Biological Implications and Functional Significance of Transglutaminase Type 2 in Nervous System Tumors
Abstract
:1. Introduction
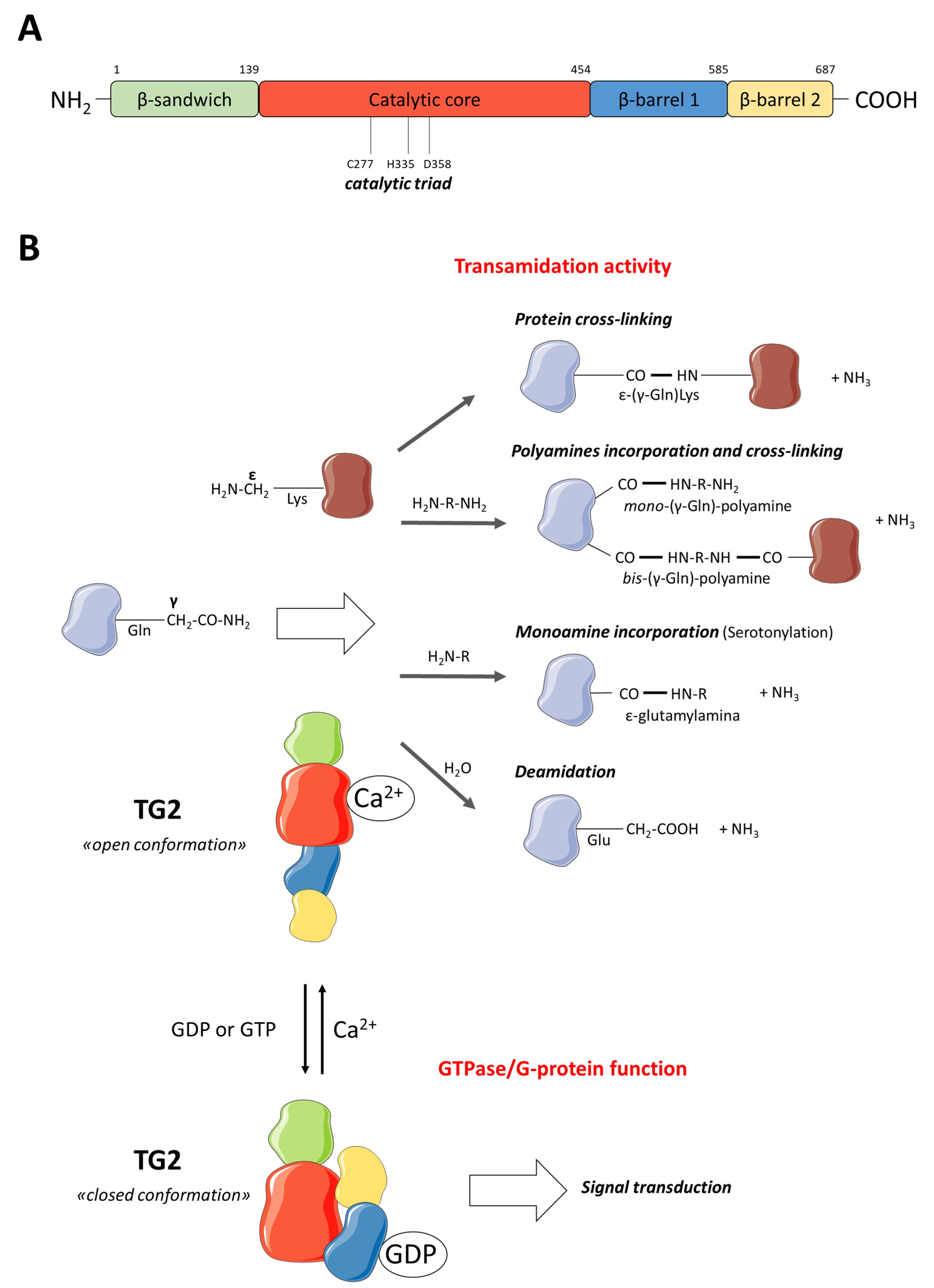
2. TG2 Description
2.1. TG2 Function
2.2. TG2 Gene Regulation
3. Physiological Role of TG2 in Nervous System
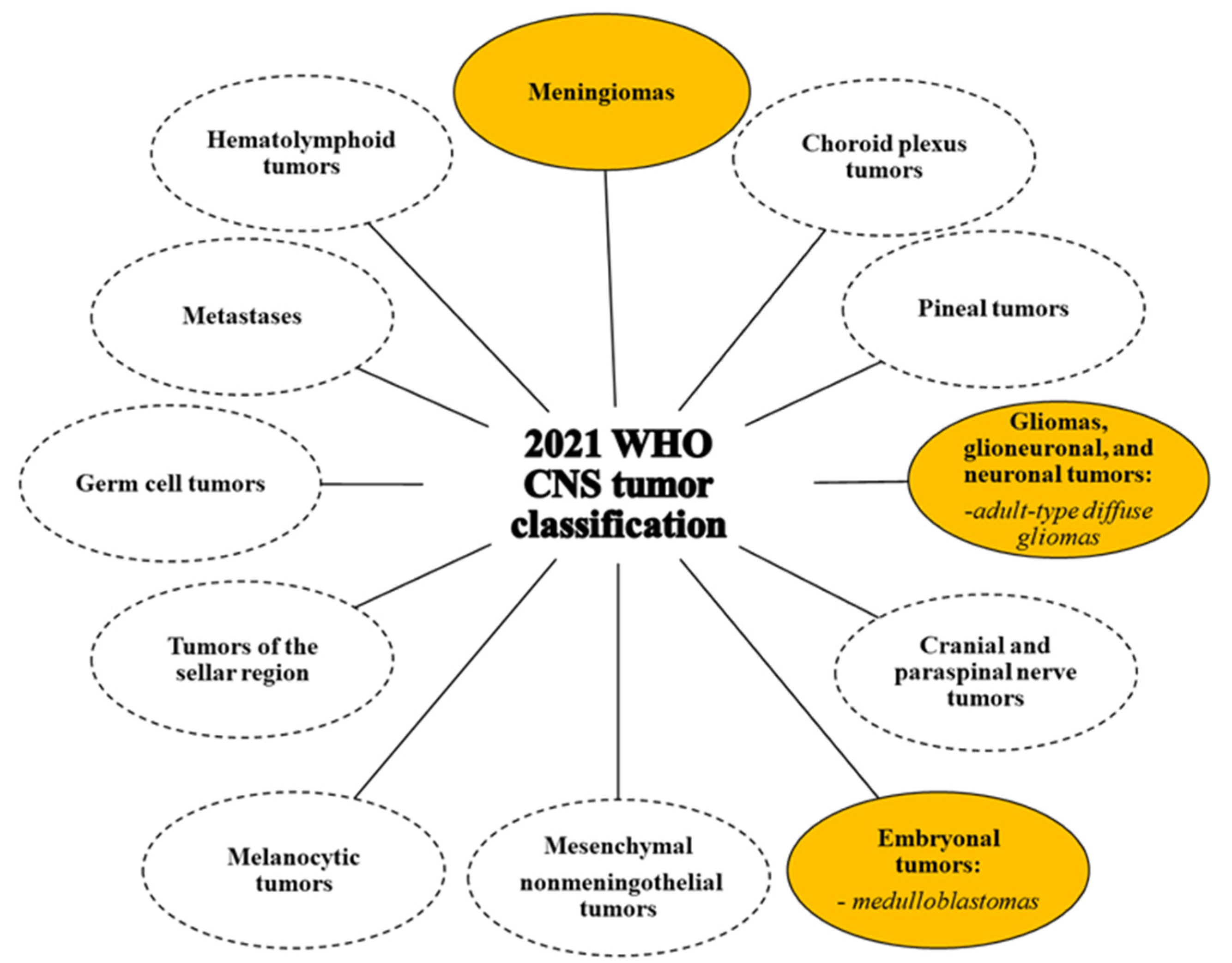
4. Nervous System Tumors: Classification and Therapy
4.1. Peripheral Nervous System Tumors
4.2. Central Nervous System Tumours
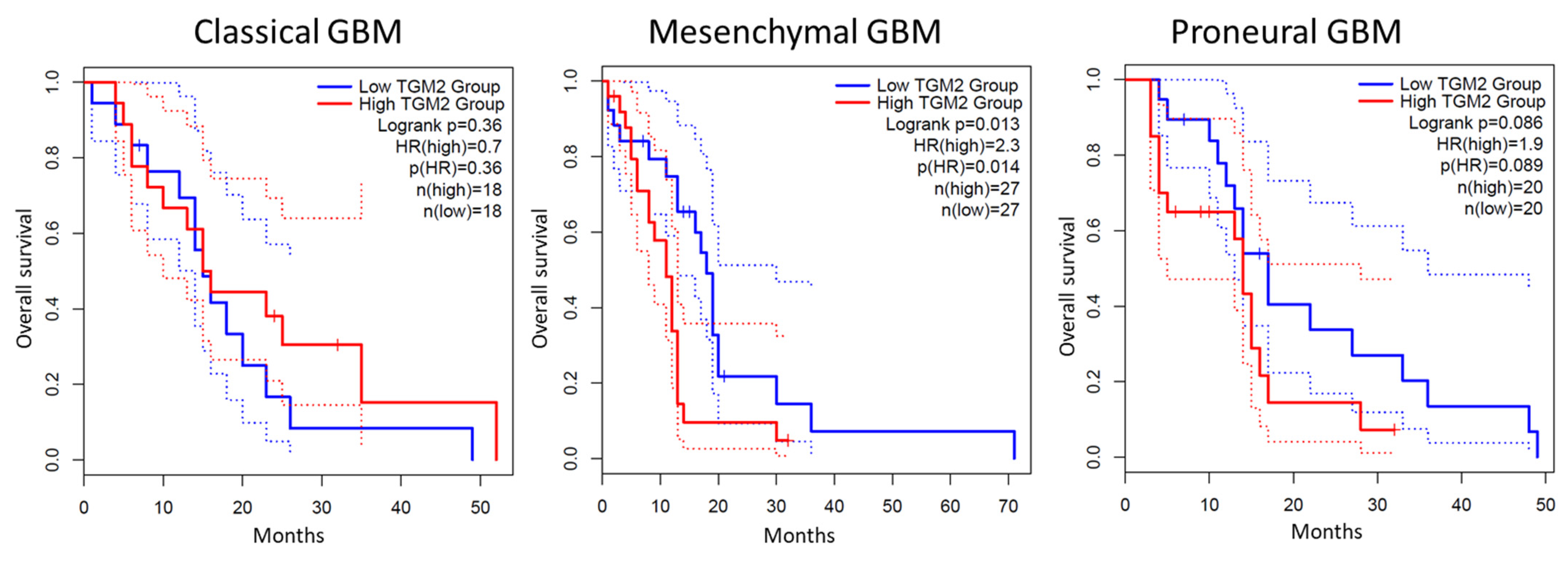
5. TG2 in Adult-Type Diffuse Gliomas
6. TG2 in Other CNS Tumors
7. TG2 in Neuroblastoma
8. Discussion and Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Liu, S.; Cerione, R.A.; Clardy, J. Structural basis for the guanine nucleotide-binding activity of tissue transglutaminase and its regulation of transamidation activity. Proc. Natl. Acad. Sci. USA 2002, 99, 2743–2747. [Google Scholar] [CrossRef]
- Savoca, M.P.; Tonoli, E.; Atobatele, A.G.; Verderio, E.A.M. Biocatalysis by Transglutaminases: A Review of Biotechnological Applications. Micromachines 2018, 9, 562. [Google Scholar] [CrossRef]
- Nurminskaya, M.V.; Belkin, A.M. Cellular functions of tissue transglutaminase. Int. Rev. Cell. Mol. Biol. 2012, 294, 1–97. [Google Scholar] [CrossRef]
- Maggio, N.; Sellitti, S.; Capano, C.P.; Papa, M. Tissue-transglutaminase in rat and human brain: Light and electron immunocytochemical analysis and in situ hybridization study. Brain Res. Bull. 2001, 56, 173–182. [Google Scholar] [CrossRef] [PubMed]
- Ruan, Q.; Johnson, G.V. Transglutaminase 2 in neurodegenerative disorders. Front. Biosci. 2007, 12, 891–904. [Google Scholar] [CrossRef]
- Yunes-Medina, L.; Feola, J.; Johnson, G.V.W. Subcellular localization patterns of transglutaminase 2 in astrocytes and neurons are differentially altered by hypoxia. Neuroreport 2017, 28, 1208–1214. [Google Scholar] [CrossRef] [PubMed]
- Rudlong, J.; Cheng, A.; Johnson, G.V.W. The role of transglutaminase 2 in mediating glial cell function and pathophysiology in the central nervous system. Anal. Biochem. 2020, 591, 113556. [Google Scholar] [CrossRef]
- Hand, D.; Perry, M.J.; Haynes, L.W. Cellular transglutaminases in neural development. Int. J. Dev. Neurosci. 1993, 11, 709–720. [Google Scholar] [CrossRef]
- Festoff, B.W.; Suo, Z.; Citron, B.A. Plasticity and stabilization of neuromuscular and CNS synapses: Interactions between thrombin protease signaling pathways and tissue transglutaminase. Int. Rev. Cytol. 2001, 211, 153–177. [Google Scholar] [CrossRef]
- Min, B.; Chung, K.C. New insight into transglutaminase 2 and link to neurodegenerative diseases. BMB Rep. 2018, 51, 5–13. [Google Scholar] [CrossRef]
- Keillor, J.W.; Johnson, G.V.W. Transglutaminase 2 as a therapeutic target for neurological conditions. Expert Opin. Ther. Targets 2021, 25, 721–731. [Google Scholar] [CrossRef] [PubMed]
- Basso, M.; Ratan, R.R. Transglutaminase is a therapeutic target for oxidative stress, excitotoxicity and stroke: A new epigenetic kid on the CNS block. J. Cereb. Blood Flow Metab. 2013, 33, 809–818. [Google Scholar] [CrossRef] [PubMed]
- Wilhelmus, M.M.; de Jager, M.; Bakker, E.N.; Drukarch, B. Tissue transglutaminase in Alzheimer’s disease: Involvement in pathogenesis and its potential as a therapeutic target. J. Alzheimers Dis. 2014, 42, S289–S303. [Google Scholar] [CrossRef]
- Verhaar, R.; Jongenelen, C.A.; Gerard, M.; Baekelandt, V.; Van Dam, A.M.; Wilhelmus, M.M.; Drukarch, B. Blockade of enzyme activity inhibits tissue transglutaminase-mediated transamidation of α-synuclein in a cellular model of Parkinson’s disease. Neurochem. Int. 2011, 58, 785–793. [Google Scholar] [CrossRef] [PubMed]
- Altuntas, S.; D’Eletto, M.; Rossin, F.; Hidalgo, L.D.; Farrace, M.G.; Falasca, L.; Piredda, L.; Cocco, S.; Mastroberardino, P.G.; Piacentini, M.; et al. Type 2 Transglutaminase, mitochondria and Huntington’s disease: Menage a trois. Mitochondrion 2014, 19, 97–104. [Google Scholar] [CrossRef]
- Lentini, A.; Abbruzzese, A.; Provenzano, B.; Tabolacci, C.; Beninati, S. Transglutaminases: Key regulators of cancer metastasis. Amino Acids 2013, 44, 25–32. [Google Scholar] [CrossRef] [PubMed]
- Eckert, R.L.; Fisher, M.L.; Grun, D.; Adhikary, G.; Xu, W.; Kerr, C. Transglutaminase is a tumor cell and cancer stem cell survival factor. Mol. Carcinog. 2015, 54, 947–958. [Google Scholar] [CrossRef] [PubMed]
- Sima, L.E.; Matei, D.; Condello, S. The Outside-In Journey of Tissue Transglutaminase in Cancer. Cells 2022, 11, 1779. [Google Scholar] [CrossRef]
- Tunici, P.; Sessa, A.; Rabellotti, E.; Calloni, A.; Perin, A. Distribution and activity of transglutaminase in rat brain carcinogenesis and in gliomas. Cancer Lett. 1999, 140, 47–51. [Google Scholar] [CrossRef]
- Iwaki, T.; Miyazono, M.; Hitotsumatsu, T.; Tateishi, J. An Immunohistochemical Study of Tissue Transglutaminase in Gliomas with Reference to Their Cell Dying Processes. Am. J. Pathol. 1994, 145, 776–781. [Google Scholar]
- Hilton, D.A.; Love, S.; Barber, R. Increased endothelial expression of transglutaminase in glioblastomas. Neuropathol. Appl. Neurobiol. 1997, 23, 507–511. [Google Scholar] [CrossRef] [PubMed]
- Yuan, L.; Behdad, A.; Siegel, M.; Khosla, C.; Higashikubo, R.; Rich, K.M. Tissue transgluaminase 2 expression in meningiomas. J. Neurooncol. 2008, 2, 125–132. [Google Scholar] [CrossRef] [PubMed]
- Dyer, L.M.; Schooler, K.P.; Ai, L.; Klop, C.; Qiu, J.; Robertson, K.D.; Brown, K.D. The transglutaminase 2 gene is aberrantly hypermethylated in glioma. J. Neurooncol. 2011, 101, 429–440. [Google Scholar] [CrossRef] [PubMed]
- Fu, J.; Yang, Q.; Sai, K.; Chen, F.; Pang, J.C.S.; Ng, H.; Kwan, A.; Chen, Z. TGM2 inhibition attenuates ID1 expression in CD44-high glioma-initiating cells. Neuro-Oncology 2013, 10, 1353–1365. [Google Scholar] [CrossRef]
- Zhang, J.; Antonyak, M.A.; Singh, G.; Cerione, R.A. A novel mechanism for the up-regulation of EGF-receptor levels in glioblastomas. Cell Rep. 2013, 3, 2008–2020. [Google Scholar] [CrossRef] [PubMed]
- Fisher, J.L.; Schwartzbaum, J.A.; Wrensch, M.; Wiemels, J.L. Epidemiology of brain tumors. Neurol. Clin. 2007, 25, 867–890. [Google Scholar] [CrossRef]
- Li, S.; Wang, C.; Chen, J.; Lan, Y.; Zhang, W.; Kang, Z.; Zheng, Y.; Zhang, R.; Yu, J.; Li, W. Signaling pathways in brain tumors and therapeutic interventions. Signal Transduct. Target Ther. 2023, 8, 8. [Google Scholar] [CrossRef]
- Mohammed, S.; Dinesan, M.; Ajayakumar, T. Survival and quality of life analysis in glioblastoma multiforme with adjuvant chemoradiotherapy: A retrospective study. Rep. Pract. Oncol. Radiother. 2022, 27, 1026–1036. [Google Scholar] [CrossRef]
- Shaffiey, S.A.; Le, H.D.; Christison-Lagay, E.; Fialkowski, E.A.; Aldrink, J.H.; Grant, C.N.; Honeyman, J.N.; Janek, K.C.; Madonna, M.B.; Rhee, D.S.; et al. Critical elements of pediatric neuroblastoma surgery. Semin. Pediatr. Surg. 2023, 32, 151338. [Google Scholar] [CrossRef]
- Martins, I.M.; Matos, M.; Costa, R.; Silva, F.; Pascoal, A.; Estevinho, L.M.; Choupina, A.B. Transglutaminases: Recent achievements and new sources. Appl. Microbiol. Biotechnol. 2014, 98, 6957–6964. [Google Scholar] [CrossRef] [PubMed]
- Folk, J.E. Transglutaminases. Annu. Rev. Biochem. 1980, 49, 517–531. [Google Scholar] [CrossRef] [PubMed]
- Alshehri, F.S.M.; Whyte, C.S.; Mutch, N.J. Factor XIII-A: An Indispensable “Factor” in Haemostasis and Wound Healing. Int. J. Mol. Sci. 2021, 22, 3055. [Google Scholar] [CrossRef] [PubMed]
- Candi, E.; Oddi, S.; Terrinoni, A.; Paradisi, A.; Ranalli, M.; Finazzi-Agró, A.; Melino, G. Transglutaminase 5 cross-links loricrin, involucrin, and small proline-rich proteins in vitro. J. Biol. Chem. 2001, 276, 35014–35023. [Google Scholar] [CrossRef] [PubMed]
- Eckert, R.L.; Sturniolo, M.T.; Broome, A.M.; Ruse, M.; Rorke, E.A. Transglutaminase function in epidermis. J. Investig. Dermatol. 2005, 124, 481–492. [Google Scholar] [CrossRef] [PubMed]
- Ye, L.; Sanders, A.J.; Jiang, W.G. Transglutaminase-4 (Prostate Transglutaminase), a Potential Biological Factor and Clinical Indicator for the Diagnosis and Prognosis of Prostate Cancer. Anticancer Res. 2023, 43, 291–296. [Google Scholar] [CrossRef] [PubMed]
- Thomas, H.; Beck, K.; Adamczyk, M.; Aeschlimann, P.; Langley, M.; Oita, R.C.; Thiebach, L.; Hils, M.; Aeschlimann, D. Transglutaminase 6: A protein associated with central nervous system development and motor function. Amino Acids 2013, 44, 161–177. [Google Scholar] [CrossRef]
- Satchwell, T.J.; Shoemark, D.K.; Sessions, R.B.; Toye, A.M. Protein 4.2: A complex linker. Blood Cells Mol. Dis. 2009, 42, 201–210. [Google Scholar] [CrossRef] [PubMed]
- Gundemir, S.; Colak, G.; Tucholski, J.; Johnson, G.V. Transglutaminase 2: A molecular Swiss army knife. Biochim. Biophys. Acta 2012, 1823, 406–419. [Google Scholar] [CrossRef]
- Tatsukawa, H.; Furutani, Y.; Hitomi, K.; Kojima, S. Transglutaminase 2 has opposing roles in the regulation of cellular functions as well as cell growth and death. Cell Death Dis. 2016, 7, e2244. [Google Scholar] [CrossRef]
- Yao, Z.; Fan, Y.; Lin, L.; Kellems, R.E.; Xia, Y. Tissue Transglutaminase: A Multifunctional and Multisite Regulator in Health and Disease. Physiol. Rev. 2024, 104, 281–325. [Google Scholar] [CrossRef]
- Jang, T.H.; Lee, D.S.; Choi, K.; Jeong, E.M.; Kim, I.G.; Kim, Y.W.; Chun, J.N.; Jeon, J.H.; Park, H.H. Crystal structure of transglutaminase 2 with GTP complex and amino acid sequence evidence of evolution of GTP binding site. PLoS ONE 2014, 9, e107005. [Google Scholar] [CrossRef] [PubMed]
- Kim, G.E.; Park, H.H. Structures of Human Transglutaminase 2: Finding Clues for Interference in Cross-linking Mediated Activity. Int. J. Mol. Sci. 2020, 21, 2225. [Google Scholar] [CrossRef] [PubMed]
- Beninati, S.; Piacentini, M.; Cocuzzi, E.T.; Autuori, F.; Folk, J.E. Covalent incorporation of polyamines as gamma-glutamyl derivatives into CHO cell protein. Biochim. Biophys. Acta 1988, 952, 325–333. [Google Scholar] [CrossRef] [PubMed]
- Keillor, J.W.; Clouthier, C.M.; Apperley, K.Y.P.; Akbar, A.; Mulani, A. Acyl transfer mechanisms of tissue transglutaminase. Bioorg. Chem. 2014, 57, 186–197. [Google Scholar] [CrossRef] [PubMed]
- Caron, N.S.; Munsie, L.N.; Keillor, J.W.; Truant, R. Using FLIM-FRET to measure conformational changes of transglutaminase type 2 in live cells. PLoS ONE 2012, 7, e44159. [Google Scholar] [CrossRef]
- Chhabra, A.; Verma, A.; Mehta, K. Tissue transglutaminase promotes or suppresses tumors depending on cell context. Anticancer Res. 2009, 29, 1909–1919. [Google Scholar] [PubMed]
- Kerr, C.; Szmacinski, H.; Fisher, M.L.; Nance, B.; Lakowicz, J.R.; Akbar, A.; Keillor, J.W.; Lok Wong, T.; Godoy-Ruiz, R.; Toth, E.A.; et al. Transamidase site-targeted agents alter the conformation of the transglutaminase cancer stem cell survival protein to reduce GTP binding activity and cancer stem cell survival. Oncogene 2017, 36, 2981–2990. [Google Scholar] [CrossRef]
- Tabolacci, C.; De Martino, A.; Mischiati, C.; Feriotto, G.; Beninati, S. The Role of Tissue Transglutaminase in Cancer Cell Initiation, Survival and Progression. Med. Sci. 2019, 25, 19. [Google Scholar] [CrossRef]
- Mastroberardino, P.G.; Farrace, M.G.; Viti, I.; Pavone, F.; Fimia, G.M.; Melino, G.; Rodolfo, C.; Piacentini, M. “Tissue” transglutaminase contributes to the formation of disulphide bridges in proteins of mitochondrial respiratory complexes. Biochim. Biophys. Acta 2006, 1757, 1357–1365. [Google Scholar] [CrossRef]
- Zemskov, E.A.; Janiak, A.; Hang, J.; Waghray, A.; Belkin, A.M. The role of tissue transglutaminase in cell-matrix interactions. Front. Biosci. 2006, 11, 1057–1076. [Google Scholar] [CrossRef]
- Tatsukawa, H.; Hitomi, K. Role of Transglutaminase 2 in Cell Death, Survival, and Fibrosis. Cells 2021, 10, 1842. [Google Scholar] [CrossRef] [PubMed]
- Nagy, L.; Saydak, M.; Shipley, N.; Lu, S.; Basilion, J.P.; Yan, Z.H.; Syka, P.; Chandraratna, R.A.; Stein, J.P.; Heyman, R.A.; et al. Identification and characterization of a versatile retinoid response element (retinoic acid receptor response element-retinoid X receptor response element) in the mouse tissue transglutaminase gene promoter. J. Biol. Chem. 1996, 271, 4355–4365. [Google Scholar] [CrossRef]
- Lentini, A.; Provenzano, B.; Tabolacci, C.; Beninati, S. Protein-polyamine conjugates by transglutaminase 2 as potential markers for antineoplastic screening of natural compounds. Amino Acids 2009, 36, 701–708. [Google Scholar] [CrossRef] [PubMed]
- Brown, K.D. Transglutaminase 2 and NF-κB: An odd couple that shapes breast cancer phenotype. Breast Cancer Res. Treat. 2013, 137, 329–336. [Google Scholar] [CrossRef]
- Lee, J.; Kim, Y.S.; Choi, D.H.; Bang, M.S.; Han, T.R.; Joh, T.H.; Kim, S.Y. Transglutaminase 2 induces nuclear factor-kappaB activation via a novel pathway in BV-2 microglia. J. Biol. Chem. 2004, 279, 53725–53735. [Google Scholar] [CrossRef] [PubMed]
- Eckert, R.L.; Kaartinen, M.T.; Nurminskaya, M.; Belkin, A.M.; Colak, G.; Johnson, G.V.; Mehta, K. Transglutaminase regulation of cell function. Physiol. Rev. 2014, 94, 383–417. [Google Scholar] [CrossRef]
- Kuncio, G.S.; Tsyganskaya, M.; Zhu, J.; Liu, S.L.; Nagy, L.; Thomazy, V.; Davies, P.J.; Zern, M.A. TNF-alpha modulates expression of the tissue transglutaminase gene in liver cells. Am. J. Physiol. 1998, 274, G240–G245. [Google Scholar] [CrossRef] [PubMed]
- Jang, G.Y.; Jeon, J.H.; Cho, S.Y.; Shin, D.M.; Kim, C.W.; Jeong, E.M.; Bae, H.C.; Kim, T.W.; Lee, S.H.; Choi, Y.; et al. Transglutaminase 2 suppresses apoptosis by modulating caspase 3 and NF-kappaB activity in hypoxic tumor cells. Oncogene 2010, 29, 356–367. [Google Scholar] [CrossRef]
- Tabolacci, C.; Lentini, A.; Provenzano, B.; Beninati, S. Evidences for a role of protein cross-links in transglutaminase-related disease. Amino Acids 2012, 42, 975–986. [Google Scholar] [CrossRef]
- Phatak, V.; Croft, S.; Setty, S.R.; Scarpellini, A.; Hughes, D.; Rees, R.; McArdle, S.; Verderio, E. Expression of transglutaminase-2 isoforms in normal human tissues and cancer cell lines: Dysregulation of alternative splicing in cancer. Amino Acids 2013, 44, 33–44. [Google Scholar] [CrossRef] [PubMed]
- Gentile, V.; Saydak, M.; Chiocca, E.A.; Akande, O.; Birckbichler, P.J.; Lee, K.N.; Stein, J.P.; Davies, P.J. Isolation and characterization of cDNA clones to mouse macrophage and human endothelial cell tissue transglutaminases. J. Biol. Chem. 1991, 266, 478–483. [Google Scholar] [CrossRef] [PubMed]
- Bianchi, N.; Beninati, S.; Bergamini, C.M. Spotlight on the transglutaminase 2 gene: A focus on genomic and transcriptional aspects. Biochem. J. 2018, 475, 1643–1667. [Google Scholar] [CrossRef] [PubMed]
- Fraij, B.M.; Birckbichler, P.J.; Patterson, M.K., Jr.; Lee, K.N.; Gonzales, R.A. A retinoic acid-inducible mRNA from human erythroleukemia cells encodes a novel tissue transglutaminase homologue. J. Biol. Chem. 1992, 267, 22616–22623. [Google Scholar] [CrossRef] [PubMed]
- Citron, B.A.; Suo, Z.; SantaCruz, K.; Davies, P.J.; Qin, F.; Festoff, B.W. Protein crosslinking, tissue transglutaminase, alternative splicing and neurodegeneration. Neurochem. Int. 2002, 40, 69–78. [Google Scholar] [CrossRef] [PubMed]
- Fraij, B.M.; Gonzales, R.A. A third human tissue transglutaminase homologue as a result of alternative gene transcripts. Biochim. Biophys. Acta 1996, 1306, 63–74. [Google Scholar] [CrossRef] [PubMed]
- Arbildi, P.; Calvo, F.; Macías, V.; Rodríguez-Camejo, C.; Sóñora, C.; Hernández, A. Study of tissue transglutaminase spliced variants expressed in THP-1 derived macrophages exhibiting distinct functional phenotypes. Immunobiology 2023, 228, 152752. [Google Scholar] [CrossRef] [PubMed]
- Lai, T.S.; Liu, Y.; Li, W.; Greenberg, C.S. Identification of two GTP-independent alternatively spliced forms of tissue transglutaminase in human leukocytes, vascular smooth muscle, and endothelial cells. FASEB J. 2007, 21, 4131–4143. [Google Scholar] [CrossRef] [PubMed]
- Sestito, C.; Brevé, J.J.P.; Killestein, J.; Teunissen, C.E.; Wilhelmus, M.M.M.; Drukarch, B.; van Dam, A.M. Differential Expression of Tissue Transglutaminase Splice Variants in Peripheral Blood Mononuclear Cells of Primary Progressive Multiple Sclerosis Patients. Med. Sci. 2018, 6, 108. [Google Scholar] [CrossRef] [PubMed]
- Minotti, L.; Baldassari, F.; Galasso, M.; Volinia, S.; Bergamini, C.M.; Bianchi, N. A long non-coding RNA inside the type 2 transglutaminase gene tightly correlates with the expression of its transcriptional variants. Amino Acids 2018, 50, 421–438. [Google Scholar] [CrossRef] [PubMed]
- Franzese, O.; Minotti, L.; Aguiari, G.; Corrà, F.; Cervellati, C.; Ferrari, C.; Volinia, S.; Bergamini, C.; Bianchi, N. Involvement of non-coding RNAs and transcription factors in the induction of Transglutaminase isoforms by ATRA. Amino Acids 2019, 51, 1273–1288. [Google Scholar] [CrossRef]
- Bergamini, C.M.; Vischioni, C.; Aguiari, G.; Grandi, C.; Terrazzan, A.; Volinia, S.; Bianchi, N.; Taccioli, C. Inhibition of the lncRNA Coded within Transglutaminase 2 Gene Impacts Several Relevant Networks in MCF-7 Breast Cancer Cells. Noncoding RNA 2021, 7, 49. [Google Scholar] [CrossRef] [PubMed]
- Aguiari, G.; Crudele, F.; Taccioli, C.; Minotti, L.; Corrà, F.; Keillor, J.W.; Grassilli, S.; Cervellati, C.; Volinia, S.; Bergamini, C.M.; et al. Dysregulation of Transglutaminase type 2 through GATA3 defines aggressiveness and Doxorubicin sensitivity in breast cancer. Int. J. Biol. Sci. 2022, 18, 1–14. [Google Scholar] [CrossRef] [PubMed]
- González-Moro, I.; Rojas-Márquez, H.; Sebastian-delaCruz, M.; Mentxaka-Salgado, J.; Olazagoitia-Garmendia, A.; Mendoza, L.M.; Lluch, A.; Fantuzzi, F.; Lambert, C.; Ares Blanco, J.; et al. A long non-coding RNA that harbors a SNP associated with type 2 diabetes regulates the expression of TGM2 gene in pancreatic beta cells. Front. Endocrinol. 2023, 14, 1101934. [Google Scholar] [CrossRef] [PubMed]
- Piacentini, M.; D’Eletto, M.; Farrace, M.G.; Rodolfo, C.; Del Nonno, F.; Ippolito, G.; Falasca, L. Characterization of distinct sub-cellular location of transglutaminase type II: Changes in intracellular distribution in physiological and pathological states. Cell Tissue Res. 2014, 358, 793–805. [Google Scholar] [CrossRef] [PubMed]
- Akimov, S.S.; Krylov, D.; Fleischman, L.F.; Belkin, A.M. Tissue transglutaminase is an integrin-binding adhesion coreceptor for fibronectin. J. Cell Biol. 2000, 148, 825–838. [Google Scholar] [CrossRef] [PubMed]
- Ji, C.; Tang, M.; Harrison, J.; Paciorkowski, A.; Johnson, G.V.W. Nuclear transglutaminase 2 directly regulates expression of cathepsin S in rat cortical neurons. Eur. J. Neurosci. 2018, 48, 3043–3051. [Google Scholar] [CrossRef]
- Basso, M.; Berlin, J.; Xia, L.; Sleiman, S.F.; Ko, B.; Haskew-Layton, R.; Kim, E.; Antonyak, M.A.; Cerione, R.A.; Iismaa, S.E.; et al. Transglutaminase inhibition protects against oxidative stress-induced neuronal death downstream of pathological ERK activation. J. Neurosci. 2012, 32, 6561–6569. [Google Scholar] [CrossRef] [PubMed]
- Citron, B.A.; Gregory, E.J.; Steigerwalt, D.S.; Qin, F.; Festoff, B.W. Regulation of the dual function tissue transglutaminase/Galpha(h) during murine neuromuscular development: Gene and enzyme isoform expression. Neurochem. Int. 2000, 37, 337–349. [Google Scholar] [CrossRef]
- Hand, D.; Campoy, F.J.; Clark, S.; Fisher, A.; Haynes, L.W. Activity and distribution of tissue transglutaminase in association with nerve-muscle synapses. J. Neurochem. 1993, 61, 1064–1072. [Google Scholar] [CrossRef] [PubMed]
- Ashton, A.C.; Dolly, J.O. Microtubules and microfilaments participate in the inhibition of synaptosomal noradrenaline release by tetanus toxin. J. Neurochem. 1997, 68, 649–658. [Google Scholar] [CrossRef]
- Facchiano, F.; Deloye, F.; Doussau, F.; Innamorati, G.; Ashton, A.C.; Dolly, J.O.; Beninati, S.; Facchiano, A.; Luini, A.; Poulain, B.; et al. Transglutaminase participates in the blockade of neurotransmitter release by tetanus toxin: Evidence for a novel biological function. Amino Acids 2010, 39, 257–269. [Google Scholar] [CrossRef]
- Hummerich, R.; Schloss, P. Serotonin--more than a neurotransmitter: Transglutaminase-mediated serotonylation of C6 glioma cells and fibronectin. Neurochem. Int. 2010, 57, 67–75. [Google Scholar] [CrossRef]
- Facchiano, F.; Facchiano, A.; Facchiano, A.M. The role of transglutaminase-2 and its substrates in human diseases. Front. Biosci. 2006, 11, 1758–1773. [Google Scholar] [CrossRef] [PubMed]
- Achyuthan, K.E.; Greenberg, C.S. Identification of a guanosine triphosphate-binding site on guinea pig liver transglutaminase. Role of GTP and calcium ions in modulating activity. J. Biol. Chem. 1987, 262, 1901–1906. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Tucholski, J.; Lesort, M.; Jope, R.S.; Johnson, G.V. Novel bimodal effects of the G-protein tissue transglutaminase on adrenoreceptor signalling. Biochem. J. 1999, 343, 541–549. [Google Scholar] [CrossRef] [PubMed]
- Şahin, M.; Öncü, G.; Yılmaz, M.A.; Özkan, D.; Saybaşılı, H. Transformation of SH-SY5Y cell line into neuron-like cells: Investigation of electrophysiological and biomechanical changes. Neurosci. Lett. 2021, 745, 135628. [Google Scholar] [CrossRef]
- Quinn, B.R.; Yunes-Medina, L.; Johnson, G.V.W. Transglutaminase 2: Friend or foe? The discordant role in neurons and astrocytes. J. Neurosci. Res. 2018, 96, 1150–1158. [Google Scholar] [CrossRef] [PubMed]
- Eitan, S.; Schwartz, M. A transglutaminase that converts interleukin-2 into a factor cytotoxic to oligodendrocytes. Science 1993, 261, 106–108. [Google Scholar] [CrossRef] [PubMed]
- Espitia Pinzon, N.; van Mierlo, H.; de Jonge, J.C.; Brevé, J.J.P.; Bol, J.G.J.M.; Drukarch, B.; van Dam, A.M.; Baron, W. Tissue Transglutaminase Promotes Early Differentiation of Oligodendrocyte Progenitor Cells. Front. Cell Neurosci. 2019, 13, 281. [Google Scholar] [CrossRef]
- Fesus, L. Transglutaminase-catalyzed protein cross-linking in the molecular program of apoptosis and its relationship to neuronal processes. Cell Mol. Neurobiol. 1998, 18, 683–694. [Google Scholar] [CrossRef]
- Sarnelli, G.; De Giorgio, R.; Gentile, F.; Calì, G.; Grandone, I.; Rocco, A.; Cosenza, V.; Cuomo, R.; D’Argenio, G. Myenteric neuronal loss in rats with experimental colitis: Role of tissue transglutaminase-induced apoptosis. Dig. Liver Dis. 2009, 41, 185–193. [Google Scholar] [CrossRef] [PubMed]
- Basso, M.; Milelli, A. Transglutaminases, neuronal cell death and neural repair: Implications for traumatic brain injury and therapeutics. Curr. Opin. Neurol. 2019, 32, 796–801. [Google Scholar] [CrossRef] [PubMed]
- Malorni, W.; Farrace, M.G.; Rodolfo, C.; Piacentini, M. Type 2 transglutaminase in neurodegenerative diseases: The mitochondrial connection. Curr. Pharm. Des. 2008, 14, 278–288. [Google Scholar] [PubMed]
- Liu, J.; Mouradian, M.M. Pathogenetic Contributions and Therapeutic Implications of Transglutaminase 2 in Neurodegenerative Diseases. Int. J. Mol. Sci. 2024, 25, 2364. [Google Scholar] [CrossRef] [PubMed]
- D’Eletto, M.; Farrace, M.G.; Falasca, L.; Reali, V.; Oliverio, S.; Melino, G.; Griffin, M.; Fimia, G.M.; Piacentini, M. Transglutaminase 2 is involved in autophagosome maturation. Autophagy 2009, 5, 1145–1154. [Google Scholar] [CrossRef] [PubMed]
- Verhaar, R.; Drukarch, B.; Bol, J.G.; Jongenelen, C.A.; Wilhelmus, M.M. Tissue transglutaminase cross-links beclin 1 and regulates autophagy in MPP+-treated human SH-SY5Y cells. Neurochem. Int. 2013, 62, 486–491. [Google Scholar] [CrossRef] [PubMed]
- Mahoney, S.A.; Perry, M.; Seddon, A.; Bohlen, P.; Haynes, L. Transglutaminase forms midkine homodimers in cerebellar neurons and modulates the neurite-outgrowth response. Biochem. Biophys. Res. Commun. 1996, 224, 147–152. [Google Scholar] [CrossRef] [PubMed]
- Emerson, J.; Delgado, T.; Girardi, P.; Johnson, G.V.W. Deletion of Transglutaminase 2 from Mouse Astrocytes Significantly Improves Their Ability to Promote Neurite Outgrowth on an Inhibitory Matrix. Int. J. Mol. Sci. 2023, 24, 6058. [Google Scholar] [CrossRef]
- Walther, D.J.; Peter, J.U.; Winter, S.; Höltje, M.; Paulmann, N.; Grohmann, M.; Vowinckel, J.; Alamo-Bethencourt, V.; Wilhelm, C.S.; Ahnert-Hilger, G.; et al. Serotonylation of small GTPases is a signal transduction pathway that triggers platelet alpha-granule release. Cell 2003, 115, 851–862. [Google Scholar] [CrossRef]
- Shi, R.X.; Liu, C.; Xu, Y.J.; Wang, Y.Y.; He, B.D.; He, X.C.; Du, H.Z.; Hu, B.; Jiao, J.; Liu, C.M.; et al. The Role and Mechanism of Transglutaminase 2 in Regulating Hippocampal Neurogenesis after Traumatic Brain Injury. Cells 2023, 12, 558. [Google Scholar] [CrossRef]
- Oono, M.; Okado-Matsumoto, A.; Shodai, A.; Ido, A.; Ohta, Y.; Abe, K.; Ayaki, T.; Ito, H.; Takahashi, R.; Taniguchi, N.; et al. Transglutaminase 2 accelerates neuroinflammation in amyotrophic lateral sclerosis through interaction with misfolded superoxide dismutase 1. J. Neurochem. 2014, 128, 403–418. [Google Scholar] [CrossRef]
- Ientile, R.; Currò, M.; Caccamo, D. Transglutaminase 2 and neuroinflammation. Amino Acids 2015, 47, 19–26. [Google Scholar] [CrossRef]
- Sestito, C.; Brevé, J.J.P.; Bol, J.G.J.M.; Wilhelmus, M.M.M.; Drukarch, B.; van Dam, A.M. Tissue Transglutaminase contributes to myelin phagocytosis in interleukin-4-treated human monocyte-derived macrophages. Cytokine 2020, 128, 155024. [Google Scholar] [CrossRef]
- Pearse, D.D.; Otero, P.A.; Diaz, A.; Pan, X.; Ghosh, M. Neuronal and Endothelial Transglutaminase-2 Expression during Experimental Autoimmune Encephalomyelitis and Multiple Sclerosis. Neuroscience 2021, 461, 140–154. [Google Scholar] [CrossRef]
- Cooper, A.J.; Jeitner, T.M.; Blass, J.P. The role of transglutaminases in neurodegenerative diseases: Overview. Neurochem. Int. 2002, 40, 1–5. [Google Scholar] [CrossRef]
- Lesort, M.; Tucholski, J.; Miller, M.L.; Johnson, G.V. Tissue transglutaminase: A possible role in neurodegenerative diseases. Prog. Neurobiol. 2000, 61, 439–463. [Google Scholar] [CrossRef]
- Grosso, H.; Mouradian, M.M. Transglutaminase 2: Biology, relevance to neurodegenerative diseases and therapeutic implications. Pharmacol. Ther. 2012, 133, 392–410. [Google Scholar] [CrossRef]
- André, W.; Nondier, I.; Valensi, M.; Guillonneau, F.; Federici, C.; Hoffner, G.; Djian, P. Identification of brain substrates of transglutaminase by functional proteomics supports its role in neurodegenerative diseases. Neurobiol. Dis. 2017, 101, 40–58. [Google Scholar] [CrossRef] [PubMed]
- Schmid, A.W.; Chiappe, D.; Pignat, V.; Grimminger, V.; Hang, I.; Moniatte, M.; Lashuel, H.A. Dissecting the mechanisms of tissue transglutaminase-induced cross-linking of alpha-synuclein: Implications for the pathogenesis of Parkinson disease. J. Biol. Chem. 2009, 284, 13128–13142. [Google Scholar] [CrossRef]
- Wilhelmus, M.M.; Verhaar, R.; Andringa, G.; Bol, J.G.; Cras, P.; Shan, L.; Hoozemans, J.J.; Drukarch, B. Presence of tissue transglutaminase in granular endoplasmic reticulum is characteristic of melanized neurons in Parkinson’s disease brain. Brain Pathol. 2011, 21, 130–139. [Google Scholar] [CrossRef]
- Hong, G.U.; Cho, J.W.; Kim, S.Y.; Shin, J.H.; Ro, J.Y. Inflammatory mediators resulting from transglutaminase 2 expressed in mast cells contribute to the development of Parkinson’s disease in a mouse model. Toxicol. Appl. Pharmacol. 2018, 358, 10–22. [Google Scholar] [CrossRef]
- Mastroberardino, P.G.; Iannicola, C.; Nardacci, R.; Bernassola, F.; De Laurenzi, V.; Melino, G.; Moreno, S.; Pavone, F.; Oliverio, S.; Fesus, L.; et al. ‘Tissue’ transglutaminase ablation reduces neuronal death and prolongs survival in a mouse model of Huntington’s disease. Cell Death Differ. 2002, 9, 873–880. [Google Scholar] [CrossRef]
- Belakhoua, S.M.; Rodriguez, F.J. Diagnostic Pathology of Tumors of Peripheral Nerve. Neurosurgery 2021, 88, 443–456. [Google Scholar] [CrossRef]
- Luksch, R.; Castellani, M.R.; Collini, P.; De Bernardi, B.; Conte, M.; Gambini, C.; Gandola, L.; Garaventa, A.; Biasoni, D.; Podda, M.; et al. Neuroblastoma (Peripheral neuroblastic tumours). Crit. Rev. Oncol. Hematol. 2016, 107, 163–181. [Google Scholar] [CrossRef]
- Swift, C.C.; Eklund, M.J.; Kraveka, J.M.; Alazraki, A.L. Updates in Diagnosis, Management, and Treatment of Neuroblastoma. Radiographics 2018, 38, 566–580. [Google Scholar] [CrossRef] [PubMed]
- Pudela, C.; Balyasny, S.; Applebaum, M.A. Nervous system: Embryonal tumors: Neuroblastoma. Atlas Genet. Cytogenet. Oncol. Haematol. 2020, 24, 284–290. [Google Scholar] [CrossRef]
- Ross, R.A.; Biedler, J.L.; Spengler, B.A. A role for distinct cell types in determining malignancy in human neuroblastoma cell lines and tumors. Cancer Lett. 2003, 197, 35–39. [Google Scholar] [CrossRef]
- Walton, J.D.; Kattan, D.R.; Thomas, S.K.; Spengler, B.A.; Guo, H.F.; Biedler, J.L.; Cheung, N.K.; Ross, R.A. Characteristics of stem cells from human neuroblastoma cell lines and in tumors. Neoplasia 2004, 6, 838–845. [Google Scholar] [CrossRef] [PubMed]
- Zeineldin, M.; Patel, A.G.; Dyer, M.A. Neuroblastoma: When differentiation goes awry. Neuron 2022, 110, 2916–2928. [Google Scholar] [CrossRef]
- Shimada, H.; Ambros, I.M.; Dehner, L.P.; Hata, J.; Joshi, V.V.; Roald, B.; Stram, D.O.; Gerbing, R.B.; Lukens, J.N.; Matthay, K.K.; et al. The International Neuroblastoma Pathology Classification (the Shimada system). Cancer 1999, 86, 364–372. [Google Scholar] [CrossRef]
- Peuchmaur, M.; d’Amore, E.S.; Joshi, V.V.; Hata, J.; Roald, B.; Dehner, L.P.; Gerbing, R.B.; Stram, D.O.; Lukens, J.N.; Matthay, K.K.; et al. Revision of the International Neuroblastoma Pathology Classification: Confirmation of favorable and unfavorable prognostic subsets in ganglioneuroblastoma, nodular. Cancer 2003, 98, 2274–2281. [Google Scholar] [CrossRef] [PubMed]
- Monclair, T.; Brodeur, G.M.; Ambros, P.F.; Brisse, H.J.; Cecchetto, G.; Holmes, K.; Kaneko, M.; London, W.B.; Matthay, K.K.; Nuchtern, J.G.; et al. The International Neuroblastoma Risk Group (INRG) staging system: An INRG Task Force report. J. Clin. Oncol. 2009, 27, 298–303. [Google Scholar] [CrossRef] [PubMed]
- Dalianis, T.; Lukoseviciute, M.; Holzhauser, S.; Kostopoulou, O.N. New Approaches Towards Targeted Therapy for Childhood Neuroblastoma. Anticancer Res. 2023, 43, 3829–3839. [Google Scholar] [CrossRef] [PubMed]
- Irwin, M.S.; Naranjo, A.; Zhang, F.F.; Cohn, S.L.; London, W.B.; Gastier-Foster, J.M.; Ramirez, N.C.; Pfau, R.; Reshmi, S.; Wagner, E.; et al. Revised Neuroblastoma Risk Classification System: A Report from the Children’s Oncology Group. J. Clin. Oncol. 2021, 39, 3229–3241. [Google Scholar] [CrossRef] [PubMed]
- Huang, M.; Weiss, W.A. Neuroblastoma and MYCN. Cold Spring Harb. Perspect. Med. 2013, 3, a014415. [Google Scholar] [CrossRef] [PubMed]
- Braoudaki, M.; Hatziagapiou, K.; Zaravinos, A.; Lambrou, G.I. MYCN in Neuroblastoma: “Old Wine into New Wineskins”. Diseases 2021, 9, 78. [Google Scholar] [CrossRef] [PubMed]
- Bartolucci, D.; Montemurro, L.; Raieli, S.; Lampis, S.; Pession, A.; Hrelia, P.; Tonelli, R. MYCN Impact on High-Risk Neuroblastoma: From Diagnosis and Prognosis to Targeted Treatment. Cancers 2022, 14, 4421. [Google Scholar] [CrossRef]
- Qadir, M.I.; Ahmed, B.; Noreen, S. Advances in the Management of Neuroblastoma. Crit. Rev. Eukaryot. Gene Expr. 2024, 34, 1–13. [Google Scholar] [CrossRef]
- National Cancer Institute. SEER Cancer Stat Facts: Brain and Other Nervous System Cancer; National Cancer Institute: Bethesda, MD, USA. Available online: https://seer.cancer.gov/statfacts/html/brain.html (accessed on 28 November 2023).
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Louis, D.N.; Perry, A.; Wesseling, P.; Brat, D.J.; Cree, I.A.; Figarella-Branger, D.; Hawkins, C.; Ng, H.K.; Pfister, S.M.; Reifenberger, G.; et al. The 2021 WHO Classification of Tumors of the Central Nervous System: A summary. Neuro-Oncology 2021, 23, 1231–1251. [Google Scholar] [CrossRef] [PubMed]
- Ostrom, Q.T.; Cioffi, G.; Waite, K.; Kruchko, C.; Barnholtz-Sloan, J.S. CBTRUS Statistical Report: Primary Brain and Other Central Nervous System Tumors Diagnosed in the United States in 2014–2018. Neuro-Oncology 2021, 23, iii1–iii105. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Holsinger, R.M.; Kruse, C.A.; Flügel, A.; Graeber, M.B. The potential for genetically altered microglia to influence glioma treatment. CNS Neurol. Disord. Drug Targets 2013, 12, 750–762. [Google Scholar] [CrossRef] [PubMed]
- Berger, T.R.; Wen, P.Y.; Lang-Orsini, M.; Chukwueke, U.N. World Health Organization 2021 Classification of Central Nervous System Tumors and Implications for Therapy for Adult-Type Gliomas: A Review. JAMA Oncol. 2022, 8, 1493–1501. [Google Scholar] [CrossRef]
- Louis, D.N.; Wesseling, P.; Aldape, K.; Brat, D.J.; Capper, D.; Cree, I.A.; Eberhart, C.; Figarella-Branger, D.; Fouladi, M.; Fuller, G.N.; et al. cIMPACT-NOW update 6: New entity and diagnostic principle recommendations of the cIMPACT-Utrecht meeting on future CNS tumor classification and grading. Brain Pathol. 2020, 30, 844–856. [Google Scholar] [CrossRef] [PubMed]
- Cotter, J.A.; Hawkins, C. Medulloblastoma: WHO 2021 and Beyond. Pediatr. Dev. Pathol. 2022, 25, 23–33. [Google Scholar] [CrossRef] [PubMed]
- Bae, J.M.; Won, J.K.; Park, S.H. Recent Advancement of the Molecular Diagnosis in Pediatric Brain Tumor. J. Korean Neurosurg. Soc. 2018, 61, 376–385. [Google Scholar] [CrossRef]
- Farooqi, A.; Li, J.; de Groot, J.; Yeboa, D.N. Current Role of Radiation Therapy in the Management of Malignant Central Nervous System Tumors. Hematol. Oncol. Clin. N. Am. 2020, 34, 13–28. [Google Scholar] [CrossRef] [PubMed]
- Patterson, J.D.; Henson, J.C.; Breese, R.O.; Bielamowicz, K.J.; Rodriguez, A. CAR T Cell Therapy for Pediatric Brain Tumors. Front. Oncol. 2020, 10, 1582. [Google Scholar] [CrossRef]
- Manoharan, N.; Liu, K.X.; Mueller, S.; Haas-Kogan, D.A.; Bandopadhayay, P. Pediatric low-grade glioma: Targeted therapeutics and clinical trials in the molecular era. Neoplasia 2023, 36, 100857. [Google Scholar] [CrossRef] [PubMed]
- De Braganca, K.C.; Packer, R.J. Treatment Options for Medulloblastoma and CNS Primitive Neuroectodermal Tumor (PNET). Curr. Treat. Options Neurol. 2013, 15, 593–606. [Google Scholar] [CrossRef]
- Fernandes, C.; Costa, A.; Osório, L.; Lago, R.C.; Linhares, P.; Carvalho, B.; Caeiro, C. Current Standards of Care in Glioblastoma Therapy. In Glioblastoma; De Vleeschouwer, S., Ed.; Codon Publications: Brisbane, Australia, 2017. [Google Scholar]
- Yin, J.; Oh, Y.T.; Kim, J.Y.; Kim, S.S.; Choi, E.; Kim, T.H.; Hong, J.H.; Chang, N.; Cho, H.J.; Sa, J.K.; et al. Transglutaminase 2 Inhibition Reverses Mesenchymal Transdifferentiation of Glioma Stem Cells by Regulating C/EBPβ Signaling. Cancer Res. 2017, 77, 4973–4984. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Tremblay, T.L.; McDermid, A.; Thibault, P.; Stanimirovic, D. Identification of Differentially Expressed Proteins in Human Glioblastoma Cell Lines and Tumors. Glia 2003, 42, 194–208. [Google Scholar] [CrossRef]
- Gundemir, S.; Monteagudo, A.; Akbar, A.; Keillor, J.W.; Johnson, G.V.W. The complex role of transglutaminase 2 in glioblastoma proliferation. Neuro-Oncology 2017, 19, 208–218. [Google Scholar] [CrossRef] [PubMed]
- Sullivan, K.E.; Rojas, K.; Cerione, R.A.; Nakano, I.; Wilson, K.F. The stem cell/cancer stem cell marker ALDH1A3 regulates the expression of the survival factor tissue transglutaminase, in mesenchymal glioma stem cells. Oncotarget 2017, 8, 22325–22343. [Google Scholar] [CrossRef] [PubMed]
- Ganesh, R.A.; Sonpatki, P.; Naik, D.; John, A.E.; Sathe, G.; Lakshmikantha, A.; Chandrachari, K.P.; Bauer, L.; Knäuper, V.; Aeschlimann, D.; et al. Multi-Omics Analysis of Glioblastoma and Glioblastoma Cell Line: Molecular Insights into the Functional Role. Front. Oncol. 2022, 12, 841890. [Google Scholar] [CrossRef]
- Verhaak, R.G.W.; Hoadley, K.A.; Purdom, E.; Wang, V.; Qi, Y.; Wilkerson, M.D.; Miller, C.R.; Ding, L.; Golub, T.; Mesirov, J.P.; et al. Integrated Genomic Analysis Identifies Clinically Relevant Subtypes of Glioblastoma Characterized by Abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell 2010, 17, 98–110. [Google Scholar] [CrossRef]
- Wang, Q.; Hu, B.; Hu, X.; Kim, H.; Squatrito, M.; Scarpace, L.; deCarvalho, A.C.; Lyu, S.; Li, P.; Li, Y.; et al. Tumor Evolution of Glioma-Intrinsic Gene Expression Subtypes Associates with Immunological Changes in the Microenvironment. Cancer Cell 2017, 32, 42–56. [Google Scholar] [CrossRef]
- Verdugo, E.; Puerto, I.; Medina, M.A. An update on the molecular biology of glioblastoma, with clinical implications and progress in its treatment. Cancer Commun. 2022, 42, 1083–1111. [Google Scholar] [CrossRef]
- Huse, J.T.; Phillips, H.S.; Brennan, C.W. Molecular Subclassification of Diffuse Gliomas: Seeing Order in the Chaos. Glia 2011, 59, 1190–1199. [Google Scholar] [CrossRef] [PubMed]
- Zheng, S.; Chheda, M.G.; Verhaak, R.G.W. Studying a Complex Tumor: Potential and Pitfalls. Cancer J. 2012, 18, 107–114. [Google Scholar] [CrossRef]
- Tang, Z.; Kang, B.; Li, C.; Chen, T.; Zhang, Z. GEPIA2: An enhanced web server for large-scale expression profiling and in-teractive analysis. Nucleic Acids Res. 2019, 47, W556–W560. [Google Scholar] [CrossRef] [PubMed]
- Katt, W.P.; Aplin, C.; Cerione, R.A. Exploring the Role of Transglutaminase in Patients with Glioblastoma: Current Perspectives. Onco Targets Ther. 2022, 15, 277–290. [Google Scholar] [CrossRef]
- Ahmadov, U.; Picard, D.; Bartl, J.; Silginer, M.; Trajkovic-Arsic, M.; Qin, N.; Blümel, L.; Wolter, M.; Lim, J.K.M.; Pauck, D.; et al. The long non-coding RNA HOTAIRM1 promotes tumor aggressiveness and radiotherapy resistance in glioblastoma. Cell Death Dis. 2021, 12, 885. [Google Scholar] [CrossRef] [PubMed]
- Berg, T.J.; Marques, C.; Pantazopoulou, V.; Johansson, E.; von Stedingk, K.; Lindgren, D.; Jeannot, P.; Pietras, E.J.; Bergström, T.; Swartling, F.J. The Irradiated Brain Microenvironment Supports Glioma Stemness and Survival via Astrocyte-Derived Transglutaminase 2. Cancer Res. 2021, 81, 2101–2115. [Google Scholar] [CrossRef] [PubMed]
- Zeng, L.; Zheng, W.; Liu, X.; Zhou, Y.; Jin, X.; Xiao, Y.; Bai, Y.; Pan, Y.; Zhang, J.; Shao, C. SDC1-TGM2-FLOT1-BHMT complex determines radiosensitivity of glioblastoma by influencing the fusion of autophagosomes with lysosomes. Theranostics 2023, 13, 3725–3743. [Google Scholar] [CrossRef] [PubMed]
- Zheng, W.; Chen, Q.; Liu, H.; Zeng, L.; Zhou, Y.; Liu, X.; Bai, Y.; Zhang, J.; Pan, Y.; Shao, C. SDC1-dependent TGM2 determines radiosensitivity in glioblastoma by coordinating EPG5-mediated fusion of autophagosomes with lysosomes. Autophagy 2023, 19, 839–857. [Google Scholar] [CrossRef] [PubMed]
- Budillon, A.; Carbone, C.; Di Gennaro, E. Tissue transglutaminase: A new target to reverse cancer drug resistance. Amino Acids 2013, 44, 63–72. [Google Scholar] [CrossRef] [PubMed]
- Yuan, L.; Choi, K.; Khosla, C.; Zheng, X.; Higashikubo, R.; Chicoine, M.R.; Rich, K.M. Tissue transglutaminase 2 inhibition promotes cell death and chemosensitivity in glioblastomas. Mol. Cancer Ther. 2005, 4, 1293–1302. [Google Scholar] [CrossRef]
- Yuan, L.; Siegel, M.; Choi, K.; Khosla, C.; Miller, C.R.; Jackson, E.N.; Piwnica-Worms, D.; Rich, K.M. Transglutaminase 2 inhibitor, KCC009, disrupts fibronectin assembly in the extracellular matrix and sensitizes orthotopic glioblastomas to chemotherapy. Oncogene 2007, 26, 2563–2573. [Google Scholar] [CrossRef]
- Su, J.; Yao, Z.; Chen, Z.; Zhou, S.; Wang, Z.; Xia, H.; Liu, S.; Wu, Y. TfR Aptamer Enhanced Blood-Brain Barrier Penetration of Biomimetic Nanocomplexes for Intracellular Transglutaminase 2 Imaging and Silencing in Glioma. Small 2022, 18, e2203448. [Google Scholar] [CrossRef]
- Marquardt, V.; Theruvath, J.; Pauck, D.; Picard, D.; Qin, N.; Blümel, L.; Maue, M.; Bartl, J.; Ahmadov, U.; Langini, M.; et al. Tacedinaline (CI-994), a class I HDAC inhibitor, targets intrinsic tumor growth and leptomeningeal dissemination in MYC-driven medulloblastoma while making them susceptible to anti-CD47-induced macrophage phagocytosis via NF-kB-TGM2 driven tumor inflammation. J. Immunother. Cancer 2023, 11, e005871. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.C.; Wei, K.C.; Chang, C.N.; Chen, P.Y.; Hsu, P.W.; Chen, C.P.; Lu, C.S.; Wang, H.L.; Gutmann, D.H.; Yeh, T.H. Transglutaminase 2 expression is increased as a function of malignancy grade and negatively regulates cell growth in meningioma. PLoS ONE 2014, 9, e108228. [Google Scholar] [CrossRef] [PubMed]
- Harb, O.A.; Elsayed, W.S.; Ismail, E.I.; Toam, M.M.; Ammar, M.G. Thioredoxin-Interacting-Protein [TXNIP] and Transglutaminase 2 [TGM2] Expression in Meningiomas of Different Grades and the Role of Their Expression in Meningioma Recurrence and Prognosis. Asian Pac. J. Cancer Prev. 2017, 18, 2299–2308. [Google Scholar] [CrossRef] [PubMed]
- Matthay, K.K.; Maris, J.M.; Schleiermacher, G.; Nakagawara, A.; Mackall, C.L.; Diller, L.; Weiss, W.A. Neuroblastoma. Nat. Rev. Dis. Primers 2016, 2, 16078. [Google Scholar] [CrossRef] [PubMed]
- Melino, G.; Farrace, M.G.; Cerù, M.P.; Piacentini, M. Correlation between transglutaminase activity and polyamine levels in human neuroblastoma cells. Effect of retinoic acid and alpha-difluoromethylornithine. Exp. Cell Res. 1988, 179, 429–445. [Google Scholar] [CrossRef]
- Piacentini, M.; Fesus, L.; Farrace, M.G.; Ghibelli, L.; Piredda, L.; Melino, G. The expression of “tissue” transglutaminase in two human cancer cell lines is related with the programmed cell death (apoptosis). Eur. J. Cell Biol. 1991, 54, 246–254. [Google Scholar]
- Piacentini, M.; Annicchiarico-Petruzzelli, M.; Oliverio, S.; Piredda, L.; Biedler, J.L.; Melino, E. Phenotype-specific “tissue” transglutaminase regulation in human neuroblastoma cells in response to retinoic acid: Correlation with cell death by apoptosis. Int. J. Cancer 1992, 52, 271–278. [Google Scholar] [CrossRef]
- Melino, G.; Annicchiarico-Petruzzelli, M.; Piredda, L.; Candi, E.; Gentile, V.; Davies, P.J.; Piacentini, M. Tissue transglutaminase and apoptosis: Sense and antisense transfection studies with human neuroblastoma cells. Mol. Cell Biol. 1994, 14, 6584–6596. [Google Scholar] [CrossRef]
- Piacentini, M.; Piredda, L.; Starace, D.T.; Annicchiarico-Petruzzelli, M.; Mattei, M.; Oliverio, S.; Farrace, M.G.; Melino, G. Differential growth of N- and S-type human neuroblastoma cells xenografted into scid mice. correlation with apoptosis. J. Pathol. 1996, 180, 415–422. [Google Scholar] [CrossRef]
- Piacentini, M.; Farrace, M.G.; Piredda, L.; Matarrese, P.; Ciccosanti, F.; Falasca, L.; Rodolfo, C.; Giammarioli, A.M.; Verderio, E.; Griffin, M.; et al. Transglutaminase overexpression sensitizes neuronal cell lines to apoptosis by increasing mitochondrial membrane potential and cellular oxidative stress. J. Neurochem. 2002, 81, 1061–1072. [Google Scholar] [CrossRef]
- Rodolfo, C.; Mormone, E.; Matarrese, P.; Ciccosanti, F.; Farrace, M.G.; Garofano, E.; Piredda, L.; Fimia, G.M.; Malorni, W.; Piacentini, M. Tissue transglutaminase is a multifunctional BH3-only protein. J. Biol. Chem. 2004, 279, 54783–54792. [Google Scholar] [CrossRef] [PubMed]
- Tucholski, J.; Lesort, M.; Johnson, G.V. Tissue transglutaminase is essential for neurite outgrowth in human neuroblastoma SH-SY5Y cells. Neuroscience 2001, 102, 481–491. [Google Scholar] [CrossRef] [PubMed]
- Tucholski, J.; Johnson, G.V. Tissue transglutaminase differentially modulates apoptosis in a stimuli-dependent manner. J. Neurochem. 2002, 81, 780–791. [Google Scholar] [CrossRef] [PubMed]
- Tucholski, J. TG2 protects neuroblastoma cells against DNA-damage-induced stress, suppresses p53 activation. Amino Acids 2010, 39, 523–532. [Google Scholar] [CrossRef] [PubMed]
- Belkin, A.M. Extracellular TG2: Emerging functions and regulation. FEBS J. 2011, 278, 4704–4716. [Google Scholar] [CrossRef]
- Aloe, L.; Rocco, M.L.; Balzamino, B.O.; Micera, A. Nerve Growth Factor: A Focus on Neuroscience and Therapy. Curr. Neuropharmacol. 2015, 13, 294–303. [Google Scholar] [CrossRef]
- Condello, S.; Caccamo, D.; Currò, M.; Ferlazzo, N.; Parisi, G.; Ientile, R. Transglutaminase 2 and NF-kappaB interplay during NGF-induced differentiation of neuroblastoma cells. Brain Res. 2008, 1207, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Algarni, A.S.; Hargreaves, A.J.; Dickenson, J.M. Activation of transglutaminase 2 by nerve growth factor in differentiating neuroblastoma cells: A role in cell survival and neurite outgrowth. Eur. J. Pharmacol. 2018, 820, 113–129. [Google Scholar] [CrossRef]
- Illendula, A.; Fultang, N.; Peethambaran, B. Retinoic acid induces differentiation in neuroblastoma via ROR1 by modulating retinoic acid response elements. Oncol. Rep. 2020, 44, 1013–1024. [Google Scholar] [CrossRef] [PubMed]
- Reynolds, C.P.; Matthay, K.K.; Villablanca, J.G.; Maurer, B.J. Retinoid therapy of high-risk neuroblastoma. Cancer Lett. 2003, 197, 185–192. [Google Scholar] [CrossRef] [PubMed]
- Lai, T.S.; Greenberg, C.S. TGM2 and implications for human disease: Role of alternative splicing. Front. Biosci. 2013, 18, 504–519. [Google Scholar] [CrossRef]
- Atobatele, A.G.; Tonoli, E.; Vadakekolathu, J.; Savoca, M.P.; Barr, M.; Kataria, Y.; Rossanese, M.; Burhan, I.; McArdle, S.; Caccamo, D.; et al. Canonical and truncated transglutaminase-2 regulate mucin-1 expression and androgen independency in prostate cancer cell lines. Cell Death Dis. 2023, 14, 317. [Google Scholar] [CrossRef] [PubMed]
- Tee, A.E.L.; Marshall, G.M.; Liu, P.Y.; Xu, N.; Haber, M.; Norris, M.D.; Iismaa, S.E.; Liu, T. Opposing effects of two tissue transglutaminase protein isoforms in neuroblastoma cell differentiation. J. Biol. Chem. 2010, 285, 3561–3567. [Google Scholar] [CrossRef] [PubMed]
- Currò, M.; Ferlazzo, N.; Giunta, M.L.; Montalto, A.S.; Russo, T.; Arena, S.; Impellizzeri, P.; Caccamo, D.; Romeo, C.; Ientile, R. Hypoxia-Dependent Expression of TG2 Isoforms in Neuroblastoma Cells as Consequence of Different MYCN Amplification Status. Int. J. Mol. Sci. 2020, 21, 1364. [Google Scholar] [CrossRef]
- Gilad, G.M.; Gilad, V.H. Cytotoxic effects of monodansylcadaverine and methylamine in primary cultures of rat cerebellar neurons. Int. J. Dev. Neurosci. 1986, 4, 401–405. [Google Scholar] [CrossRef]
- Pardin, C.; Roy, I.; Lubell, W.D.; Keillor, J.W. Reversible and competitive cinnamoyl triazole inhibitors of tissue transglutaminase. Chem. Biol. Drug Des. 2008, 72, 189–196. [Google Scholar] [CrossRef]
- Apperley, K.Y.P.; Roy, I.; Saucier, V.; Brunet-Filion, N.; Piscopo, S.P.; Pardin, C.; De Francesco, É.; Hao, C.; Keillor, J.W. Development of new scaffolds as reversible tissue transglutaminase inhibitors, with improved potency or resistance to glutathione addition. Medchemcomm 2016, 8, 338–345. [Google Scholar] [CrossRef]
- Kim, S.Y. New Insights into Development of Transglutaminase 2 Inhibitors as Pharmaceutical Lead Compounds. Med. Sci. 2018, 6, 87. [Google Scholar] [CrossRef] [PubMed]
- Case, A.; Stein, R.L. Kinetic analysis of the interaction of tissue transglutaminase with a nonpeptidic slow-binding inhibitor. Biochemistry 2007, 46, 1106–1115. [Google Scholar] [CrossRef] [PubMed]
- Katt, W.P.; Antonyak, M.A.; Cerione, R.A. The diamond anniversary of tissue transglutaminase: A protein of many talents. Drug Discov. Today 2018, 23, 575–591. [Google Scholar] [CrossRef] [PubMed]
- Jeitner, T.M.; Pinto, J.T.; Cooper, A.J.L. Cystamine and cysteamine as inhibitors of transglutaminase activity in vivo. Biosci. Rep. 2018, 38, BSR20180691. [Google Scholar] [CrossRef] [PubMed]
- Arbez, N.; Roby, E.; Akimov, S.; Eddings, C.; Ren, M.; Wang, X.; Ross, C.A. Cysteamine Protects Neurons from Mutant Huntingtin Toxicity. J. Huntingt. Dis. 2019, 8, 129–143. [Google Scholar] [CrossRef] [PubMed]
- Almami, I.S.; Aldubayan, M.A.; Felemban, S.G.; Alyamani, N.; Howden, R.; Robinson, A.J.; Pearson, T.D.Z.; Boocock, D.; Algarni, A.S.; Garner, A.C.; et al. Neurite outgrowth inhibitory levels of organophosphates induce tissue transglutaminase activity in differentiating N2a cells: Evidence for covalent adduct formation. Arch. Toxicol. 2020, 94, 3861–3875. [Google Scholar] [CrossRef]
- Aldubayan, M.A.; Almami, I.S.; Felemban, S.G.; Alhowail, A.H.; Bonner, P.L.R.; Hargreaves, A.J. Organophosphates modulate tissue transglutaminase activity in differentiated C6 neural cells. Eur. Rev. Med. Pharmacol. Sci. 2022, 26, 168–182. [Google Scholar] [CrossRef]
- Schuppan, D.; Mäki, M.; Lundin, K.E.A.; Isola, J.; Friesing-Sosnik, T.; Taavela, J.; Popp, A.; Koskenpato, J.; Langhorst, J.; Hovde, Ø.; et al. A Randomized Trial of a Transglutaminase 2 Inhibitor for Celiac Disease. N. Engl. J. Med. 2021, 385, 35–45. [Google Scholar] [CrossRef]
- Zhang, J.; Ding, Y.R.; Wang, R. Inhibition of tissue transglutaminase promotes Abeta-induced apoptosis in SH-SY5Y cells. Acta Pharmacol. Sin. 2016, 37, 1534–1542. [Google Scholar] [CrossRef] [PubMed]
- van Strien, M.E.; de Vries, H.E.; Chrobok, N.L.; Bol, J.G.J.M.; Breve, J.J.P.; van der Pol, S.M.P.; Kooij, G.; van Buul, J.D.; Karpuj, M.; Steinman, L.; et al. Tissue Transglutaminase contributes to experimental multiple sclerosis pathogenesis and clinical outcome by promoting macrophage migration. Brain Behav. Immun. 2015, 50, 141–154. [Google Scholar] [CrossRef] [PubMed]
- Katt, W.P.; Blobel, N.J.; Komarova, S.; Antonyak, M.A.; Nakano, I.; Cerione, R.A. A small molecule regulator of tissue transglutaminase conformation inhibits the malignant phenotype of cancer cells. Oncotarget 2018, 9, 34379–34397. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Perez, M.; Caja, S.; Melino, G.; Johnson, T.S.; Lindfors, K.; Griffin, M. A novel extracellular role for tissue transglutaminase in matrix-bound VEGF-mediated angiogenesis. Cell Death Dis. 2013, 4, e808. [Google Scholar] [CrossRef]
- Martini, M.; de Pascalis, I.; D’Alessandris, Q.G.; Fiorentino, V.; Pierconti, F.; Marei, H.E.; Ricci-Vitiani, L.; Pallini, R.; Larocca, L.M. VEGF-121 plasma level as biomarker for response to anti-angiogenetic therapy in recurrent glioblastoma. BMC Cancer 2018, 18, 553. [Google Scholar] [CrossRef]
- Buccarelli, M.; Castellani, G.; Ricci-Vitiani, L. Glioblastoma-Specific Strategies of Vascularization: Implications in Anti-Angiogenic Therapy Resistance. J. Pers. Med. 2022, 12, 1625. [Google Scholar] [CrossRef]
| Inhibitor | Type | References |
|---|---|---|
| MDC | Pseudo-substrate; competitive | [187] |
| CP4d | Reversible | [188] |
| GK921 | Reversible | [143,189,190] |
| LDN-27219 | Reversible | [191] |
| TTGM5826 | Reversible | [192] |
| Cystamine | Pseudo-substrate; irreversible | [193,194] |
| KCC009 | Peptidomimetic; irreversible | [14,22,160,161] |
| Z-DON (Z006) | Peptidomimetic; irreversible | [14,180,192,195,196] |
| ZED1227 | Peptidomimetic; irreversible | [197] |
| NTU283 (D003) | Irreversible | [198] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Buccarelli, M.; Castellani, G.; Fiorentino, V.; Pizzimenti, C.; Beninati, S.; Ricci-Vitiani, L.; Scattoni, M.L.; Mischiati, C.; Facchiano, F.; Tabolacci, C. Biological Implications and Functional Significance of Transglutaminase Type 2 in Nervous System Tumors. Cells 2024, 13, 667. https://doi.org/10.3390/cells13080667
Buccarelli M, Castellani G, Fiorentino V, Pizzimenti C, Beninati S, Ricci-Vitiani L, Scattoni ML, Mischiati C, Facchiano F, Tabolacci C. Biological Implications and Functional Significance of Transglutaminase Type 2 in Nervous System Tumors. Cells. 2024; 13(8):667. https://doi.org/10.3390/cells13080667
Chicago/Turabian StyleBuccarelli, Mariachiara, Giorgia Castellani, Vincenzo Fiorentino, Cristina Pizzimenti, Simone Beninati, Lucia Ricci-Vitiani, Maria Luisa Scattoni, Carlo Mischiati, Francesco Facchiano, and Claudio Tabolacci. 2024. "Biological Implications and Functional Significance of Transglutaminase Type 2 in Nervous System Tumors" Cells 13, no. 8: 667. https://doi.org/10.3390/cells13080667







