A Novel Interaction of Slug (SNAI2) and Nuclear Actin
Abstract
1. Introduction
2. Materials and Methods
2.1. The Yeast Two-Hybrid System
2.2. Cell Culture
2.3. Apoptosis Assay
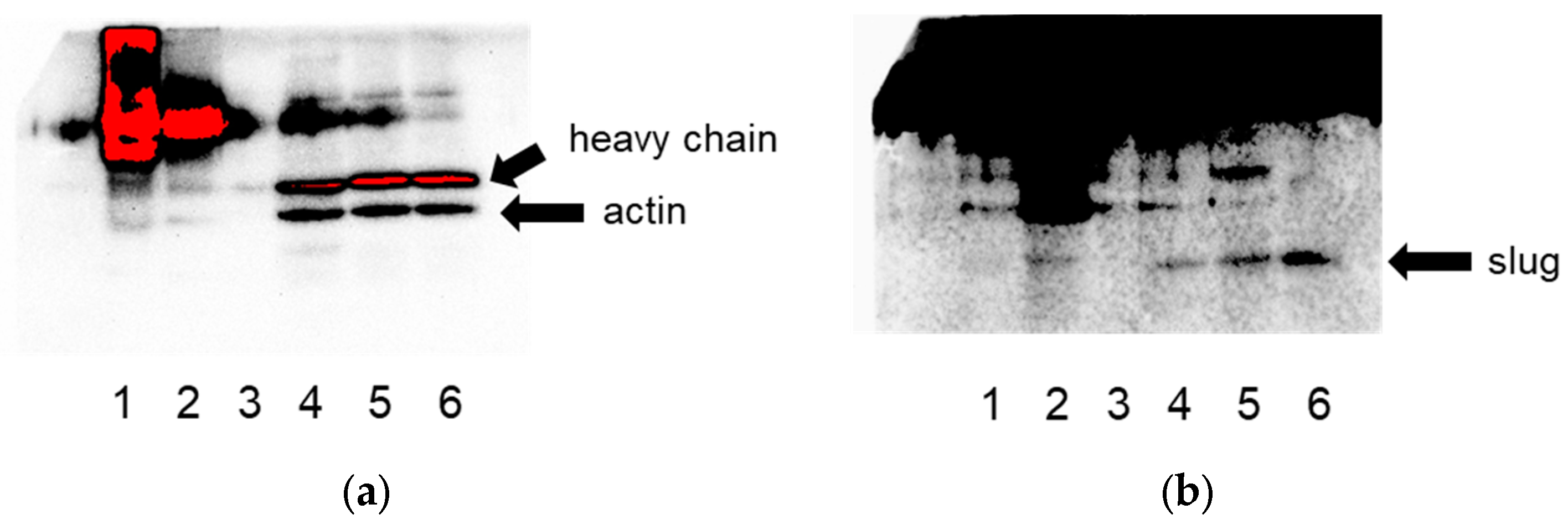
2.4. Immunoprecipitation
2.5. Western Blot
2.6. Transfection
2.7. Confocal Microscopy
2.8. Liquid Chromatography–Mass Spectrometry Analysis
2.9. Statistics
3. Results
3.1. Actin Interacts with Slug
3.2. Slug and Actin Mutually Influence Each Other
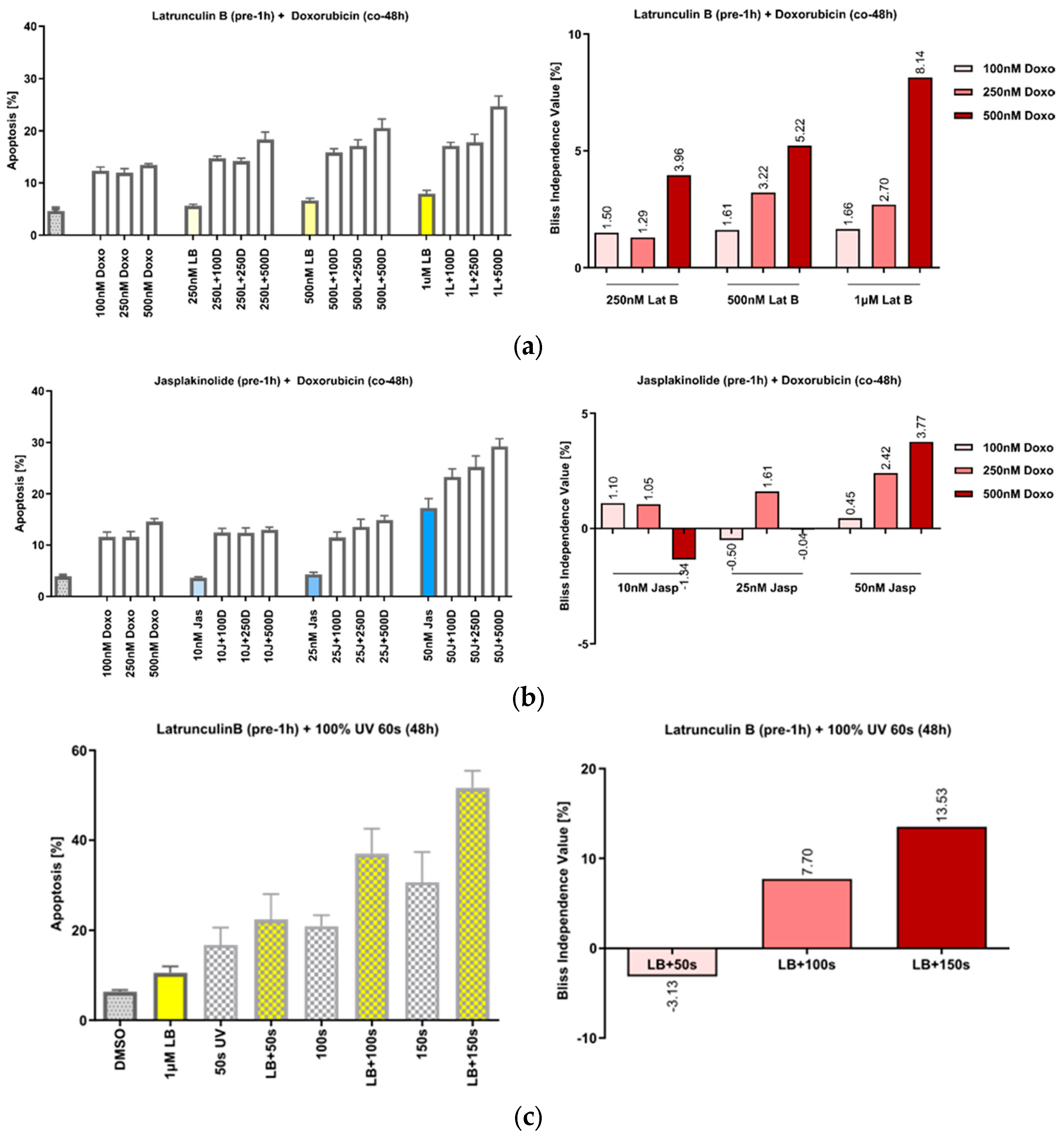
3.3. The Manipulation of Actin Acts Synergistically with DNA Damage by Doxorubicin or UVA Irradiation
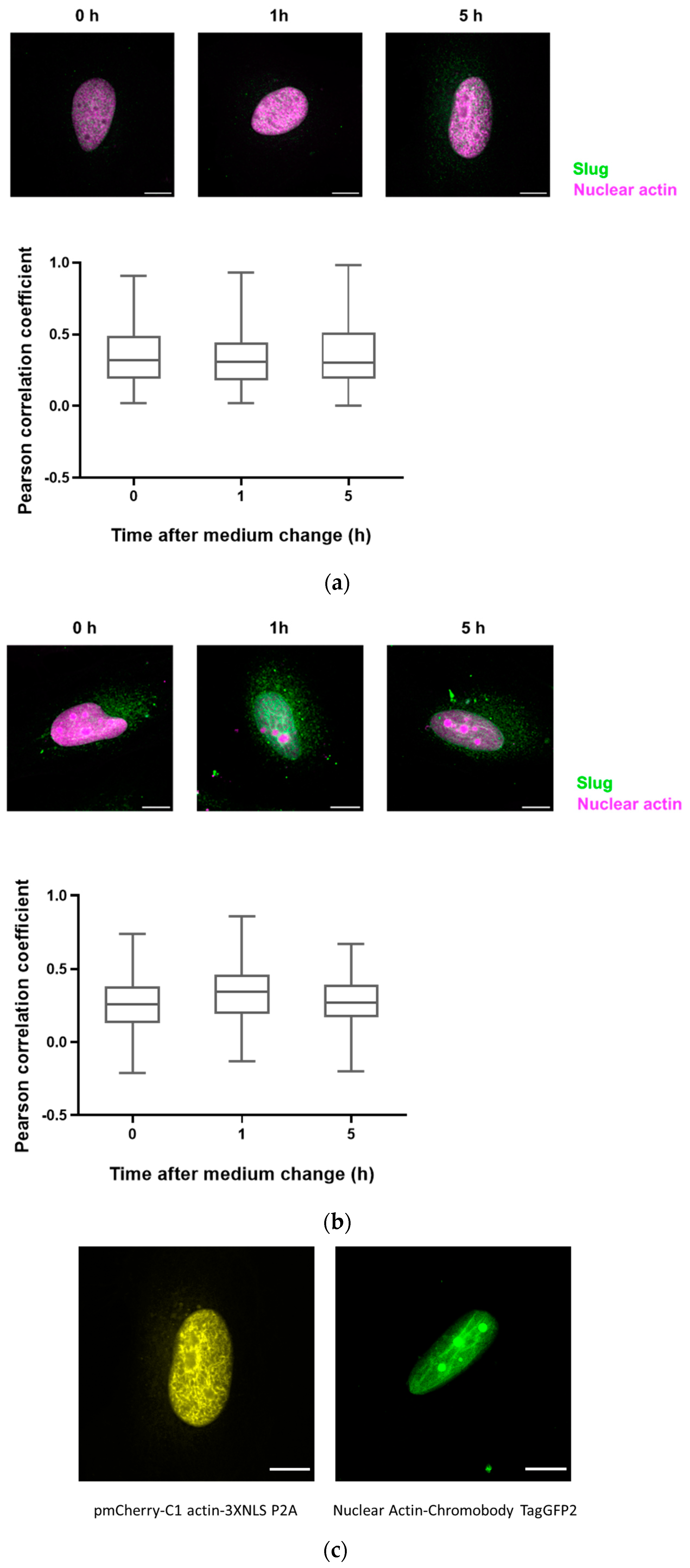
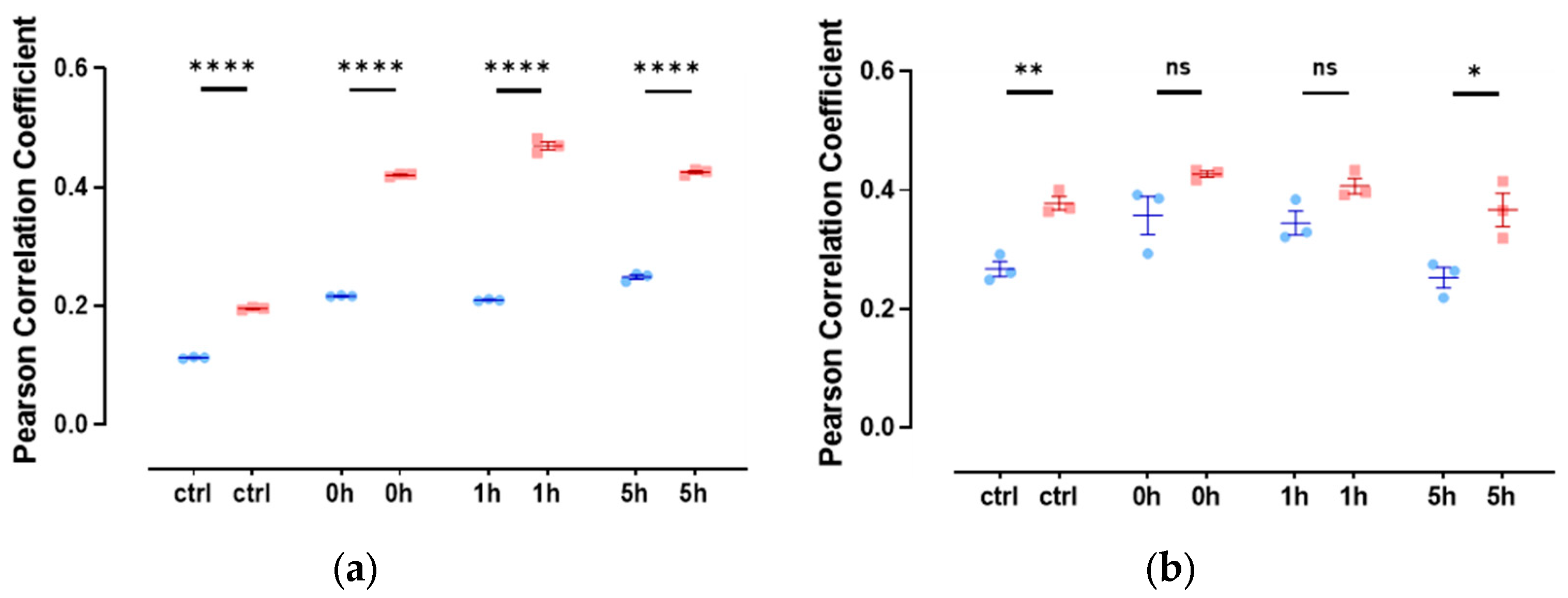
3.4. Slug and Actin Fused with an NLS Show Weak Spatial Correlation during DNA Damage Repair
3.5. The Colocalization of Slug with Other Proteins of DNA Repair Complexes Changes during Repair and Depending on the Levels of Actin with an NLS
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Pollard, T.D. Actin and Actin-Binding Proteins. Cold Spring Harb. Perspect. Biol. 2016, 8, a018226. [Google Scholar] [CrossRef] [PubMed]
- Merino, F.; Pospich, S.; Raunser, S. Towards a structural understanding of the remodeling of the actin cytoskeleton. Semin. Cell Dev. Biol. 2020, 102, 51–64. [Google Scholar] [CrossRef]
- Wang, S.; Crevenna, A.H.; Ugur, I.; Marion, A.; Antes, I.; Kazmaier, U.; Hoyer, M.; Lamb, D.C.; Gegenfurtner, F.; Kliesmete, Z.; et al. Actin stabilizing compounds show specific biological effects due to their binding mode. Sci. Rep. 2019, 9, 9731. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Gegenfurtner, F.A.; Crevenna, A.H.; Ziegenhain, C.; Kliesmete, Z.; Enard, W.; Muller, R.; Vollmar, A.M.; Schneider, S.; Zahler, S. Chivosazole A Modulates Protein-Protein Interactions of Actin. J. Nat. Prod. 2019, 82, 1961–1970. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Meixner, M.; Yu, L.; Zhuo, L.; Karmann, L.; Kazmaier, U.; Vollmar, A.M.; Antes, I.; Zahler, S. Turning the Actin Nucleating Compound Miuraenamide into Nucleation Inhibitors. ACS Omega 2021, 6, 22165–22172. [Google Scholar] [CrossRef] [PubMed]
- Hyrskyluoto, A.; Vartiainen, M.K. Regulation of nuclear actin dynamics in development and disease. Curr. Opin. Cell Biol. 2020, 64, 18–24. [Google Scholar] [CrossRef]
- Serebryannyy, L.; de Lanerolle, P. Nuclear actin: The new normal. Mutat. Res. 2020, 821, 111714. [Google Scholar] [CrossRef] [PubMed]
- Kloc, M.; Chanana, P.; Vaughn, N.; Uosef, A.; Kubiak, J.Z.; Ghobrial, R.M. New Insights into Cellular Functions of Nuclear Actin. Biology 2021, 10, 304. [Google Scholar] [CrossRef] [PubMed]
- Wollscheid, H.P.; Ulrich, H.D. Chromatin meets the cytoskeleton: The importance of nuclear actin dynamics and associated motors for genome stability. DNA Repair 2023, 131, 103571. [Google Scholar] [CrossRef]
- Zahler, S. Nuclear actin in cancer biology. Int. Rev. Cell Mol. Biol. 2020, 355, 53–66. [Google Scholar] [CrossRef]
- Phillips, S.; Kuperwasser, C. SLUG: Critical regulator of epithelial cell identity in breast development and cancer. Cell Adh. Migr. 2014, 8, 578–587. [Google Scholar] [CrossRef] [PubMed]
- Kajita, M.; McClinic, K.N.; Wade, P.A. Aberrant expression of the transcription factors snail and slug alters the response to genotoxic stress. Mol. Cell Biol. 2004, 24, 7559–7566. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Yao, J.; Suyama, K.; Qian, X.; Qian, B.Z.; Bandyopadhyay, S.; Loudig, O.; De Leon-Rodriguez, C.; Zhou, Z.N.; Segall, J.; et al. Slug promotes survival during metastasis through suppression of Puma-mediated apoptosis. Cancer Res. 2014, 74, 3695–3706. [Google Scholar] [CrossRef] [PubMed]
- Perez-Caro, M.; Bermejo-Rodriguez, C.; Gonzalez-Herrero, I.; Sanchez-Beato, M.; Piris, M.A.; Sanchez-Garcia, I. Transcriptomal profiling of the cellular response to DNA damage mediated by Slug (Snai2). Br. J. Cancer 2008, 98, 480–488. [Google Scholar] [CrossRef]
- Murakoshi, H.; Lee, S.J.; Yasuda, R. Highly sensitive and quantitative FRET-FLIM imaging in single dendritic spines using improved non-radiative YFP. Brain Cell Biol. 2008, 36, 31–42. [Google Scholar] [CrossRef] [PubMed]
- Nicoletti, I.; Migliorati, G.; Pagliacci, M.C.; Grignani, F.; Riccardi, C. A rapid and simple method for measuring thymocyte apoptosis by propidium iodide staining and flow cytometry. J. Immunol. Methods 1991, 139, 271–279. [Google Scholar] [CrossRef] [PubMed]
- Belin, B.J.; Lee, T.; Mullins, R.D. DNA damage induces nuclear actin filament assembly by Formin -2 and Spire-(1/2) that promotes efficient DNA repair. Elife 2015, 4, e07735. [Google Scholar] [CrossRef]
- Demichev, V.; Messner, C.B.; Vernardis, S.I.; Lilley, K.S.; Ralser, M. DIA-NN: Neural networks and interference correction enable deep proteome coverage in high throughput. Nat. Methods 2020, 17, 41–44. [Google Scholar] [CrossRef]
- Tyanova, S.; Temu, T.; Sinitcyn, P.; Carlson, A.; Hein, M.Y.; Geiger, T.; Mann, M.; Cox, J. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods 2016, 13, 731–740. [Google Scholar] [CrossRef]
- Pomeroy, A.E.; Schmidt, E.V.; Sorger, P.K.; Palmer, A.C. Drug independence and the curability of cancer by combination chemotherapy. Trends Cancer 2022, 8, 915–929. [Google Scholar] [CrossRef]
- Ryall, K.A.; Tan, A.C. Systems biology approaches for advancing the discovery of effective drug combinations. J. Cheminform. 2015, 7, 7. [Google Scholar] [CrossRef] [PubMed]
- Pfitzer, L.; Moser, C.; Gegenfurtner, F.; Arner, A.; Foerster, F.; Atzberger, C.; Zisis, T.; Kubisch-Dohmen, R.; Busse, J.; Smith, R.; et al. Targeting actin inhibits repair of doxorubicin-induced DNA damage: A novel therapeutic approach for combination therapy. Cell Death Dis. 2019, 10, 302. [Google Scholar] [CrossRef] [PubMed]
- Gross, K.M.; Zhou, W.; Breindel, J.L.; Ouyang, J.; Jin, D.X.; Sokol, E.S.; Gupta, P.B.; Huber, K.; Zou, L.; Kuperwasser, C. Loss of Slug Compromises DNA Damage Repair and Accelerates Stem Cell Aging in Mammary Epithelium. Cell Rep. 2019, 28, 394–407.e396. [Google Scholar] [CrossRef] [PubMed]
- Emadi Baygi, M.; Soheili, Z.S.; Essmann, F.; Deezagi, A.; Engers, R.; Goering, W.; Schulz, W.A. Slug/SNAI2 regulates cell proliferation and invasiveness of metastatic prostate cancer cell lines. Tumour Biol. 2010, 31, 297–307. [Google Scholar] [CrossRef] [PubMed]
- Morita, T.; Mayanagi, T.; Sobue, K. Dual roles of myocardin-related transcription factors in epithelial mesenchymal transition via slug induction and actin remodeling. J. Cell Biol. 2007, 179, 1027–1042. [Google Scholar] [CrossRef] [PubMed]
- Zagelbaum, J.; Schooley, A.; Zhao, J.; Schrank, B.R.; Callen, E.; Zha, S.; Gottesman, M.E.; Nussenzweig, A.; Rabadan, R.; Dekker, J.; et al. Multiscale reorganization of the genome following DNA damage facilitates chromosome translocations via nuclear actin polymerization. Nat. Struct. Mol. Biol. 2023, 30, 99–106. [Google Scholar] [CrossRef] [PubMed]
- Melak, M.; Plessner, M.; Grosse, R. Actin visualization at a glance. J. Cell Sci. 2017, 130, 525–530. [Google Scholar] [CrossRef] [PubMed]
- Samper-Martin, B.; Sarrias, A.; Lazaro, B.; Perez-Montero, M.; Rodriguez-Rodriguez, R.; Ribeiro, M.P.C.; Banon, A.; Wolfgeher, D.; Jessen, H.J.; Alsina, B.; et al. Polyphosphate degradation by Nudt3-Zn(2+) mediates oxidative stress response. Cell Rep. 2021, 37, 110004. [Google Scholar] [CrossRef]
- Chang, J.S.; Henry, K.; Wolf, B.L.; Geli, M.; Lemmon, S.K. Protein phosphatase-1 binding to scd5p is important for regulation of actin organization and endocytosis in yeast. J. Biol. Chem. 2002, 277, 48002–48008. [Google Scholar] [CrossRef]
- Lee, J.; Beliakoff, J.; Sun, Z. The novel PIAS-like protein hZimp10 is a transcriptional co-activator of the p53 tumor suppressor. Nucleic Acids Res. 2007, 35, 4523–4534. [Google Scholar] [CrossRef]
- Horikoshi, N.; Pandita, R.K.; Mujoo, K.; Hambarde, S.; Sharma, D.; Mattoo, A.R.; Chakraborty, S.; Charaka, V.; Hunt, C.R.; Pandita, T.K. beta2-spectrin depletion impairs DNA damage repair. Oncotarget 2016, 7, 33557–33570. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Zhang, R.; Liu, C.; Niu, Y.; Jing, Y.; Zhang, H.; Wang, J.; Yang, J.; Zen, K.; Zhang, J.; Zhang, C.Y.; et al. MicroRNA-128-3p regulates mitomycin C-induced DNA damage response in lung cancer cells through repressing SPTAN1. Oncotarget 2017, 8, 58098–58107. [Google Scholar] [CrossRef] [PubMed]
- Mitra, A.; Yan, J.; Xia, X.; Zhou, S.; Chen, J.; Mishra, L.; Li, S. IL6-mediated inflammatory loop reprograms normal to epithelial-mesenchymal transition(+) metastatic cancer stem cells in preneoplastic liver of transforming growth factor beta-deficient beta2-spectrin(+/−) mice. Hepatology 2017, 65, 1222–1236. [Google Scholar] [CrossRef] [PubMed]


| Insert Identity | Frequency |
|---|---|
| 3′-phosphoadenosine 5′-phosphosulfate synthase 1 (PAPSS1) | 1 |
| BRX1, biogenesis of ribosomes (BRIX1) | 1 |
| chromosome 9 open reading frame 131 (C9orf131) | 1 |
| cofilin 2 (CFL2) | 16 |
| COP9 signalosome subunit 5 (COPS5) | 8 |
| cyclase-associated actin cytoskeleton regulatory protein 2 (CAP2) | 164 |
| cysteine sulfinic acid decarboxylase (CSAD) | 3 |
| double zinc ribbon and ankyrin repeat domains 1 (DZANK1) | 1 |
| dual-specificity phosphatase 5 (DUSP5) | 3 |
| endoplasmic reticulum lectin 1 (ERLEC1) | 1 |
| ENY2, transcription and export complex 2 subunit (ENY2) | 1 |
| epidermal growth factor receptor pathway substrate 8 (EPS8) | 1 |
| F-box and WD repeat domain-containing 7 (FBXW7) | 1 |
| gamma-aminobutyric acid type A receptor alpha4 subunit (GABRA4) | 1 |
| integrin subunit alpha 8 (ITGA8) | 2 |
| late endosomal/lysosomal adaptor, MAPK and MTOR activator 5 (LAMTOR5) | 8 |
| lysine acetyltransferase 14 (KAT14) | 10 |
| NLR family pyrin domain-containing 1 (NLRP1) | 13 |
| nuclear receptor binding factor 2 (NRBF2) | 2 |
| phosphodiesterase 1C (PDE1C) | 1 |
| RAD51 associated protein 2 (RAD51AP2) | 62 |
| ribosomal protein L26 (RPL26) | 1 |
| snail family transcriptional repressor 2 (SNAI2) | 1 |
| SNAP-associated protein (SNAPIN) | 9 |
| translation machinery associated 7 homolog (TMA7) | 11 |
| VHL-binding protein 1 (VBP1) | 3 |
| zinc finger CCHC-type containing 4 (ZCCHC4) | 2 |
| zinc finger protein 148 (ZNF148) | 3 |
| Genes | p-Value | Q-Value | log2-Fold Change |
|---|---|---|---|
| GEMIN7 | 0.000386 | 0.044 | −1.9 |
| NUDT3 | 0.000135 | 0.027 | −0.6 |
| SC5D | 0.00012 | 0.036 | −1.8 |
| SPTAN1 | 0.00042 | 0.045778 | 0.4 |
| SPTBN1 | 9.41 × 10−5 | 0.054 | 0.5 |
| VPS33A | 0.000353 | 0.058667 | −0.3 |
| ZMIZ1 | 0.000495 | 0.0412 | −2.8 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhuo, L.; Stöckl, J.B.; Fröhlich, T.; Moser, S.; Vollmar, A.M.; Zahler, S. A Novel Interaction of Slug (SNAI2) and Nuclear Actin. Cells 2024, 13, 696. https://doi.org/10.3390/cells13080696
Zhuo L, Stöckl JB, Fröhlich T, Moser S, Vollmar AM, Zahler S. A Novel Interaction of Slug (SNAI2) and Nuclear Actin. Cells. 2024; 13(8):696. https://doi.org/10.3390/cells13080696
Chicago/Turabian StyleZhuo, Ling, Jan B. Stöckl, Thomas Fröhlich, Simone Moser, Angelika M. Vollmar, and Stefan Zahler. 2024. "A Novel Interaction of Slug (SNAI2) and Nuclear Actin" Cells 13, no. 8: 696. https://doi.org/10.3390/cells13080696
APA StyleZhuo, L., Stöckl, J. B., Fröhlich, T., Moser, S., Vollmar, A. M., & Zahler, S. (2024). A Novel Interaction of Slug (SNAI2) and Nuclear Actin. Cells, 13(8), 696. https://doi.org/10.3390/cells13080696







