchez, R.; Simon-Ebert, T.; Schroeder, T.; Götz, M.; Berninger, B. Generation of Subtype-Specific Neurons from Postnatal Astroglia of the Mouse Cerebral Cortex. Nat. Protoc. 2011, 6, 214–228. [Google Scholar] [CrossRef]
- Papadimitriou, E.; Koutsoudaki, P.N.; Thanou, I.; Karagkouni, D.; Karamitros, T.; Chroni-Tzartou, D.; Gaitanou, M.; Gkemisis, C.; Margariti, M.; Xingi, E.; et al. A miR-124-Mediated Post-Transcriptional Mechanism Controlling the Cell Fate Switch of Astrocytes to Induced Neurons. Stem Cell Rep. 2023, 18, 915–935. [Google Scholar] [CrossRef]
- Qian, H.; Kang, X.; Hu, J.; Zhang, D.; Liang, Z.; Meng, F.; Zhang, X.; Xue, Y.; Maimon, R.; Dowdy, S.F.; et al. Reversing a Model of Parkinson’s Disease with In Situ Converted Nigral Neurons. Nature 2020, 582, 550–556. [Google Scholar] [CrossRef] [PubMed]
- Spassky, N.; Meunier, A. The Development and Functions of Multiciliated Epithelia. Nat. Rev. Mol. Cell Biol. 2017, 18, 423–436. [Google Scholar] [CrossRef] [PubMed]
- Llorens-Bobadilla, E.; Chell, J.M.; Le Merre, P.; Wu, Y.; Zamboni, M.; Bergenstråhle, J.; Stenudd, M.; Sopova, E.; Lundeberg, J.; Shupliakov, O.; et al. A Latent Lineage Potential in Resident Neural Stem Cells Enables Spinal Cord Repair. Science 2020, 370, eabb8795. [Google Scholar] [CrossRef]
- Stenudd, M.; Sabelström, H.; Llorens-Bobadilla, E.; Zamboni, M.; Blom, H.; Brismar, H.; Zhang, S.; Basak, O.; Clevers, H.; Göritz, C.; et al. Identification of a Discrete Subpopulation of Spinal Cord Ependymal Cells with Neural Stem Cell Properties. Cell Rep. 2022, 38, 110440. [Google Scholar] [CrossRef]
- Kazanis, I.; Ffrench-Constant, C. The Number of Stem Cells in the Subependymal Zone of the Adult Rodent Brain Is Correlated with the Number of Ependymal Cells and Not with the Volume of the Niche. Stem Cells Dev. 2012, 21, 1090–1096. [Google Scholar] [CrossRef]
- Sawamoto, K.; Wichterle, H.; Gonzalez-Perez, O.; Cholfin, J.A.; Yamada, M.; Spassky, N.; Murcia, N.S.; Garcia-Verdugo, J.M.; Marin, O.; Rubenstein, J.L.R.; et al. New Neurons Follow the Flow of Cerebrospinal Fluid in the Adult Brain. Science 2006, 311, 629–632. [Google Scholar] [CrossRef]
- Young, C.C.; van der Harg, J.M.; Lewis, N.J.; Brooks, K.J.; Buchan, A.M.; Szele, F.G. Ependymal Ciliary Dysfunction and Reactive Astrocytosis in a Reorganized Subventricular Zone after Stroke. Cereb. Cortex 2013, 23, 647–659. [Google Scholar] [CrossRef]
- Dimitrakopoulos, D.; Dimitriou, C.; McClenahan, F.; Franklin, R.J.M.; Kazanis, I. Author Spotlight: Collecting Neural Stem and Progenitor Cells from Live Animals Using a Novel Brain Milking Protocol. JoVE J. Vis. Exp. 2024, 204, e65308. [Google Scholar] [CrossRef]
- McClenahan, F.; Dimitriou, C.; Koutsakis, C.; Dimitrakopoulos, D.; Arampatzis, A.; Kakouri, P.; Kourla, M.; Oikonomou, S.; Andreopoulou, E.; Patsonis, M.; et al. Isolation of Neural Stem and Oligodendrocyte Progenitor Cells from the Brain of Live Rats. Stem Cell Rep. 2021, 16, 2534–2547. [Google Scholar] [CrossRef]
- Bernal, A.; Arranz, L. Nestin-Expressing Progenitor Cells: Function, Identity and Therapeutic Implications. Cell Mol. Life Sci. 2018, 75, 2177–2195. [Google Scholar] [CrossRef]
- Suzuki, S.; Namiki, J.; Shibata, S.; Mastuzaki, Y.; Okano, H. The Neural Stem/Progenitor Cell Marker Nestin Is Expressed in Proliferative Endothelial Cells, but Not in Mature Vasculature. J. Histochem. Cytochem. 2010, 58, 721–730. [Google Scholar] [CrossRef]
- Graham, V.; Khudyakov, J.; Ellis, P.; Pevny, L. SOX2 Functions to Maintain Neural Progenitor Identity. Neuron 2003, 39, 749–765. [Google Scholar] [CrossRef]
- Kazanis, I.; Evans, K.A.; Andreopoulou, E.; Dimitriou, C.; Koutsakis, C.; Karadottir, R.T.; Franklin, R.J.M. Subependymal Zone-Derived Oligodendroblasts Respond to Focal Demyelination but Fail to Generate Myelin in Young and Aged Mice. Stem Cell Rep. 2017, 8, 685–700. [Google Scholar] [CrossRef]
- Wu, Y.; Chen, X.; Xi, G.; Zhou, X.; Pan, S.; Ying, Q.-L. Long-Term Self-Renewal of Naïve Neural Stem Cells in a Defined Condition. Biochim. Biophys. Acta BBA Mol. Cell Res. 2019, 1866, 971–977. [Google Scholar] [CrossRef]
- Codega, P.; Silva-Vargas, V.; Paul, A.; Maldonado-Soto, A.R.; DeLeo, A.M.; Pastrana, E.; Doetsch, F. Prospective Identification and Purification of Quiescent Adult Neural Stem Cells from Their In Vivo Niche. Neuron 2014, 82, 545–559. [Google Scholar] [CrossRef]
- Luo, J.; Shook, B.A.; Daniels, S.B.; Conover, J.C. Subventricular Zone-Mediated Ependyma Repair in the Adult Mammalian Brain. J. Neurosci. 2008, 28, 3804–3813. [Google Scholar] [CrossRef]
- Oliveira, J.F.; Sardinha, V.M.; Guerra-Gomes, S.; Araque, A.; Sousa, N. Do Stars Govern Our Actions? Astrocyte Involvement in Rodent Behavior. Trends Neurosci. 2015, 38, 535–549. [Google Scholar] [CrossRef]
- Alvarez-Buylla, A.; García-Verdugo, J.M.; Tramontin, A.D. A Unified Hypothesis on the Lineage of Neural Stem Cells. Nat. Rev. Neurosci. 2001, 2, 287–293. [Google Scholar] [CrossRef]
- Heinrich, C.; Spagnoli, F.M.; Berninger, B. In Vivo Reprogramming for Tissue Repair. Nat. Cell Biol. 2015, 17, 204–211. [Google Scholar] [CrossRef]
- Lange, S.C.; Bak, L.K.; Waagepetersen, H.S.; Schousboe, A.; Norenberg, M.D. Primary Cultures of Astrocytes: Their Value in Understanding Astrocytes in Health and Disease. Neurochem. Res. 2012, 37, 2569–2588. [Google Scholar] [CrossRef]
- Sharif, N.; Calzolari, F.; Berninger, B. Direct In Vitro Reprogramming of Astrocytes into Induced Neurons. In Neural Reprogramming: Methods and Protocols; Ahlenius, H., Ed.; Methods in Molecular Biology; Springer: New York, NY, USA, 2021; pp. 13–29. ISBN 978-1-07-161601-7. [Google Scholar]
- Willaime-Morawek, S.; Seaberg, R.M.; Batista, C.; Labbé, E.; Attisano, L.; Gorski, J.A.; Jones, K.R.; Kam, A.; Morshead, C.M.; van der Kooy, D. Embryonic Cortical Neural Stem Cells Migrate Ventrally and Persist as Postnatal Striatal Stem Cells. J. Cell Biol. 2006, 175, 159–168. [Google Scholar] [CrossRef]
- Song, B.; Feldmann, J.W.; Cao, S.; Feitosa, M.; Kong, Y.; Kim, W.; Schweitzer, A.; Leblanc, P.; Schweitzer, J.S.; Kim, K.-S. A Pitx3-Deficient Developmental Mouse Model for Fine Motor, Olfactory, and Gastrointestinal Symptoms of Parkinson’s Disease. Neurobiol. Dis. 2022, 170, 105777. [Google Scholar] [CrossRef]
- Mourtzi, T.; Dimitrakopoulos, D.; Kakogiannis, D.; Salodimitris, C.; Botsakis, K.; Meri, D.K.; Anesti, M.; Dimopoulou, A.; Charalampopoulos, I.; Gravanis, A.; et al. Characterization of Substantia Nigra Neurogenesis in Homeostasis and Dopaminergic Degeneration: Beneficial Effects of the Microneurotrophin BNN-20. Stem Cell Res. Ther. 2021, 12, 335. [Google Scholar] [CrossRef] [PubMed]
- Gajera, C.R.; Emich, H.; Lioubinski, O.; Christ, A.; Beckervordersandforth-Bonk, R.; Yoshikawa, K.; Bachmann, S.; Christensen, E.I.; Götz, M.; Kempermann, G.; et al. LRP2 in Ependymal Cells Regulates BMP Signaling in the Adult Neurogenic Niche. J. Cell Sci. 2010, 123, 1922–1930. [Google Scholar] [CrossRef] [PubMed]
- Dulken, B.W.; Leeman, D.S.; Boutet, S.C.; Hebestreit, K.; Brunet, A. Single-Cell Transcriptomic Analysis Defines Heterogeneity and Transcriptional Dynamics in the Adult Neural Stem Cell Lineage. Cell Rep. 2017, 18, 777–790. [Google Scholar] [CrossRef] [PubMed]
- Götz, M.; Sirko, S.; Beckers, J.; Irmler, M. Reactive Astrocytes as Neural Stem or Progenitor Cells: In Vivo Lineage, In Vitro Potential, and Genome-Wide Expression Analysis. Glia 2015, 63, 1452–1468. [Google Scholar] [CrossRef] [PubMed]
- Calzolari, F.; Michel, J.; Baumgart, E.V.; Theis, F.; Götz, M.; Ninkovic, J. Fast Clonal Expansion and Limited Neural Stem Cell Self-Renewal in the Adult Subependymal Zone. Nat. Neurosci. 2015, 18, 490–492. [Google Scholar] [CrossRef] [PubMed]
- Obernier, K.; Cebrian-Silla, A.; Thomson, M.; Parraguez, J.I.; Anderson, R.; Guinto, C.; Rodriguez, J.R.; Garcia-Verdugo, J.-M.; Alvarez-Buylla, A. Adult Neurogenesis Is Sustained by Symmetric Self-Renewal and Differentiation. Cell Stem Cell 2018, 22, 221–234.e8. [Google Scholar] [CrossRef] [PubMed]
- Delgado, A.C.; Maldonado-Soto, A.R.; Silva-Vargas, V.; Mizrak, D.; von Känel, T.; Tan, K.R.; Paul, A.; Madar, A.; Cuervo, H.; Kitajewski, J.; et al. Release of Stem Cells from Quiescence Reveals Gliogenic Domains in the Adult Mouse Brain. Science 2021, 372, 1205–1209. [Google Scholar] [CrossRef] [PubMed]
- Marqués-Torrejón, M.Á.; Williams, C.A.C.; Southgate, B.; Alfazema, N.; Clements, M.P.; Garcia-Diaz, C.; Blin, C.; Arranz-Emparan, N.; Fraser, J.; Gammoh, N.; et al. LRIG1 Is a Gatekeeper to Exit from Quiescence in Adult Neural Stem Cells. Nat. Commun. 2021, 12, 2594. [Google Scholar] [CrossRef] [PubMed]
- Machado, L.; Esteves De Lima, J.; Fabre, O.; Proux, C.; Legendre, R.; Szegedi, A.; Varet, H.; Ingerslev, L.R.; Barrès, R.; Relaix, F.; et al. In Situ Fixation Redefines Quiescence and Early Activation of Skeletal Muscle Stem Cells. Cell Rep. 2017, 21, 1982–1993. [Google Scholar] [CrossRef]
- van Velthoven, C.T.J.; Rando, T.A. Stem Cell Quiescence: Dynamism, Restraint, and Cellular Idling. Cell Stem Cell 2019, 24, 213–225. [Google Scholar] [CrossRef]
- Willaime-Morawek, S.; Van Der Kooy, D. Cortex- and Striatum- Derived Neural Stem Cells Produce Distinct Progeny in the Olfactory Bulb and Striatum. Eur. J. Neurosci. 2008, 27, 2354–2362. [Google Scholar] [CrossRef] [PubMed]
- Anesti, M.; Magkafa, S.; Prantikou, E.; Kazanis, I. Divergence between Neuronal and Oligodendroglial Cell Fate, in Postnatal Brain Neural Stem Cells, Leads to Divergent Properties in Polymorphic In Vitro Assays. Cells 2022, 11, 1743. [Google Scholar] [CrossRef] [PubMed]
- Lalioti, M.-E.; Kaplani, K.; Lokka, G.; Georgomanolis, T.; Kyrousi, C.; Dong, W.; Dunbar, A.; Parlapani, E.; Damianidou, E.; Spassky, N.; et al. GemC1 Is a Critical Switch for Neural Stem Cell Generation in the Postnatal Brain. Glia 2019, 67, 2360–2373. [Google Scholar] [CrossRef] [PubMed]
- Douet, V.; Kerever, A.; Arikawa-Hirasawa, E.; Mercier, F. Fractone-Heparan Sulphates Mediate FGF-2 Stimulation of Cell Proliferation in the Adult Subventricular Zone. Cell Prolif. 2013, 46, 137–145. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Van Gulden, S.; McGuire, T.L.; Fleming, A.C.; Oka, C.; Kessler, J.A.; Peng, C.-Y. BMP-Responsive Protease HtrA1 Is Differentially Expressed in Astrocytes and Regulates Astrocytic Development and Injury Response. J. Neurosci. 2018, 38, 3840–3857. [Google Scholar] [CrossRef] [PubMed]
- Lin, R.; Cai, J.; Kenyon, L.; Iozzo, R.; Rosenwasser, R.; Iacovitti, L. Systemic Factors Trigger Vasculature Cells to Drive Notch Signaling and Neurogenesis in Neural Stem Cells in the Adult Brain. Stem Cells 2019, 37, 395–406. [Google Scholar] [CrossRef] [PubMed]
- Sanai, N.; Tramontin, A.D.; Quiñones-Hinojosa, A.; Barbaro, N.M.; Gupta, N.; Kunwar, S.; Lawton, M.T.; McDermott, M.W.; Parsa, A.T.; Manuel-García Verdugo, J.; et al. Unique Astrocyte Ribbon in Adult Human Brain Contains Neural Stem Cells but Lacks Chain Migration. Nature 2004, 427, 740–744. [Google Scholar] [CrossRef]
- Guan, J.; Wang, G.; Wang, J.; Zhang, Z.; Fu, Y.; Cheng, L.; Meng, G.; Lyu, Y.; Zhu, J.; Li, Y.; et al. Chemical Reprogramming of Human Somatic Cells to Pluripotent Stem Cells. Nature 2022, 605, 325–331. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).