Quantitative Phase Imaging as Sensitive Screening Method for Nanoparticle-Induced Cytotoxicity Assessment
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Lines and Cell Cultivation
2.2. Nanomaterials
2.3. Cell Preparation for Multimodal Cytotoxicity Assessment Experiments
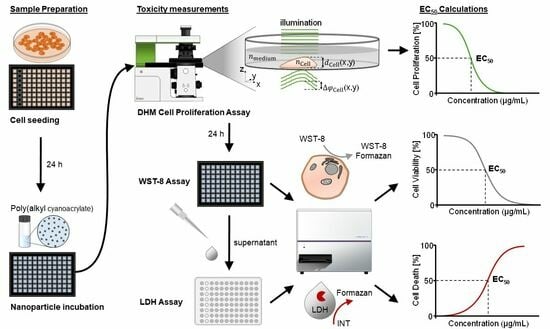
2.4. Experimental Workflow of DHM with Downstream WST-8 and LDH Assays
2.5. DHM QPI Cell Proliferation Assay
2.6. WST-8 Cell Viability Assay
2.7. LDH Release Cell Death Assay
2.8. Calculation of EC50 Values and Statistical Analysis
3. Results
3.1. Qualitative Analysis of Cell Proliferation and Morphology Alterations in DHM QPI Images after Incubation with PACA Nanoparticles
3.2. Multimodal Cytotoxicity Assessment by DHM with Downstream WST-8 and LDH
4. Discussion
5. Conclusions and Future Perspectives
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sun, L.; Liu, H.; Ye, Y.; Lei, Y.; Islam, R.; Tan, S.; Tong, R.; Miao, Y.B.; Cai, L. Smart nanoparticles for cancer therapy. Signal Transduct. Target. Ther. 2023, 8, 418. [Google Scholar] [CrossRef] [PubMed]
- Mazzotta, E.; De Santo, M.; Lombardo, D.; Leggio, A.; Pasqua, L. Mesoporous silicas in materials engineering: Nanodevices for bionanotechnologies. Mater. Today Bio 2022, 17, 100472. [Google Scholar] [CrossRef] [PubMed]
- Domingues, C.; Santos, A.; Alvarez-Lorenzo, C.; Concheiro, A.; Jarak, I.; Veiga, F.; Figueiras, A. Where is nano today and where is it headed? A review of nanomedicine and the dilemma of nanotoxicology. ACS Nano 2022, 16, 9994–10041. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.J.; Lee, Y.H.; Hsu, Y.H.; Liao, C.T.; Lin, Y.F.; Chiu, H.W. Current strategies in assessment of nanotoxicity: Alternatives to in vivo animal testing. Int. J. Mol. Sci. 2021, 22, 4216. [Google Scholar] [CrossRef] [PubMed]
- OECD. OECD Guidelines for the Testing of Chemicals; OECD: Paris, France, 1994. [Google Scholar]
- Drasler, B.; Sayre, P.; Steinhäuser, K.G.; Petri-Fink, A.; Rothen-Rutishauser, B. In vitro approaches to assess the hazard of nanomaterials. NanoImpact 2017, 8, 99–116. [Google Scholar] [CrossRef]
- Monteiro-Riviere, N.A.; Inman, A.O.; Zhang, L.W. Limitations and relative utility of screening assays to assess engineered nanoparticle toxicity in a human cell line. Toxicol. Appl. Pharmacol. 2009, 234, 222–235. [Google Scholar] [CrossRef] [PubMed]
- Park, M.V.; Lankveld, D.P.; van Loveren, H.; de Jong, W.H. The status of in vitro toxicity studies in the risk assessment of nanomaterials. Nanomedicine 2009, 4, 669–685. [Google Scholar] [CrossRef] [PubMed]
- Armand, L.; Tarantini, A.; Beal, D.; Biola-Clier, M.; Bobyk, L.; Sorieul, S.; Pernet-Gallay, K.; Marie-Desvergne, C.; Lynch, I.; Herlin-Boime, N.; et al. Long-term exposure of A549 cells to titanium dioxide nanoparticles induces DNA damage and sensitizes cells towards genotoxic agents. Nanotoxicology 2016, 10, 913–923. [Google Scholar] [CrossRef] [PubMed]
- Anguissola, S.; Garry, D.; Salvati, A.; O’Brien, P.J.; Dawson, K.A. High content analysis provides mechanistic insights on the pathways of toxicity induced by amine-modified polystyrene nanoparticles. PLoS ONE 2014, 9, e108025. [Google Scholar] [CrossRef] [PubMed]
- Farcal, L.; Torres Andón, F.; Di Cristo, L.; Rotoli, B.M.; Bussolati, O.; Bergamaschi, E.; Mech, A.; Hartmann, N.B.; Rasmussen, K.; Riego-Sintes, J.; et al. Comprehensive in vitro toxicity testing of a panel of representative oxide nanomaterials: First steps towards an intelligent testing strategy. PLoS ONE 2015, 10, e0127174. [Google Scholar] [CrossRef] [PubMed]
- Rampersad, S.N. Multiple applications of Alamar Blue as an indicator of metabolic function and cellular health in cell viability bioassays. Sensors 2012, 12, 12347–12360. [Google Scholar] [CrossRef] [PubMed]
- Hu, W.; Culloty, S.; Darmody, G.; Lynch, S.; Davenport, J.; Ramirez-Garcia, S.; Dawson, K.; Lynch, I.; Doyle, H.; Sheehan, D. Neutral red retention time assay in determination of toxicity of nanoparticles. Mar. Environ. Res. 2015, 111, 158–161. [Google Scholar] [CrossRef] [PubMed]
- Luo, Y.; Wang, C.; Qiao, Y.; Hossain, M.; Ma, L.; Su, M. In vitro cytotoxicity of surface modified bismuth nanoparticles. J. Mater. Sci. Mater. Med. 2012, 23, 2563–2573. [Google Scholar] [CrossRef] [PubMed]
- Goode, A.E.; Carter, D.A.G.; Motskin, M.; Pienaar, I.S.; Chen, S.; Hu, S.; Ruenraroengsak, P.; Ryan, M.P.; Shaffer, M.S.P.; Dexter, D.T.; et al. High resolution and dynamic imaging of biopersistence and bioreactivity of extra and intracellular MWNTs exposed to microglial cells. Biomaterials 2015, 70, 57–70. [Google Scholar] [CrossRef] [PubMed]
- Strober, W. Trypan blue exclusion test of cell viability. Curr. Protoc. Immunol. 1997, 21, A-3B. [Google Scholar] [CrossRef] [PubMed]
- Stoehr, L.C.; Gonzalez, E.; Stampfl, A.; Casals, E.; Duschl, A.; Puntes, V.; Oostingh, G.J. Shape matters: Effects of silver nanospheres and wires on human alveolar epithelial cells. Part. Fibre Toxicol. 2011, 8, 36. [Google Scholar] [CrossRef] [PubMed]
- Kumar, P.; Nagarajan, A.; Uchil, P.D. Analysis of cell viability by the lactate dehydrogenase assay. Cold Spring Harb. Protoc. 2018, 6, 465–468. [Google Scholar] [CrossRef] [PubMed]
- Olive, P.L.; Banáth, J.P. The comet assay: A method to measure DNA damage in individual cells. Nat. Protoc. 2006, 1, 23–29. [Google Scholar] [CrossRef] [PubMed]
- Vandeputte, C.; Guizon, I.; Genestie-Denis, I.; Vannier, B.; Lorenzon, G. A microtiter plate assay for total glutathione and glutathione disulfide contents in cultured/isolated cells: Performance study of a new miniaturized protocol. Cell Biol. Toxicol. 1994, 10, 415–421. [Google Scholar] [CrossRef] [PubMed]
- Kocbach, A.; Totlandsdal, A.I.; Låg, M.; Refsnes, M.; Schwarze, P.E. Differential binding of cytokines to environmentally relevant particles: A possible source for misinterpretation of in vitro results? Toxicol. Lett. 2008, 176, 131–137. [Google Scholar] [CrossRef] [PubMed]
- Kemper, B.; Von Bally, G. Digital holographic microscopy for live cell applications and technical inspection. Appl. Opt. 2008, 47, A52–A61. [Google Scholar] [CrossRef] [PubMed]
- Park, Y.; Depeursinge, C.; Popescu, G. Quantitative phase imaging in biomedicine. Nat. Photonics 2018, 12, 578–589. [Google Scholar] [CrossRef]
- Kühn, J.; Shaffer, E.; Mena, J.; Breton, B.; Parent, J.; Rappaz, B.; Chambon, M.; Emery, Y.; Magistretti, P.; Depeursinge, C.; et al. Label-Free Cytotoxicity Screening Assay by Digital Holographic Microscopy. Assay Drug Dev. Technol. 2013, 11, 101–107. [Google Scholar] [CrossRef]
- Mugnano, M.; Memmolo, P.; Miccio, L.; Grilli, S.; Merola, F.; Calabuig, A.; Bramanti, A.; Mazzon, E.; Ferraro, P. In vitro cytotoxicity evaluation of cadmium by label-free holographic microscopy. J. Biophotonics 2018, 11, e201800099. [Google Scholar] [CrossRef]
- Makdasi, E.; Laskar, O.; Milrot, E.; Schuster, O.; Shmaya, S.; Yitzhaki, S. Whole-Cell Multiparameter Assay for Ricin and Abrin Activity-Based Digital Holographic Microscopy. Toxins 2019, 11, 174. [Google Scholar] [CrossRef]
- Janicke, B.; Kårsnäs, A.; Egelberg, P.; Alm, K. Label-Free High Temporal Resolution Assessment of Cell Proliferation Using Digital Holographic Microscopy. Cytom. Part A 2017, 91, 460–469. [Google Scholar] [CrossRef]
- Eder, K.M.; Marzi, A.; Barroso, Á.; Ketelhut, S.; Kemper, B.; Schnekenburger, J. Label-free digital holographic microscopy for in vitro cytotoxic effect quantification of organic nanoparticles. Cells 2022, 11, 644. [Google Scholar] [CrossRef]
- Vauthier, C.; Dubernet, C.; Fattal, E.; Pinto-Alphandary, H.; Couvreur, P. Poly (alkylcyanoacrylates) as biodegradable materials for biomedical applications. Adv. Drug Deliv. Rev. 2003, 55, 519–548. [Google Scholar] [CrossRef] [PubMed]
- Sulheim, E.; Iversen, T.G.; To Nakstad, V.; Klinkenberg, G.; Sletta, H.; Schmid, R.; Hatletveit, A.R.; Wågbø, A.M.; Sundan, A.; Skotland, T.; et al. Cytotoxicity of poly (alkyl cyanoacrylate) nanoparticles. Int. J. Mol. Sci. 2017, 18, 2454. [Google Scholar] [CrossRef]
- Øverbye, A.; Torgersen, M.L.; Sønstevold, T.; Iversen, T.G.; Mørch, Ý.; Skotland, T.; Sandvig, K. Cabazitaxel-loaded poly (alkyl cyanoacrylate) nanoparticles: Toxicity and changes in the proteome of breast, colon and prostate cancer cells. Nanotoxicology 2021, 15, 865–884. [Google Scholar] [CrossRef] [PubMed]
- Sudji, I.R.; Subburaj, Y.; Frenkel, N.; García-Sáez, A.J.; Wink, M. Membrane disintegration caused by the steroid saponin digitonin is related to the presence of cholesterol. Molecules 2015, 20, 20146–20160. [Google Scholar] [CrossRef] [PubMed]
- Meibohm, B.; Derendorf, H. Basic concepts of pharmacokinetic/pharmacodynamic (PK/PD) modelling. Int. J. Clin. Pharmacol. Ther. 1997, 35, 401–413. [Google Scholar] [PubMed]
- Holford, N.H.; Sheiner, L.B. Kinetics of pharmacologic response. Pharmacol. Ther. 1982, 16, 143–166. [Google Scholar] [CrossRef] [PubMed]
- Kroll, A.; Dierker, C.; Rommel, C.; Hahn, D.; Wohlleben, W.; Schulze-Isfort, C.; Göbbert, C.; Voetz, M.; Hardinghaus, F.; Schnekenburger, J. Cytotoxicity screening of 23 engineered nanomaterials using a test matrix of ten cell lines and three different assays. Part. Fibre Toxicol. 2011, 8, 9. [Google Scholar] [CrossRef] [PubMed]
- Kemper, B.; Carl, D.; Schnekenburger, J.; Bredebusch, I.; Schäfer, M.; Domschke, W.; von Bally, G. Investigation of living pancreas tumor cells by digital holographic microscopy. J. Biomed. Opt. 2006, 11, 34005. [Google Scholar] [CrossRef] [PubMed]
- Min, J.; Yao, B.; Trendafilova, V.; Ketelhut, S.; Kastl, L.; Greve, B.; Kemper, B. Quantitative phase imaging of cells in a flow cytometry arrangement utilizing Michelson interferometer-based off-axis digital holographic microscopy. J. Biophotonics 2019, 12, e201900085. [Google Scholar] [CrossRef] [PubMed]
- Kemper, B.; Barroso, Á.; Eder, K.; Marzi, A.; Ritz, S.; Schnekenburger, J.; Ketelhut, S. Reduction of the influence of parasitic interferences and coherence induced disturbances in digital holography-based quantitative phase microscopy by modulation of the object illumination with an electrically focus tunable lens. Proc. SPIE 2021, 11653, 116530A. [Google Scholar]
- Sternberg, S.R. Biomedical image processing. Computer 1983, 16, 22–34. [Google Scholar] [CrossRef]
- Popescu, G.; Park, Y.; Lue, N.; Best-Popescu, C.; Deflores, L.; Dasari, R.R.; Feld, M.S.; Badizadegan, K. Optical imaging of cell mass and growth dynamics. Am. J. Physiol. -Cell Physiol. 2008, 295, C538–C544. [Google Scholar] [CrossRef] [PubMed]
- Barer, R. Refractometry and interferometry of living cells. JOSA 1957, 47, 545–556. [Google Scholar] [CrossRef] [PubMed]
- Barer, R. Determination of dry mass, thickness, solid and water concentration in living cells. Nature 1953, 172, 1097–1098. [Google Scholar] [CrossRef] [PubMed]
- Eder, K.M.; Marzi, A.; Wågbø, A.M.; Vermeulen, J.P.; de la Fonteyne-Blankestijn, L.J.; Rösslein, M.; Ossig, R.; Klinkenberg, G.; Vandebriel, R.J.; Schnekenburger, J. Standardization of an in vitro assay matrix to assess cytotoxicity of organic nanocarriers: A pilot interlaboratory comparison. Drug Deliv. Transl. Res. 2022, 12, 2187–2206. [Google Scholar] [CrossRef] [PubMed]
- Sohaebuddin, S.K.; Thevenot, P.T.; Baker, D.; Eaton, J.W.; Tang, L. Nanomaterial cytotoxicity is composition, size, and cell type dependent. Part. Fibre Toxicol. 2010, 7, 22. [Google Scholar] [CrossRef] [PubMed]
- Woźniak, A.; Malankowska, A.; Nowaczyk, G.; Grześkowiak, B.F.; Tuśnio, K.; Słomski, R.; Zaleska-Medynska, A.; Jurga, S. Size and shape-dependent cytotoxicity profile of gold nanoparticles for biomedical applications. J. Mater. Sci. Mater. Med. 2017, 28, 92. [Google Scholar] [CrossRef]
- Patra, H.K.; Banerjee, S.; Chaudhuri, U.; Lahiri, P.; Dasgupta, A.K. Cell selective response to gold nanoparticles. Nanomed. Nanotechnol. Biol. Med. 2007, 3, 111–119. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.A.; Åberg, C.; Salvati, A.; Dawson, K.A. Role of cell cycle on the cellular uptake and dilution of nanoparticles in a cell population. Nat. Nanotechnol. 2012, 7, 62–68. [Google Scholar] [CrossRef] [PubMed]
- Jeynes, J.C.; Jeynes, C.; Merchant, M.J.; Kirkby, K.J. Measuring and modelling cell-to-cell variation in uptake of gold nanoparticles. Analyst 2013, 138, 7070–7074. [Google Scholar] [CrossRef] [PubMed]
- Weyermann, J.; Lochmann, D.; Zimmer, A. A practical note on the use of cytotoxicity assays. Int. J. Pharm. 2005, 288, 369–376. [Google Scholar] [CrossRef] [PubMed]
- Adan, A.; Kiraz, Y.; Baran, Y. Cell Proliferation and Cytotoxicity Assays. Curr. Pharm. Biotechnol. 2016, 17, 1213–1221. [Google Scholar] [CrossRef] [PubMed]
- Riss, T.L.; Moravec, R.A. Use of Multiple Assay Endpoints to Investigate the Effects of Incubation Time, Dose of Toxin, and Plating Density in Cell-Based Cytotoxicity Assays. Assay Drug Dev. Technol. 2004, 2, 51–62. [Google Scholar] [CrossRef] [PubMed]
- Kante, B.; Couvreur, P.; Dubois-Krack, G.; de Meester, C.; Guiot, P.; Roland, M.; Mercier, M.; Speiser, P. Toxicity of polyalkylcyanoacrylate nanoparticles I: Free nanoparticles. J. Pharm. Sci. 1982, 71, 786–790. [Google Scholar] [CrossRef]
- Boutros, M.; Heigwer, F.; Laufer, C. Microscopy-based high-content screening. Cell 2015, 163, 1314–1325. [Google Scholar] [CrossRef] [PubMed]
- Perlman, Z.E.; Slack, M.D.; Feng, Y.; Mitchison, T.J.; Wu, L.F.; Altschuler, S.J. Multidimensional drug profiling by automated microscopy. Science 2004, 306, 1194–1198. [Google Scholar] [CrossRef] [PubMed]
- Lai, S.; Centi, S.; Borri, C.; Ratto, F.; Cavigli, L.; Michelettia, F.; Kemper, B.; Ketelhut, S.; Kozyreva, T.; Gonnelli, L.; et al. A multifunctional organosilica cross-linker for the bio-conjugation of gold nanorods. Colloids Surf. B Biointerfaces 2017, 157, 174–181. [Google Scholar] [CrossRef] [PubMed]
- Eder, K.; Kutscher, T.; Marzi, A.; Barroso, Á.; Schnekenburger, J.; Kemper, B. Automated Detection of Macrophages in Quantitative Phase Images by Deep Learning Using a Mask Region-Based Convolutional Neural Network. Proc. SPIE 2021, 116551, 116551K. [Google Scholar]
- Girshovitz, P.; Shaked, N.T. Generalized cell morphological parameters based on interferometric phase microscopy and their application to cell life cycle characterization. Biomed. Opt. Express 2012, 3, 1757–1773. [Google Scholar] [CrossRef] [PubMed]
- Belashov, A.V.; Zhikhoreva, A.A.; Belyaeva, T.N.; Salova, A.V.; Kornilova, E.S.; Semenova, I.V.; Vasyutinskii, O.S. Learning Assisted Classification of Cell Lines and Cell States on Quantitative Phase Images. Cells 2021, 10, 2587. [Google Scholar] [CrossRef]
- Vrignaud, P.; Semiond, D.; Lejeune, P.; Bouchard, H.; Calvet, L.; Combeau, C.; Riou, J.F.; Commercon, A.; Lavelle, F.; Bissery, M.C. Preclinical antitumor activity of cabazitaxel, a semisynthetic taxane active in taxane-resistant tumors. Clin. Cancer Res. 2013, 19, 2973–2983. [Google Scholar] [CrossRef] [PubMed]
- Fan, F.; Wierda, D.; Rozman, K.K. Effects of 2,3,7,8-tetrachlorodibenzo-p-dioxin on humoral and cell-mediated immunity in Sprague-Dawley rats. Toxicology 1996, 106, 221–228. [Google Scholar] [CrossRef] [PubMed]
- Palanza, P.; Parmigiani, S.; vom Saal, F.S. Effects of prenatal exposure to low doses of diethylstilbestrol, o, p‘DDT, and methoxychlor on postnatal growth and neurobehavioral development in male and female mice. Horm. Behav. 2001, 40, 252–265. [Google Scholar] [CrossRef]
- Vandenberg, L.N. Low-dose effects of hormones and endocrine disruptors. Vitam. Horm. 2014, 94, 129–165. [Google Scholar] [PubMed]
- Chen, R.; Cheng, Q.; Owusu-Ansah, K.G.; Chen, J.; Song, G.; Xie, H.; Zhou, L.; Xu, X.; Jiang, D.; Zheng, S. Cabazitaxel, a novel chemotherapeutic alternative for drug-resistant hepatocellular carcinoma. Am. J. Cancer Res. 2018, 8, 1297–1306. [Google Scholar] [PubMed]
- Mita, A.C.; Figlin, R.; Mita, M.M. Cabazitaxel: More Than a New Taxane for Metastatic Castrate-Resistant Prostate Cancer? Clin. Cancer Res. 2012, 18, 6574–6579. [Google Scholar] [CrossRef]
- Dumontet, C.; Jordan, M.A. Microtubule-binding agents: A dynamic field of cancer therapeutics. Nat. Rev. Drug Discov. 2010, 9, 790–803. [Google Scholar] [CrossRef] [PubMed]
- Fusser, M.; Øverbye, A.; Pandya, A.D.; Mørch, Ý.; Borgos, S.E.; Kildal, W.; Snipstad, S.; Sulheim, E.; Fleten, K.G.; Askautrud, H.A.; et al. Cabazitaxel-loaded Poly (2-ethylbutyl cyanoacrylate) nanoparticles improve treatment efficacy in a patient derived breast cancer xenograft. J. Control. Release 2019, 293, 183–192. [Google Scholar] [CrossRef] [PubMed]
- Szwed, M.; Sønstevold, T.; Øverbye, A.; Engedal, N.; Grallert, B.; Mørch, Ý.; Sulheim, E.; Iversen, T.G.; Skotland, T.; Sandvig, K.; et al. Small Variations in Nanoparticle Structure Dictate Differential Cellular Stress Responses and Mode of Cell Death. Nanotoxicology 2019, 13, 1–22. [Google Scholar] [CrossRef] [PubMed]




| EC50 [µg/mL] | |||
|---|---|---|---|
| DHM | WST-8 | LDH | |
| RAW 264.7 | 9 ± 1 | 14 ± 1 | 22 ± 4 |
| NIH-3T3 | 15 ± 8 | 40 ± 4 | 311 ± 116 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Marzi, A.; Eder, K.M.; Barroso, Á.; Kemper, B.; Schnekenburger, J. Quantitative Phase Imaging as Sensitive Screening Method for Nanoparticle-Induced Cytotoxicity Assessment. Cells 2024, 13, 697. https://doi.org/10.3390/cells13080697
Marzi A, Eder KM, Barroso Á, Kemper B, Schnekenburger J. Quantitative Phase Imaging as Sensitive Screening Method for Nanoparticle-Induced Cytotoxicity Assessment. Cells. 2024; 13(8):697. https://doi.org/10.3390/cells13080697
Chicago/Turabian StyleMarzi, Anne, Kai Moritz Eder, Álvaro Barroso, Björn Kemper, and Jürgen Schnekenburger. 2024. "Quantitative Phase Imaging as Sensitive Screening Method for Nanoparticle-Induced Cytotoxicity Assessment" Cells 13, no. 8: 697. https://doi.org/10.3390/cells13080697
APA StyleMarzi, A., Eder, K. M., Barroso, Á., Kemper, B., & Schnekenburger, J. (2024). Quantitative Phase Imaging as Sensitive Screening Method for Nanoparticle-Induced Cytotoxicity Assessment. Cells, 13(8), 697. https://doi.org/10.3390/cells13080697