ESCRT Machinery in HBV Life Cycle: Dual Roles in Autophagy and Membrane Dynamics for Viral Pathogenesis
Abstract
1. Introduction
2. ESCRT-Dependent MVB Biogenesis
3. Autophagy
4. Crosstalk Between Autophagy and ESCRT Pathways
5. ESCRT and Lysosome
6. ESCRT in Diseases
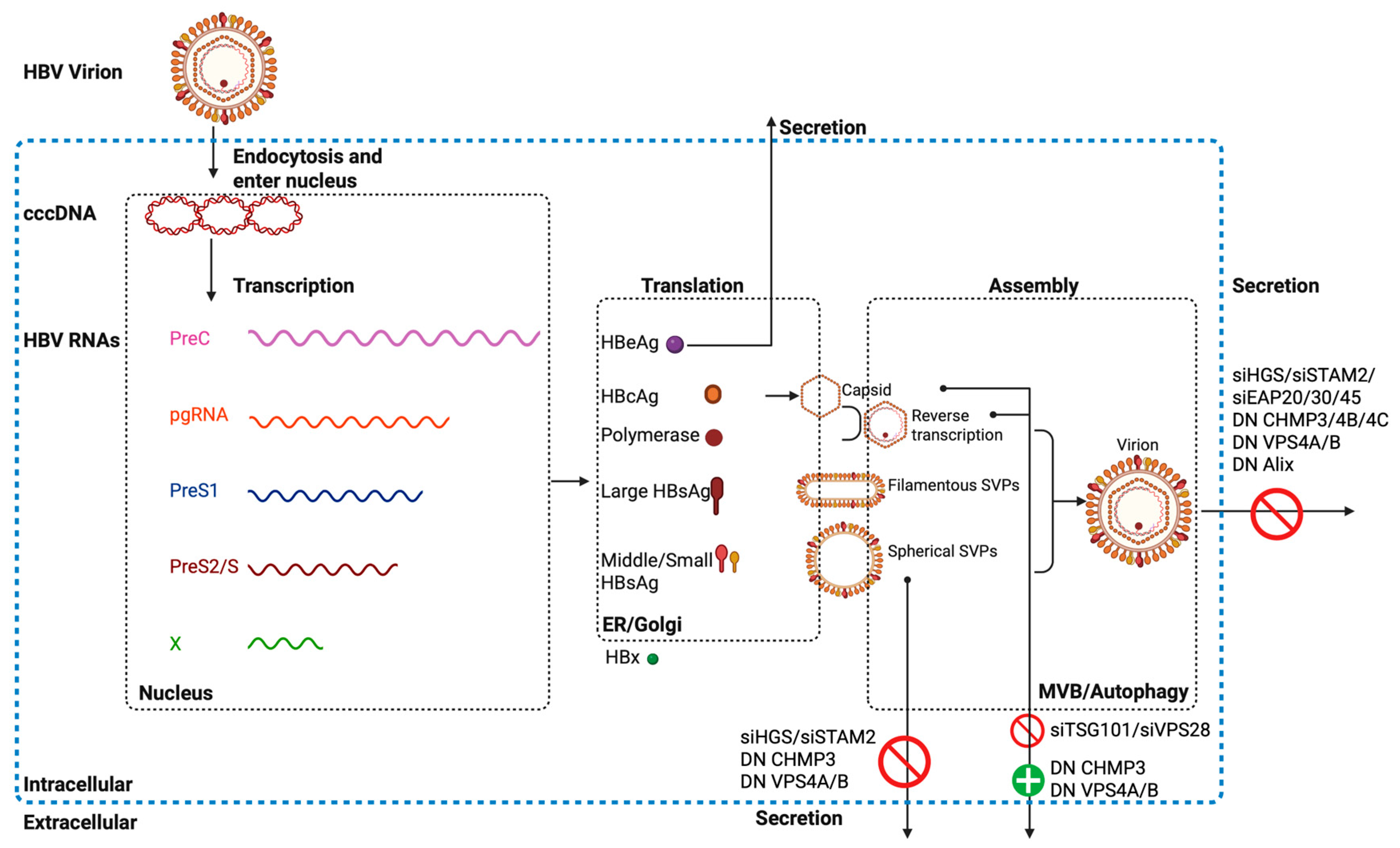
7. ESCRT and HBV
8. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hurley, J.H. The ESCRT complexes. Crit. Rev. Biochem. Mol. Biol. 2010, 45, 463–487. [Google Scholar] [CrossRef] [PubMed]
- Hurley, J.H. ESCRTs are everywhere. EMBO J. 2015, 34, 2398–2407. [Google Scholar] [CrossRef] [PubMed]
- Vietri, M.; Radulovic, M.; Stenmark, H. The many functions of ESCRTs. Nat. Rev. Mol. Cell Biol. 2020, 21, 25–42. [Google Scholar] [CrossRef] [PubMed]
- Kroemer, G.; Marino, G.; Levine, B. Autophagy and the integrated stress response. Mol. Cell 2010, 40, 280–293. [Google Scholar] [CrossRef]
- Chang, C.; Jensen, L.E.; Hurley, J.H. Autophagosome biogenesis comes out of the black box. Nat. Cell Biol. 2021, 23, 450–456. [Google Scholar] [CrossRef]
- Rusten, T.E.; Simonsen, A. ESCRT functions in autophagy and associated disease. Cell Cycle 2008, 7, 1166–1172. [Google Scholar] [CrossRef]
- WHO. Hepatitis B. 2023. Available online: https://www.who.int/news-room/fact-sheets/detail/hepatitis-b (accessed on 9 April 2024).
- Li, J.; Lin, Y.; Wang, X.; Lu, M. Interconnection of cellular autophagy and endosomal vesicle trafficking and its role in hepatitis B virus replication and release. Virol. Sin. 2024, 39, 24–30. [Google Scholar] [CrossRef]
- Catalano, M.; O’Driscoll, L. Inhibiting extracellular vesicles formation and release: A review of EV inhibitors. J. Extracell. Vesicles 2020, 9, 1703244. [Google Scholar] [CrossRef]
- Kaur, S.; Verma, H.; Dhiman, M.; Tell, G.; Gigli, G.L.; Janes, F.; Mantha, A.K. Brain Exosomes: Friend or Foe in Alzheimer’s Disease? Mol. Neurobiol. 2021, 58, 6610–6624. [Google Scholar] [CrossRef]
- Wollert, T.; Hurley, J.H. Molecular mechanism of multivesicular body biogenesis by ESCRT complexes. Nature 2010, 464, 864–869. [Google Scholar] [CrossRef]
- Raiborg, C.; Bremnes, B.; Mehlum, A.; Gillooly, D.J.; D’Arrigo, A.; Stang, E.; Stenmark, H. FYVE and coiled-coil domains determine the specific localisation of Hrs to early endosomes. J. Cell Sci. 2001, 114 Pt 12, 2255–2263. [Google Scholar] [CrossRef] [PubMed]
- Sundquist, W.I.; Schubert, H.L.; Kelly, B.N.; Hill, G.C.; Holton, J.M.; Hill, C.P. Ubiquitin recognition by the human TSG101 protein. Mol. Cell 2004, 13, 783–789. [Google Scholar] [CrossRef] [PubMed]
- Kostelansky, M.S.; Sun, J.; Lee, S.; Kim, J.; Ghirlando, R.; Hierro, A.; Emr, S.D.; Hurley, J.H. Structural and functional organization of the ESCRT-I trafficking complex. Cell 2006, 125, 113–126. [Google Scholar] [CrossRef]
- Christ, L.; Wenzel, E.M.; Liestol, K.; Raiborg, C.; Campsteijn, C.; Stenmark, H. ALIX and ESCRT-I/II function as parallel ESCRT-III recruiters in cytokinetic abscission. J. Cell Biol. 2016, 212, 499–513. [Google Scholar] [CrossRef]
- Schoneberg, J.; Pavlin, M.R.; Yan, S.; Righini, M.; Lee, I.H.; Carlson, L.A.; Bahrami, A.H.; Goldman, D.H.; Ren, X.; Hummer, G.; et al. ATP-dependent force generation and membrane scission by ESCRT-III and Vps4. Science 2018, 362, 1423–1428. [Google Scholar] [CrossRef]
- Azad, K.; Guilligay, D.; Boscheron, C.; Maity, S.; De Franceschi, N.; Sulbaran, G.; Effantin, G.; Wang, H.; Kleman, J.P.; Bassereau, P.; et al. Structural basis of CHMP2A-CHMP3 ESCRT-III polymer assembly and membrane cleavage. Nat. Struct. Mol. Biol. 2023, 30, 81–90. [Google Scholar] [CrossRef]
- Galluzzi, L.; Baehrecke, E.H.; Ballabio, A.; Boya, P.; Bravo-San Pedro, J.M.; Cecconi, F.; Choi, A.M.; Chu, C.T.; Codogno, P.; Colombo, M.I.; et al. Molecular definitions of autophagy and related processes. EMBO J. 2017, 36, 1811–1836. [Google Scholar] [CrossRef]
- Habib, E.; Cook, A.; Mathavarajah, S.; Dellaire, G. Adding Some “Splice” to Stress Eating: Autophagy, ESCRT and Alternative Splicing Orchestrate the Cellular Stress Response. Genes 2021, 12, 1196. [Google Scholar] [CrossRef]
- Yla-Anttila, P.; Vihinen, H.; Jokitalo, E.; Eskelinen, E.L. 3D tomography reveals connections between the phagophore and endoplasmic reticulum. Autophagy 2009, 5, 1180–1185. [Google Scholar] [CrossRef]
- Hailey, D.W.; Rambold, A.S.; Satpute-Krishnan, P.; Mitra, K.; Sougrat, R.; Kim, P.K.; Lippincott-Schwartz, J. Mitochondria supply membranes for autophagosome biogenesis during starvation. Cell 2010, 141, 656–667. [Google Scholar] [CrossRef]
- Ravikumar, B.; Moreau, K.; Jahreiss, L.; Puri, C.; Rubinsztein, D.C. Plasma membrane contributes to the formation of pre-autophagosomal structures. Nat. Cell Biol. 2010, 12, 747–757. [Google Scholar] [CrossRef] [PubMed]
- Oshima, R.; Hasegawa, T.; Tamai, K.; Sugeno, N.; Yoshida, S.; Kobayashi, J.; Kikuchi, A.; Baba, T.; Futatsugi, A.; Sato, I.; et al. ESCRT-0 dysfunction compromises autophagic degradation of protein aggregates and facilitates ER stress-mediated neurodegeneration via apoptotic and necroptotic pathways. Sci. Rep. 2016, 6, 24997. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Miao, G.; Shen, J.L.; Fortier, T.M.; Baehrecke, E.H. ESCRT dysfunction compromises endoplasmic reticulum maturation and autophagosome biogenesis in Drosophila. Curr. Biol. 2022, 32, 1262–1274.e4. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, Y.; He, H.; Tang, Z.; Hattori, T.; Liu, Y.; Young, M.M.; Serfass, J.M.; Chen, L.; Gebru, M.; Chen, C.; et al. An autophagy assay reveals the ESCRT-III component CHMP2A as a regulator of phagophore closure. Nat. Commun. 2018, 9, 2855. [Google Scholar] [CrossRef]
- Zhen, Y.; Spangenberg, H.; Munson, M.J.; Brech, A.; Schink, K.O.; Tan, K.W.; Sorensen, V.; Wenzel, E.M.; Radulovic, M.; Engedal, N.; et al. ESCRT-mediated phagophore sealing during mitophagy. Autophagy 2020, 16, 826–841. [Google Scholar] [CrossRef]
- Zhou, F.; Wu, Z.; Zhao, M.; Murtazina, R.; Cai, J.; Zhang, A.; Li, R.; Sun, D.; Li, W.; Zhao, L.; et al. Rab5-dependent autophagosome closure by ESCRT. J. Cell Biol. 2019, 218, 1908–1927. [Google Scholar] [CrossRef]
- Jahreiss, L.; Menzies, F.M.; Rubinsztein, D.C. The itinerary of autophagosomes: From peripheral formation to kiss-and-run fusion with lysosomes. Traffic 2008, 9, 574–587. [Google Scholar] [CrossRef]
- Nara, A.; Mizushima, N.; Yamamoto, A.; Kabeya, Y.; Ohsumi, Y.; Yoshimori, T. SKD1 AAA ATPase-dependent endosomal transport is involved in autolysosome formation. Cell Struct. Funct. 2002, 27, 29–37. [Google Scholar] [CrossRef]
- Filimonenko, M.; Stuffers, S.; Raiborg, C.; Yamamoto, A.; Malerod, L.; Fisher, E.M.; Isaacs, A.; Brech, A.; Stenmark, H.; Simonsen, A. Functional multivesicular bodies are required for autophagic clearance of protein aggregates associated with neurodegenerative disease. J. Cell Biol. 2007, 179, 485–500. [Google Scholar] [CrossRef]
- Lee, J.A.; Beigneux, A.; Ahmad, S.T.; Young, S.G.; Gao, F.B. ESCRT-III dysfunction causes autophagosome accumulation and neurodegeneration. Curr. Biol. 2007, 17, 1561–1567. [Google Scholar] [CrossRef]
- Djeddi, A.; Michelet, X.; Culetto, E.; Alberti, A.; Barois, N.; Legouis, R. Induction of autophagy in ESCRT mutants is an adaptive response for cell survival in C. elegans. J. Cell Sci. 2012, 125 Pt 3, 685–694. [Google Scholar] [CrossRef] [PubMed]
- Lefebvre, C.; Legouis, R.; Culetto, E. ESCRT and autophagies: Endosomal functions and beyond. Semin. Cell Dev. Biol. 2018, 74, 21–28. [Google Scholar] [CrossRef] [PubMed]
- Murrow, L.; Malhotra, R.; Debnath, J. ATG12-ATG3 interacts with Alix to promote basal autophagic flux and late endosome function. Nat. Cell Biol. 2015, 17, 300–310. [Google Scholar] [CrossRef]
- Wrobel, M.; Cendrowski, J.; Szymanska, E.; Grebowicz-Maciukiewicz, M.; Budick-Harmelin, N.; Macias, M.; Szybinska, A.; Mazur, M.; Kolmus, K.; Goryca, K.; et al. ESCRT-I fuels lysosomal degradation to restrict TFEB/TFE3 signaling via the Rag-mTORC1 pathway. Life Sci. Alliance 2022, 5, e202101239. [Google Scholar] [CrossRef]
- Radulovic, M.; Schink, K.O.; Wenzel, E.M.; Nahse, V.; Bongiovanni, A.; Lafont, F.; Stenmark, H. ESCRT-mediated lysosome repair precedes lysophagy and promotes cell survival. EMBO J. 2018, 37, e99753. [Google Scholar] [CrossRef]
- Keeley, O.; Coyne, A.N. Nuclear and degradative functions of the ESCRT-III pathway: Implications for neurodegenerative disease. Nucleus 2024, 15, 2349085. [Google Scholar] [CrossRef]
- Willen, K.; Edgar, J.R.; Hasegawa, T.; Tanaka, N.; Futter, C.E.; Gouras, G.K. Abeta accumulation causes MVB enlargement and is modelled by dominant negative VPS4A. Mol. Neurodegener. 2017, 12, 61. [Google Scholar] [CrossRef]
- Benyair, R.; Giridharan, S.S.P.; Rivero-Rios, P.; Hasegawa, J.; Bristow, E.; Eskelinen, E.L.; Shmueli, M.D.; Fishbain-Yoskovitz, V.; Merbl, Y.; Sharkey, L.M.; et al. Upregulation of the ESCRT pathway and multivesicular bodies accelerates degradation of proteins associated with neurodegeneration. Autophagy Rep. 2023, 2, 2166722. [Google Scholar] [CrossRef]
- Zhang, W.; Yang, J.; Wang, B.; Lu, Y.; Yang, J.; Zhong, W.; Yu, Z.; Qin, Z.; Xiao, B.; Wang, K.; et al. HRS mediates tumor immune evasion by regulating proteostasis-associated interferon pathway activation. Cell Rep. 2023, 42, 113352. [Google Scholar] [CrossRef]
- Toyoshima, M.; Tanaka, N.; Aoki, J.; Tanaka, Y.; Murata, K.; Kyuuma, M.; Kobayashi, H.; Ishii, N.; Yaegashi, N.; Sugamura, K. Inhibition of tumor growth and metastasis by depletion of vesicular sorting protein Hrs: Its regulatory role on E-cadherin and beta-catenin. Cancer Res. 2007, 67, 5162–5171. [Google Scholar] [CrossRef]
- Zhu, G.; Gilchrist, R.; Borley, N.; Chng, H.W.; Morgan, M.; Marshall, J.F.; Camplejohn, R.S.; Muir, G.H.; Hart, I.R. Reduction of TSG101 protein has a negative impact on tumor cell growth. Int. J. Cancer 2004, 109, 541–547. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Belogortseva, N.; Porter, D.; Park, M. Chmp1A functions as a novel tumor suppressor gene in human embryonic kidney and ductal pancreatic tumor cells. Cell Cycle 2008, 7, 2886–2893. [Google Scholar] [CrossRef] [PubMed]
- Sadler, J.B.A.; Wenzel, D.M.; Strohacker, L.K.; Guindo-Martinez, M.; Alam, S.L.; Mercader, J.M.; Torrents, D.; Ullman, K.S.; Sundquist, W.I.; Martin-Serrano, J. A cancer-associated polymorphism in ESCRT-III disrupts the abscission checkpoint and promotes genome instability. Proc. Natl. Acad. Sci. USA 2018, 115, E8900–E8908. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.J.; Zhan, S.T.; Pan, Y.Q.; Bao, W.; Yang, Y. The role of Vps4 in cancer development. Front. Oncol. 2023, 13, 1203359. [Google Scholar] [CrossRef]
- Usami, Y.; Popov, S.; Popova, E.; Inoue, M.; Weissenhorn, W.; Göttlinger, H.G. The ESCRT pathway and HIV-1 budding. Biochem. Soc. Trans. 2009, 37 Pt 1, 181–184. [Google Scholar] [CrossRef]
- Lippincott-Schwartz, J.; Freed, E.O.; van Engelenburg, S.B. A Consensus View of ESCRT-Mediated Human Immunodeficiency Virus Type 1 Abscission. Annu. Rev. Virol. 2017, 4, 309–325. [Google Scholar] [CrossRef]
- Gordon, T.B.; Hayward, J.A.; Marsh, G.A.; Baker, M.L.; Tachedjian, G. Host and Viral Proteins Modulating Ebola and Marburg Virus Egress. Viruses 2019, 11, 25. [Google Scholar] [CrossRef]
- Prange, R. Hepatitis B virus movement through the hepatocyte: An update. Biol. Cell 2022, 114, 325–348. [Google Scholar] [CrossRef]
- Votteler, J.; Sundquist, W.I. Virus budding and the ESCRT pathway. Cell Host Microbe 2013, 14, 232–241. [Google Scholar] [CrossRef]
- Inoue, J.; Sato, K.; Ninomiya, M.; Masamune, A. Envelope Proteins of Hepatitis B Virus: Molecular Biology and Involvement in Carcinogenesis. Viruses 2021, 13, 1124. [Google Scholar] [CrossRef]
- Hu, J.; Liu, K. Complete and Incomplete Hepatitis B Virus Particles: Formation, Function, and Application. Viruses 2017, 9, 56. [Google Scholar] [CrossRef] [PubMed]
- Kian Chua, P.; Lin, M.H.; Shih, C. Potent inhibition of human Hepatitis B virus replication by a host factor Vps4. Virology 2006, 354, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Lambert, C.; Doring, T.; Prange, R. Hepatitis B virus maturation is sensitive to functional inhibition of ESCRT-III, Vps4, and gamma 2-adaptin. J. Virol. 2007, 81, 9050–9060. [Google Scholar] [CrossRef]
- Jiang, B.; Himmelsbach, K.; Ren, H.; Boller, K.; Hildt, E. Subviral Hepatitis B Virus Filaments, like Infectious Viral Particles, Are Released via Multivesicular Bodies. J. Virol. 2015, 90, 3330–3341. [Google Scholar] [CrossRef]
- Bardens, A.; Doring, T.; Stieler, J.; Prange, R. Alix regulates egress of hepatitis B virus naked capsid particles in an ESCRT-independent manner. Cell Microbiol. 2011, 13, 602–619. [Google Scholar] [CrossRef]
- Stieler, J.T.; Prange, R. Involvement of ESCRT-II in hepatitis B virus morphogenesis. PLoS ONE 2014, 9, e91279. [Google Scholar] [CrossRef]
- Chou, S.F.; Tsai, M.L.; Huang, J.Y.; Chang, Y.S.; Shih, C. The Dual Role of an ESCRT-0 Component HGS in HBV Transcription and Naked Capsid Secretion. PLoS Pathog. 2015, 11, e1005123. [Google Scholar] [CrossRef]
- Zheng, Y.; Wang, M.; Li, S.; Bu, Y.; Xu, Z.; Zhu, G.; Wu, C.; Zhao, K.; Li, A.; Chen, Q.; et al. Hepatitis B virus hijacks TSG101 to facilitate egress via multiple vesicle bodies. PLoS Pathog. 2023, 19, e1011382. [Google Scholar] [CrossRef]
- Shen, S.; Cai, D.; Liang, H.; Zeng, G.; Liu, W.; Yan, R.; Yu, X.; Zhang, H.; Liu, S.; Li, W.; et al. NEDD4 family ubiquitin ligase AIP4 interacts with Alix to enable HBV naked capsid egress in an Alix ubiquitination-independent manner. PLoS Pathog. 2024, 20, e1012485. [Google Scholar] [CrossRef]
- Ninomiya, M.; Inoue, J.; Krueger, E.W.; Chen, J.; Cao, H.; Masamune, A.; McNiven, M.A. The Exosome-Associated Tetraspanin CD63 Contributes to the Efficient Assembly and Infectivity of the Hepatitis B Virus. Hepatol. Commun. 2021, 5, 1238–1251. [Google Scholar] [CrossRef]
| Complex | Mammals | Yeast (S. cerevisiae) | Drosophila |
|---|---|---|---|
| ESCRT-0 | HGS (HRS), STAM1/2 | Vps27, Hse1 | Hrs, Stam |
| ESCRT-I | TSG101, VPS28, VPS37A/B/C/D/, MVB12 | Vps23, Vps28, Vps37, MVB12 | Tsg101, Vps28, Vps37 |
| ESCRT-II | EAP45, EAP30, EAP20 | Vps22, Vps25, Vps36 | Vps22, Vps25, Vps36 |
| ESCRT-III | CHMP1A/B, CHMP2A/B, CHMP3, CHMP4A/B/C, CHMP5, CHMP6, CHMP7, CHMP8/IST1 | Did2, Vps2, Vps24, Snf7, Vps60, Vps20, Chm7, Ist1 | Shrub (CHMP4), CHMP2, CHMP3, CHMP1, IST1 |
| VPS4 ATPase System | VPS4A/B, LIP5 | Vps4, Vta1 | Vps4, CG7913 (Vta1-like) |
| Accessory Proteins | Alix (PDCD6IP) | Bro1, Doa4 | Alix (homolog?), Bro1 |
| Target | Model | Results | Conclusion | Reference |
|---|---|---|---|---|
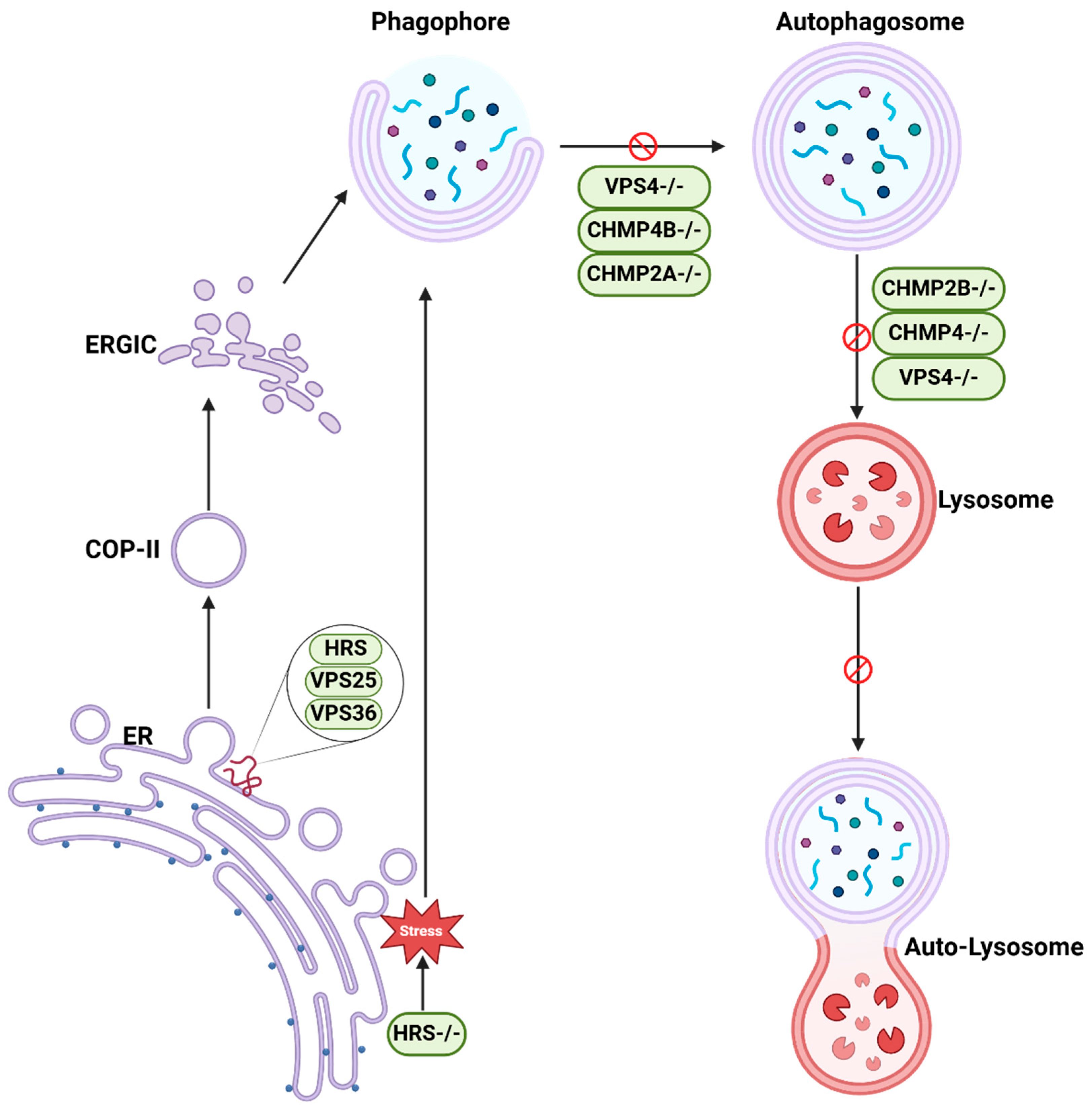
| HRS | Primary hippocampal neurons and mice brain | HRS-deficient cells induce ER stress activation and subsequent JNK signaling and increase LC3 and p62. | Silencing of HRS impairs the late stage of autophagic flux. | [23] |
| Hrs Vps25/32/13D | Drosophila intestine cells | Cells lacking either ESCRTs influence ER maturation, COPII trafficking, ER-Golgi intermediate compartment assembly, and autophagosome formation. | ESCRTs regulates COPII vesicle formation that influences autophagy. | [24] |
| CHMP2A | HeLa cells and U-2 OS cells | CHMP2A deficiency results in phagophore accumulation. | CHMP2A translocates to the phagophore and regulates the separation of the inner and outer autophagosomal membranes to form double-membrane autophagosomes. | [25] |
| CHMP2A/4B | Human retinal pigment epithelial cells | CHMP4B is recruited transiently to nascent autophagosomes, and depletion of CHMP2A inhibited phagophore sealing during mitophagy. | CHMP2A and CHMP4B mediate phagophore closure. | [26] |
| Snf7 Vps4 | Yeast cells | Depletion of Snf7 and the Vps4 causes late autophagy defects and accumulation of autophagosomes. | Rab5 controlls Atg17-Snf7 interaction leading to recruitment of ESCRT to open autophagosomes and catalyzing their closure. | [27] |
| VPS4 | Mouse | VPS4/SKD1 dominant-negative mutant causes a defect in autophagy-dependent bulk protein degradation. | VPS4/SKD1 is required for formation of autolysosomes. | [29] |
| CHMP2B | Neuron cells | CHMP2B mutants lead to accumulation of protein aggregates. | Efficient autophagic degradation requires functional MVBs. | [30] |
| Snf7 CHMP2B | Cortical neurons and flies | The loss of mSnf7-2 or CHMP2B(Intron5) expression cause the accumulation of autophagosomes. | ESCRT-III dysfunction is associated with the autophagy pathway. | [31] |
| VPS27/32 VPS4 | Caenorhabditis elegans | All ESCRT mutants present an accumulation of abnormal endosomes and autophagosomes. | The accumulation of autophagosomes is secondary to the formation of enlarged endosomes and is due to the induction of the autophagic flux. | [32] |
| VPS4 | HeLa cells | The inhibition of the AAA-ATPase VPS4 activity impairs autophagosome completion. | ESCRT machinery acts in the final step of autophagosome formation. | [25] |
| Alix | Mouse embryonic fibroblasts | Alix depletion leads to a reduction in basal autophagy. | The interactions between ATG12-ATG3 and Alix promote basal autophagy. | [34] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, J.; Prange, R.; Lu, M. ESCRT Machinery in HBV Life Cycle: Dual Roles in Autophagy and Membrane Dynamics for Viral Pathogenesis. Cells 2025, 14, 603. https://doi.org/10.3390/cells14080603
Li J, Prange R, Lu M. ESCRT Machinery in HBV Life Cycle: Dual Roles in Autophagy and Membrane Dynamics for Viral Pathogenesis. Cells. 2025; 14(8):603. https://doi.org/10.3390/cells14080603
Chicago/Turabian StyleLi, Jia, Reinhild Prange, and Mengji Lu. 2025. "ESCRT Machinery in HBV Life Cycle: Dual Roles in Autophagy and Membrane Dynamics for Viral Pathogenesis" Cells 14, no. 8: 603. https://doi.org/10.3390/cells14080603
APA StyleLi, J., Prange, R., & Lu, M. (2025). ESCRT Machinery in HBV Life Cycle: Dual Roles in Autophagy and Membrane Dynamics for Viral Pathogenesis. Cells, 14(8), 603. https://doi.org/10.3390/cells14080603







