Predicting Tolerance to Cow’s Milk Allergy in Children Using IgE and IgG4 Peptide Binding Profiles
Abstract
1. Introduction
2. Materials and Methods
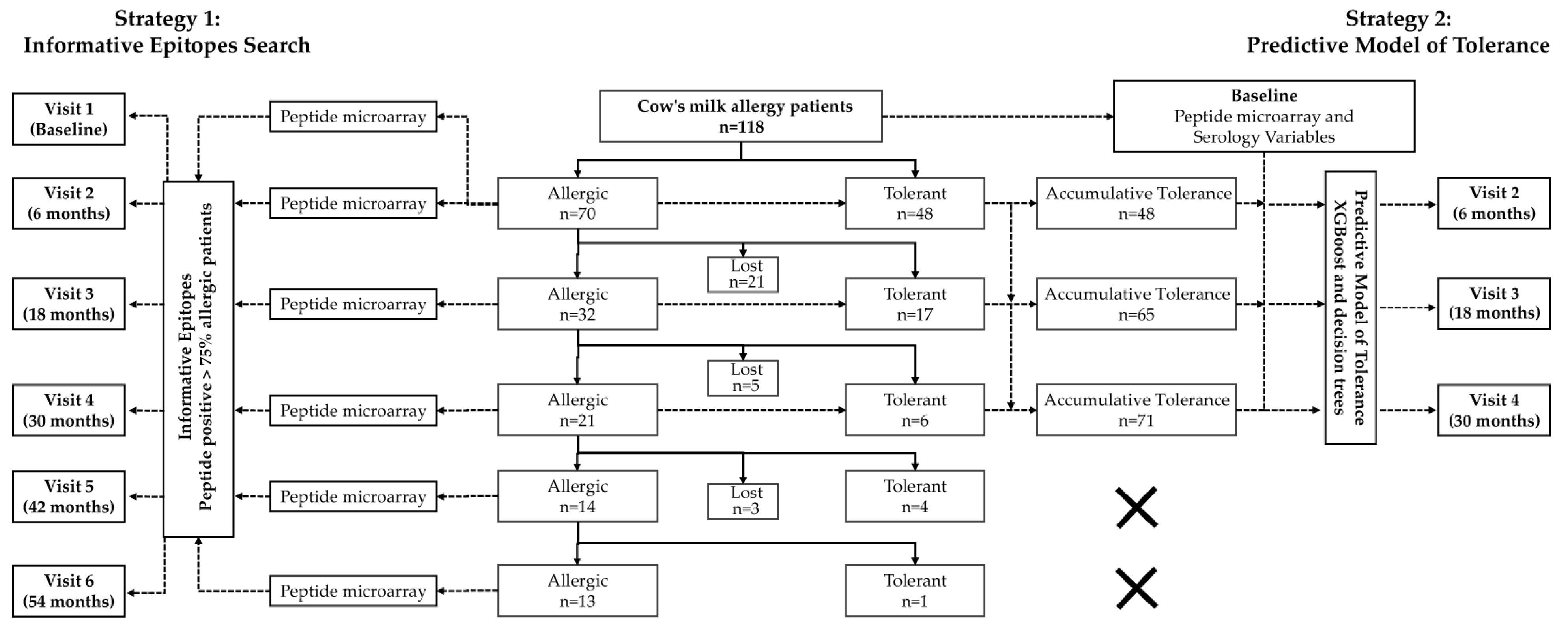
2.1. Design and Setting
2.2. Peptide Microarray Immunoassay
2.3. Data Analysis
3. Results
3.1. Characteristics of the Patients
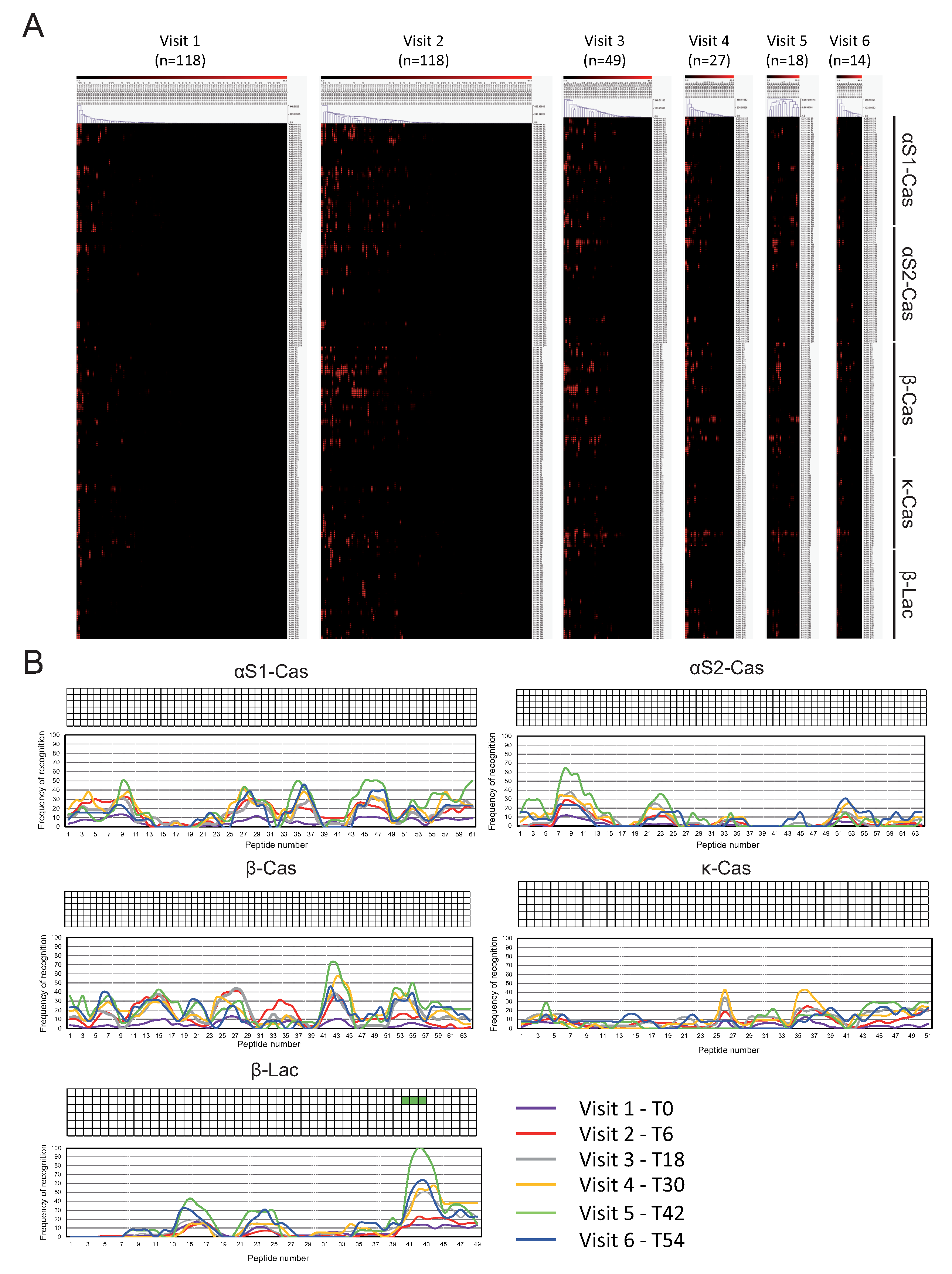
3.2. IgE and IgG4 Antibody Binding to Sequential Epitopes Reveled Four Informative Epitopes Recognized in All-Time Points
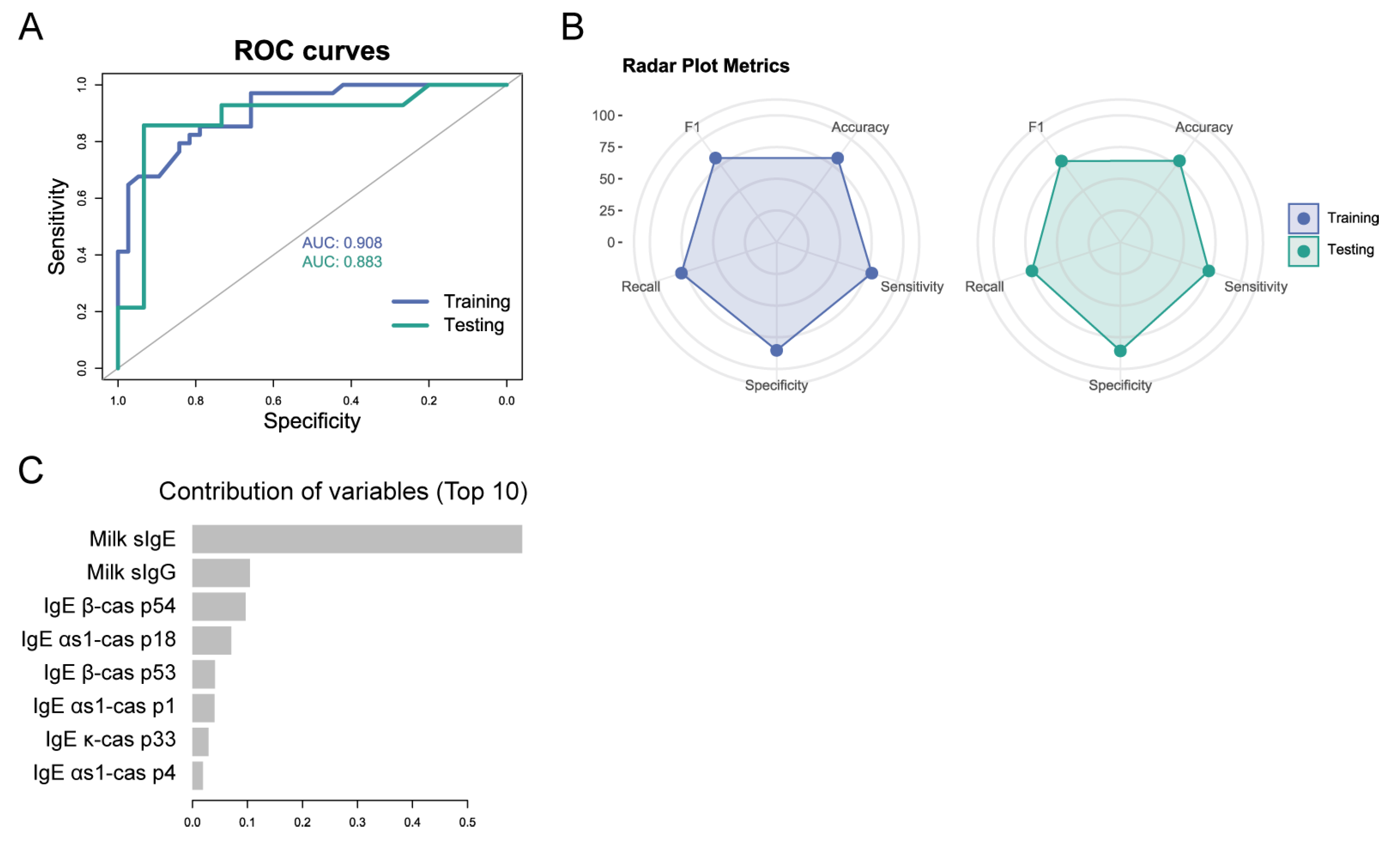
3.3. Combining Serological Variables and Peptide Biomarkers at Baseline Can Predict Tolerability at Later Time Points
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| BLG | β-Lactoglobulin |
| CAS | Casein |
| CM | Cow’s milk |
| CMA | Cow’s milk allergy |
| DBPCFC | Double-blind, placebo-controlled food challenge |
| IQR | Interquartile Range |
| SPT | Skin prick test |
| AUC | Area under the curve |
| XGBoost | Extreme gradient boosting |
References
- Switkowski, K.M.; Oken, E.; Rifas-Shiman, S.L.; Camargo, C.A., Jr.; Gold, D.R.; Sordillo, J.E.; Lightdale, J.R. Timing of Cow’s Milk Protein Introduction and Childhood Adverse Reactions to Cow’s Milk. J. Allergy Clin. Immunol. 2022, 10, 2713–2721.e2. [Google Scholar] [CrossRef] [PubMed]
- Fiocchi, A.; Schunemann, H.J.; Brozek, J.; Restani, P.; Beyer, K.; Troncone, R.; Martelli, A.; Terracciano, L.; Bahna, S.L.; Rance, F.; et al. Diagnosis and Rationale for Action Against Cow’s Milk Allergy (DRACMA): A summary report. J. Allergy Clin. Immunol. 2010, 126, 1119–1128.e12. [Google Scholar] [CrossRef] [PubMed]
- Wal, J.M.; Bernard, H.; Creminon, C.; Hamberger, C.; David, B.; Peltre, G. Cow’s milk allergy: The humoral immune response to eight purified allergens. Adv. Exp. Med. Biol. 1995, 371B, 879–881. [Google Scholar] [PubMed]
- Cerecedo, I.; Zamora, J.; Shreffler, W.G.; Lin, J.; Bardina, L.; Dieguez, M.C.; Wang, J.; Muriel, A.; de la Hoz, B.; Sampson, H.A. Mapping of the IgE and IgG4 sequential epitopes of milk allergens with a peptide microarray-based immunoassay. J. Allergy Clin. Immunol. 2008, 122, 589–594. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.; Bruni, F.M.; Fu, Z.; Maloney, J.; Bardina, L.; Boner, A.L.; Gimenez, G.; Sampson, H.A. A bioinformatics approach to identify patients with symptomatic peanut allergy using peptide microarray immunoassay. J. Allergy Clin. Immunol. 2012, 129, 1321–1328.e5. [Google Scholar] [CrossRef]
- Sanchez-Ruano, L.; de la Hoz, B.; Martinez-Botas, J. Clinical utility of microarray B-cell epitope mapping in food allergies: A systematic review. Pediatr. Allergy Immunol. 2020, 31, 175–185. [Google Scholar] [CrossRef]
- Chatchatee, P.; Jarvinen, K.M.; Bardina, L.; Beyer, K.; Sampson, H.A. Identification of IgE- and IgG-binding epitopes on alpha(s1)-casein: Differences in patients with persistent and transient cow’s milk allergy. J. Allergy Clin. Immunol. 2001, 107, 379–383. [Google Scholar] [CrossRef]
- Vila, L.; Beyer, K.; Jarvinen, K.M.; Chatchatee, P.; Bardina, L.; Sampson, H.A. Role of conformational and linear epitopes in the achievement of tolerance in cow’s milk allergy. Clin. Exp. Allergy 2001, 31, 1599–1606. [Google Scholar] [CrossRef]
- Jarvinen, K.M.; Beyer, K.; Vila, L.; Chatchatee, P.; Busse, P.J.; Sampson, H.A. B-cell epitopes as a screening instrument for persistent cow’s milk allergy. J. Allergy Clin. Immunol. 2002, 110, 293–297. [Google Scholar] [CrossRef]
- Matsumoto, N.; Okochi, M.; Matsushima, M.; Kato, R.; Takase, T.; Yoshida, Y.; Kawase, M.; Isobe, K.; Kawabe, T.; Honda, H. Peptide array-based analysis of the specific IgE and IgG4 in cow’s milk allergens and its use in allergy evaluation. Peptides 2009, 30, 1840–1847. [Google Scholar] [CrossRef]
- Savilahti, E.M.; Rantanen, V.; Lin, J.S.; Karinen, S.; Saarinen, K.M.; Goldis, M.; Makela, M.J.; Hautaniemi, S.; Savilahti, E.; Sampson, H.A. Early recovery from cow’s milk allergy is associated with decreasing IgE and increasing IgG4 binding to cow’s milk epitopes. J. Allergy Clin. Immunol. 2010, 125, 1315–1321.e9. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Lin, J.; Bardina, L.; Goldis, M.; Nowak-Wegrzyn, A.; Shreffler, W.G.; Sampson, H.A. Correlation of IgE/IgG4 milk epitopes and affinity of milk-specific IgE antibodies with different phenotypes of clinical milk allergy. J. Allergy Clin. Immunol. 2010, 125, 695–702, 702.e1–702.e6. [Google Scholar] [CrossRef] [PubMed]
- Tomicic, S.; Norrman, G.; Falth-Magnusson, K.; Jenmalm, M.C.; Devenney, I.; Bottcher, M.F. High levels of IgG4 antibodies to foods during infancy are associated with tolerance to corresponding foods later in life. Pediatr. Allergy Immunol. 2009, 20, 35–41. [Google Scholar] [CrossRef]
- Savilahti, E.M.; Saarinen, K.M.; Savilahti, E. Duration of clinical reactivity in cow’s milk allergy is associated with levels of specific immunoglobulin G4 and immunoglobulin A antibodies to beta-lactoglobulin. Clin. Exp. Allergy 2010, 40, 251–256. [Google Scholar] [CrossRef]
- Ruiter, B.; Knol, E.F.; van Neerven, R.J.; Garssen, J.; Bruijnzeel-Koomen, C.A.; Knulst, A.C.; van Hoffen, E. Maintenance of tolerance to cow’s milk in atopic individuals is characterized by high levels of specific immunoglobulin G4. Clin. Exp. Allergy 2007, 37, 1103–1110. [Google Scholar] [CrossRef]
- Qin, L.; Tang, L.F.; Cheng, L.; Wang, H.Y. The clinical significance of allergen-specific IgG4 in allergic diseases. Front. Immunol. 2022, 13, 1032909. [Google Scholar] [CrossRef]
- Manso, L.; Pineda, R.; Huertas, B.; Fernandez-Rivas, M.; Dieguez, M.C.; Cerecedo, I.; Muriel, A.; Fernandez, F.B.; DunnGalvin, A.; Antolin-Amerigo, D.; et al. Validation of the Spanish Version of the Food Allergy Quality of Life Questionnaire-Parent Form (S-FAQLQ-PF). J. Investig. Allergol. Clin. Immunol. 2017, 27, 363–369. [Google Scholar] [CrossRef]
- Hoffmann-Sommergruber, K. The SAFE project: ‘plant food allergies: Field to table strategies for reducing their incidence in Europe’ an EC-funded study. Allergy 2005, 60, 436–442. [Google Scholar] [CrossRef]
- Martinez-Botas, J.; Rodriguez-Alvarez, M.; Cerecedo, I.; Vlaicu, C.; Dieguez, M.C.; Gomez-Coronado, D.; Fernandez-Rivas, M.; de la Hoz, B. Identification of novel peptide biomarkers to predict safety and efficacy of cow’s milk oral immunotherapy by peptide microarray. Clin. Exp. Allergy 2015, 45, 1071–1084. [Google Scholar] [CrossRef]
- Lin, J.; Bardina, L.; Shreffler, W.G.; Andreae, D.A.; Ge, Y.; Wang, J.; Bruni, F.M.; Fu, Z.; Han, Y.; Sampson, H.A. Development of a novel peptide microarray for large-scale epitope mapping of food allergens. J. Allergy Clin. Immunol. 2009, 124, 315–322, 322.e1–322.e3. [Google Scholar] [CrossRef]
- Lin, J.; Bardina, L.; Shreffler, W.G. Microarrayed allergen molecules for diagnostics of allergy. Methods Mol. Biol. 2009, 524, 259–272. [Google Scholar] [PubMed]
- Chen, T.; Guestrin, C. XGBoost: A scalable tree boosting system. arXiv 2016, arXiv:1603.02754. [Google Scholar]
- Kuhn, M. Building Predictive Models in R Using the caret Package. J. Stat. Softw. 2008, 28, 1–26. [Google Scholar] [CrossRef]
- Skripak, J.M.; Matsui, E.C.; Mudd, K.; Wood, R.A. The natural history of IgE-mediated cow’s milk allergy. J. Allergy Clin. Immunol. 2007, 120, 1172–1177. [Google Scholar] [CrossRef]
- Santos, A.F.; Riggioni, C.; Agache, I.; Akdis, C.A.; Akdis, M.; Alvarez-Perea, A.; Alvaro-Lozano, M.; Ballmer-Weber, B.; Barni, S.; Beyer, K.; et al. EAACI guidelines on the management of IgE-mediated food allergy. Allergy 2025, 80, 14–36. [Google Scholar] [CrossRef]
- Caubet, J.C.; Lin, J.; Ahrens, B.; Gimenez, G.; Bardina, L.; Niggemann, B.; Sampson, H.A.; Beyer, K. Natural tolerance development in cow’s milk allergic children: IgE and IgG4 epitope binding. Allergy 2017, 72, 1677–1685. [Google Scholar] [CrossRef]
- Koike, Y.; Sato, S.; Yanagida, N.; Asaumi, T.; Ogura, K.; Ohtani, K.; Imai, T.; Ebisawa, M. Predictors of Persistent Milk Allergy in Children: A Retrospective Cohort Study. Int. Arch. Allergy Immunol. 2018, 175, 177–180. [Google Scholar] [CrossRef]
- Rambo, I.M.; Kronfel, C.M.; Rivers, A.R.; Swientoniewski, L.T.; McBride, J.K.; Cheng, H.; Simon, R.J.; Ryan, R.; Tilles, S.A.; Nesbit, J.B.; et al. IgE and IgG4 epitopes of the peanut allergens shift following oral immunotherapy. Front. Allergy 2023, 4, 1279290. [Google Scholar] [CrossRef]
- Vickery, B.P.; Lin, J.; Kulis, M.; Fu, Z.; Steele, P.H.; Jones, S.M.; Scurlock, A.M.; Gimenez, G.; Bardina, L.; Sampson, H.A.; et al. Peanut oral immunotherapy modifies IgE and IgG4 responses to major peanut allergens. J. Allergy Clin. Immunol. 2013, 131, 128–134.e1-3. [Google Scholar] [CrossRef]
- Garcia-Ara, M.C.; Boyano-Martinez, M.T.; Diaz-Pena, J.M.; Martin-Munoz, M.F.; Martin-Esteban, M. Cow’s milk-specific immunoglobulin E levels as predictors of clinical reactivity in the follow-up of the cow’s milk allergy infants. Clin. Exp. Allergy 2004, 34, 866–870. [Google Scholar] [CrossRef]
- Jarvinen, K.M.; Chatchatee, P.; Bardina, L.; Beyer, K.; Sampson, H.A. IgE and IgG binding epitopes on alpha-lactalbumin and beta-lactoglobulin in cow’s milk allergy. Int. Arch. Allergy Immunol. 2001, 126, 111–118. [Google Scholar] [CrossRef] [PubMed]
- Fiocchi, A.; Bouygue, G.R.; Albarini, M.; Restani, P. Molecular diagnosis of cow’s milk allergy. Curr. Opin. Allergy Clin. Immunol. 2011, 11, 216–221. [Google Scholar] [CrossRef] [PubMed]
- Valenta, R.; Ferreira, F.; Focke-Tejkl, M.; Linhart, B.; Niederberger, V.; Swoboda, I.; Vrtala, S. From allergen genes to allergy vaccines. Annu. Rev. Immunol. 2010, 28, 211–241. [Google Scholar] [CrossRef] [PubMed]



| Characteristics | Number |
|---|---|
| Total number of patients | 118 |
| Age at initial visit, median (IQR) | 6.2 mo. (4.8–7.44) |
| Sex, n (%) | |
| Female | 58 (49.2%) |
| Male | 60 (50.8%) |
| Ethnicity, n (%) | |
| Caucasians | 107 (90.68%) |
| Hispanics | 9 (7.62%) |
| Others | 2 (1.68%) |
| Atopic dermatitis, n (%) | 52 (44.44%) |
| Breastfeeding, n (%) | 118 (100%) |
| Neonatal bottle, n (%) | 55 (46.61%) |
| Initial symptoms of CMA, n (%) | |
| Oral | 29 (24.58%) |
| Cutaneous | 102 (86.44%) |
| Gastrointestinal | 54 (45.76%) |
| Respiratory | 9 (7.63%) |
| Cardiovascular | 2 (1.69%) |
| Neurological | 1 (0.85%) |
| Total IgE, kU/L, median (IQR) | 14.35 (7.88–28.03) |
| Specific IgE, kU/L, median (IQR) | |
| Milk | 0.89 (0.35–2.64) |
| α-Lactalbumin | 0.35 (0.17–0.80) |
| β-Lactoglobulin | 0.39 (0.30–1.26) |
| Casein | 0.34 (0.13–0.72) |
| Specific IgG4, mg/L, median (IQR) | |
| α-Lactalbumin | 0.08 (0.01–0.35) |
| β-Lactoglobulin | 0.16 (0.07–0.82) |
| Casein | 0.11 (0.05–0.26) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fernández-Lozano, C.; Olmos-Piñero, S.; Sánchez-Ruano, L.; Terrados, S.; Diéguez, M.d.C.; Fernández-Rivas, M.; Vlaicu, C.; Cerecedo, I.; Gonzalo-Fernandez, A.; de la Hoz, B.; et al. Predicting Tolerance to Cow’s Milk Allergy in Children Using IgE and IgG4 Peptide Binding Profiles. Cells 2025, 14, 344. https://doi.org/10.3390/cells14050344
Fernández-Lozano C, Olmos-Piñero S, Sánchez-Ruano L, Terrados S, Diéguez MdC, Fernández-Rivas M, Vlaicu C, Cerecedo I, Gonzalo-Fernandez A, de la Hoz B, et al. Predicting Tolerance to Cow’s Milk Allergy in Children Using IgE and IgG4 Peptide Binding Profiles. Cells. 2025; 14(5):344. https://doi.org/10.3390/cells14050344
Chicago/Turabian StyleFernández-Lozano, Carlos, Sergio Olmos-Piñero, Laura Sánchez-Ruano, Soledad Terrados, Mª del Carmen Diéguez, Montserrat Fernández-Rivas, Cristina Vlaicu, Inmaculada Cerecedo, Alejandro Gonzalo-Fernandez, Belén de la Hoz, and et al. 2025. "Predicting Tolerance to Cow’s Milk Allergy in Children Using IgE and IgG4 Peptide Binding Profiles" Cells 14, no. 5: 344. https://doi.org/10.3390/cells14050344
APA StyleFernández-Lozano, C., Olmos-Piñero, S., Sánchez-Ruano, L., Terrados, S., Diéguez, M. d. C., Fernández-Rivas, M., Vlaicu, C., Cerecedo, I., Gonzalo-Fernandez, A., de la Hoz, B., & Martínez-Botas, J. (2025). Predicting Tolerance to Cow’s Milk Allergy in Children Using IgE and IgG4 Peptide Binding Profiles. Cells, 14(5), 344. https://doi.org/10.3390/cells14050344








