Emerging Concepts and Functions of Autophagy as a Regulator of Synaptic Components and Plasticity
Abstract
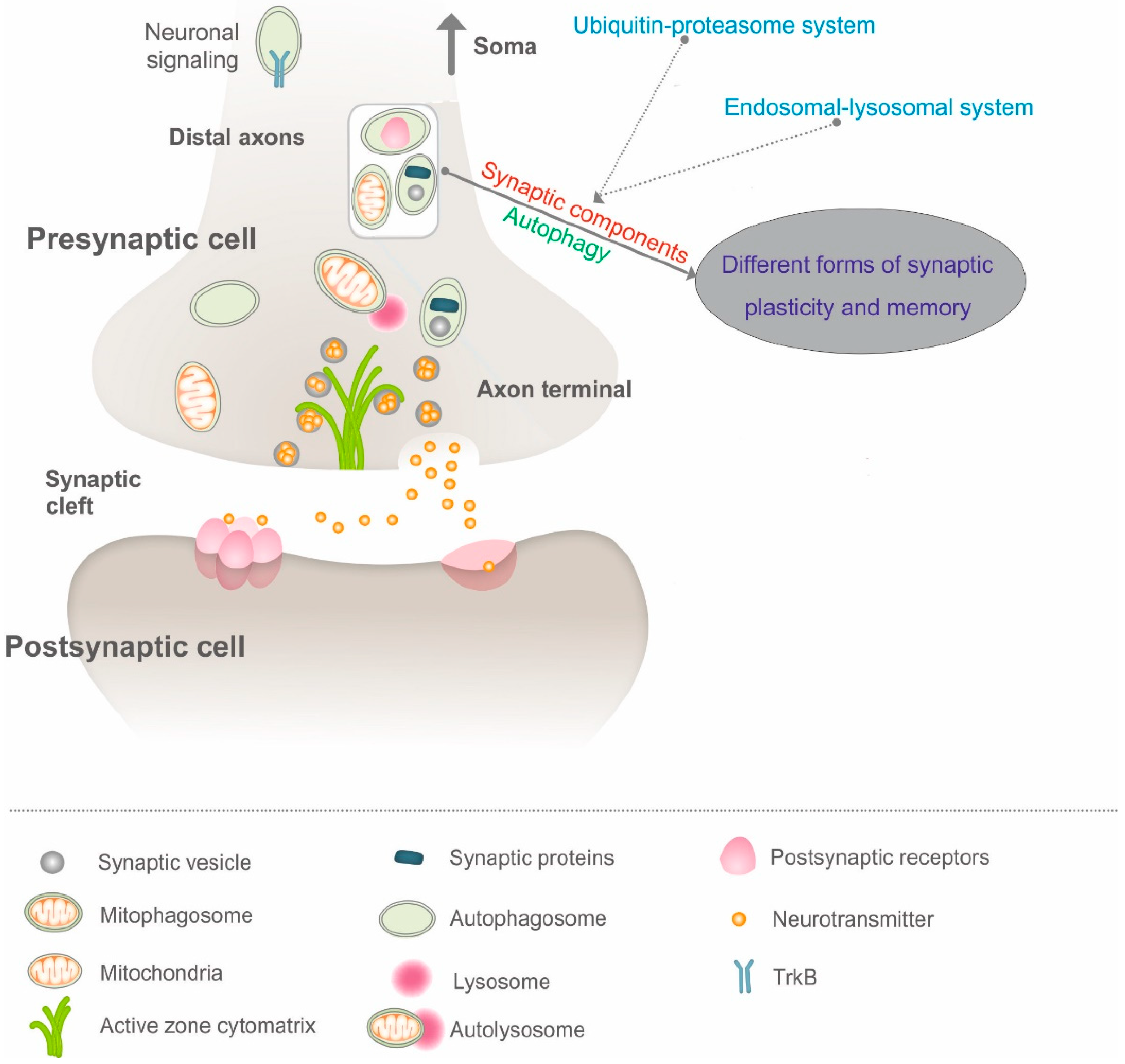
1. Introduction
2. Compartment-Specific Regulation of Autophagy in Neurons: A Non-Canonical Role for Autophagosomes in Mediating Neuronal Signaling
3. Described and Emerging Synaptic Components of Autophagy
4. Intersection of Autophagy and Synaptic Plasticity: Emerging Concepts and Relevance?
5. Conclusions
6. Future Perspective
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Wilhelm, B.G.; Mandad, S.; Truckenbrodt, S.; Krohnert, K.; Schafer, C.; Rammner, B.; Koo, S.J.; Classen, G.A.; Krauss, M.; Haucke, V.; et al. Composition of isolated synaptic boutons reveals the amounts of vesicle trafficking proteins. Science 2014, 344, 1023–1028. [Google Scholar] [CrossRef] [PubMed]
- Dieterich, D.C.; Kreutz, M.R. Proteomics of the Synapse—A Quantitative Approach to Neuronal Plasticity. Mol. Cell Proteom. 2016, 15, 368–381. [Google Scholar] [CrossRef] [PubMed]
- Patrizio, A.; Specht, C.G. Counting numbers of synaptic proteins: Absolute quantification and single molecule imaging techniques. Neurophotonics 2016, 3, 041805. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Eriksson, P.S.; Perfilieva, E.; Björk-Eriksson, T.; Alborn, A.-M.; Nordborg, C.; Peterson, D.A.; Gage, F.H. Neurogenesis in the adult human hippocampus. Nat. Med. 1998, 4, 1313–1317. [Google Scholar] [CrossRef] [PubMed]
- Spalding, K.L.; Bergmann, O.; Alkass, K.; Bernard, S.; Salehpour, M.; Huttner, H.B.; Boström, E.; Westerlund, I.; Vial, C.; Buchholz, B.A.; et al. Dynamics of Hippocampal Neurogenesis in Adult Humans. Cell 2013, 153, 1219–1227. [Google Scholar] [CrossRef] [PubMed]
- Ernst, A.; Alkass, K.; Bernard, S.; Salehpour, M.; Perl, S.; Tisdale, J.; Possnert, G.; Druid, H.; Frisén, J. Neurogenesis in the Striatum of the Adult Human Brain. Cell 2014, 156, 1072–1083. [Google Scholar] [CrossRef] [PubMed]
- Sorrells, S.F.; Paredes, M.F.; Cebrian-Silla, A.; Sandoval, K.; Qi, D.; Kelley, K.W.; James, D.; Mayer, S.; Chang, J.; Auguste, K.I.; et al. Human hippocampal neurogenesis drops sharply in children to undetectable levels in adults. Nature 2018, 555, 377–381. [Google Scholar] [CrossRef]
- Magrassi, L.; Leto, K.; Rossi, F. Lifespan of neurons is uncoupled from organismal lifespan. Proc. Natl. Acad. Sci. USA 2013, 110, 4374–4379. [Google Scholar] [CrossRef]
- Castillo-Morales, A.; Monzón-Sandoval, J.; Urrutia, A.O.; Gutiérrez, H. Postmitotic cell longevity–associated genes: A transcriptional signature of postmitotic maintenance in neural tissues. Neurobiol. Aging 2019, 74, 147–160. [Google Scholar] [CrossRef]
- Lilienbaum, A. Relationship between the proteasomal system and autophagy. Int. J. Biochem. Mol. Biol. 2013, 4, 1–26. [Google Scholar]
- Dikic, I. Proteasomal and Autophagic Degradation Systems. Annu. Rev. Biochem. 2017, 86, 193–224. [Google Scholar] [CrossRef] [PubMed]
- Nam, T.; Han, J.H.; Devkota, S.; Lee, H.-W. Emerging Paradigm of Crosstalk between Autophagy and the Ubiquitin-Proteasome System. Mol. Cells 2017, 40, 897–905. [Google Scholar] [CrossRef] [PubMed]
- Kaushik, S.; Cuervo, A.M. Chaperone-mediated autophagy: A unique way to enter the lysosome world. Trends Cell Biol. 2012, 22, 407–417. [Google Scholar] [CrossRef] [PubMed]
- Mijaljica, D.; Prescott, M.; Devenish, R.J. Microautophagy in mammalian cells: Revisiting a 40-year-old conundrum. Autophagy 2011, 7, 673–682. [Google Scholar] [CrossRef] [PubMed]
- Mizushima, N.; Komatsu, M. Autophagy: Renovation of cells and tissues. Cell 2011, 147, 728–741. [Google Scholar] [CrossRef] [PubMed]
- Hara, T.; Nakamura, K.; Matsui, M.; Yamamoto, A.; Nakahara, Y.; Suzuki-Migishima, R.; Yokoyama, M.; Mishima, K.; Saito, I.; Okano, H.; et al. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature 2006, 441, 885–889. [Google Scholar] [CrossRef] [PubMed]
- Komatsu, M.; Waguri, S.; Chiba, T.; Murata, S.; Iwata, J.; Tanida, I.; Ueno, T.; Koike, M.; Uchiyama, Y.; Kominami, E.; et al. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature 2006, 441, 880–884. [Google Scholar] [CrossRef]
- Juhász, G.; Erdi, B.; Sass, M.; Neufeld, T.P. Atg7-dependent autophagy promotes neuronal health, stress tolerance, and longevity but is dispensable for metamorphosis in Drosophila. Genes Dev. 2007, 21, 3061–3066. [Google Scholar] [CrossRef]
- Fimia, G.M.; Stoykova, A.; Romagnoli, A.; Giunta, L.; Di Bartolomeo, S.; Nardacci, R.; Corazzari, M.; Fuoco, C.; Ucar, A.; Schwartz, P.; et al. Ambra1 regulates autophagy and development of the nervous system. Nature 2007, 447, 1121–1125. [Google Scholar] [CrossRef]
- Liang, C.-C.; Wang, C.; Peng, X.; Gan, B.; Guan, J.-L. Neural-specific deletion of FIP200 leads to cerebellar degeneration caused by increased neuronal death and axon degeneration. J. Biol. Chem. 2010, 285, 3499–3509. [Google Scholar] [CrossRef]
- Maday, S. Mechanisms of neuronal homeostasis: Autophagy in the axon. Brain Res. 2016, 1649, 143–150. [Google Scholar] [CrossRef] [PubMed]
- Ramesh, N.; Pandey, U.B. Autophagy Dysregulation in ALS: When Protein Aggregates Get Out of Hand. Front. Mol. Neurosci. 2017, 10, 263. [Google Scholar] [CrossRef]
- Tang, G.; Gudsnuk, K.; Kuo, S.-H.; Cotrina, M.L.; Rosoklija, G.; Sosunov, A.; Sonders, M.S.; Kanter, E.; Castagna, C.; Yamamoto, A.; et al. Loss of mTOR-dependent macroautophagy causes autistic-like synaptic pruning deficits. Neuron 2014, 83, 1131–1143. [Google Scholar] [CrossRef]
- Dragich, J.M.; Kuwajima, T.; Hirose-Ikeda, M.; Yoon, M.S.; Eenjes, E.; Bosco, J.R.; Fox, L.M.; Lystad, A.H.; Oo, T.F.; Yarygina, O.; et al. Autophagy linked FYVE (Alfy/WDFY3) is required for establishing neuronal connectivity in the mammalian brain. Elife 2016, 5. [Google Scholar] [CrossRef] [PubMed]
- Stavoe, A.K.H.; Hill, S.E.; Hall, D.H.; Colón-Ramos, D.A. KIF1A/UNC-104 Transports ATG-9 to Regulate Neurodevelopment and Autophagy at Synapses. Dev. Cell 2016, 38, 171–185. [Google Scholar] [CrossRef] [PubMed]
- Rudnick, N.D.; Griffey, C.J.; Guarnieri, P.; Gerbino, V.; Wang, X.; Piersaint, J.A.; Tapia, J.C.; Rich, M.M.; Maniatis, T. Distinct roles for motor neuron autophagy early and late in the SOD1G93A mouse model of ALS. Proc. Natl. Acad. Sci. USA 2017, 114, E8294–E8303. [Google Scholar] [CrossRef] [PubMed]
- Maday, S.; Wallace, K.E.; Holzbaur, E.L.F. Autophagosomes initiate distally and mature during transport toward the cell soma in primary neurons. J. Cell Biol. 2012, 196, 407–417. [Google Scholar] [CrossRef]
- Maday, S.; Holzbaur, E.L.F. Compartment-Specific Regulation of Autophagy in Primary Neurons. J. Neurosci. 2016, 36, 5933–5945. [Google Scholar] [CrossRef] [PubMed]
- Kononenko, N.L.; Claßen, G.A.; Kuijpers, M.; Puchkov, D.; Maritzen, T.; Tempes, A.; Malik, A.R.; Skalecka, A.; Bera, S.; Jaworski, J.; et al. Retrograde transport of TrkB-containing autophagosomes via the adaptor AP-2 mediates neuronal complexity and prevents neurodegeneration. Nat. Commun. 2017, 8, 14819. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.-C.; Lauwers, E.; Verstreken, P. Presynaptic protein homeostasis and neuronal function. Curr. Opin. Genet. Dev. 2017, 44, 38–46. [Google Scholar] [CrossRef]
- Vijayan, V.; Verstreken, P. Autophagy in the presynaptic compartment in health and disease. J. Cell Biol. 2017, 216, 1895–1906. [Google Scholar] [CrossRef] [PubMed]
- Jin, E.J.; Kiral, F.R.; Hiesinger, P.R. The where, what, and when of membrane protein degradation in neurons. Dev. Neurobiol. 2018, 78, 283–297. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y.; Sigrist, S. Autophagy and proteostasis in the control of synapse aging and disease. Curr. Opin. Neurobiol. 2018, 48, 113–121. [Google Scholar] [CrossRef] [PubMed]
- Terenzio, M.; Schiavo, G.; Fainzilber, M. Compartmentalized Signaling in Neurons: From Cell Biology to Neuroscience. Neuron 2017, 96, 667–679. [Google Scholar] [CrossRef]
- Gumy, L.F.; Hoogenraad, C.C. Local mechanisms regulating selective cargo entry and long-range trafficking in axons. Curr. Opin. Neurobiol. 2018, 51, 23–28. [Google Scholar] [CrossRef] [PubMed]
- Kulkarni, V.V.; Maday, S. Compartment-specific dynamics and functions of autophagy in neurons: Dynamics and Functions of Neuronal Autophagy. Dev. Neurobiol. 2018, 78, 298–310. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Martin, S.; Papadopulos, A.; Harper, C.B.; Mavlyutov, T.A.; Niranjan, D.; Glass, N.R.; Cooper-White, J.J.; Sibarita, J.-B.; Choquet, D.; et al. Control of autophagosome axonal retrograde flux by presynaptic activity unveiled using botulinum neurotoxin type a. J. Neurosci. 2015, 35, 6179–6194. [Google Scholar] [CrossRef]
- Wang, T.; Martin, S.; Nguyen, T.H.; Harper, C.B.; Gormal, R.S.; Martínez-Mármol, R.; Karunanithi, S.; Coulson, E.J.; Glass, N.R.; Cooper-White, J.J.; et al. Flux of signalling endosomes undergoing axonal retrograde transport is encoded by presynaptic activity and TrkB. Nat. Commun. 2016, 7, 12976. [Google Scholar] [CrossRef]
- Birdsall, V.; Waites, C.L. Autophagy at the synapse. Neurosci. Lett. 2018. [Google Scholar] [CrossRef]
- Nikoletopoulou, V.; Tavernarakis, N. Regulation and Roles of Autophagy at Synapses. Trends Cell Biol. 2018, 28, 646–661. [Google Scholar] [CrossRef]
- Schlager, M.A.; Hoogenraad, C.C. Basic mechanisms for recognition and transport of synaptic cargos. Mol. Brain 2009, 2, 25. [Google Scholar] [CrossRef] [PubMed]
- Nirschl, J.J.; Ghiretti, A.E.; Holzbaur, E.L.F. The impact of cytoskeletal organization on the local regulation of neuronal transport. Nat. Rev. Neurosci. 2017, 18, 585–597. [Google Scholar] [CrossRef] [PubMed]
- Deng, Z.; Purtell, K.; Lachance, V.; Wold, M.S.; Chen, S.; Yue, Z. Autophagy Receptors and Neurodegenerative Diseases. Trends Cell Biol. 2017, 27, 491–504. [Google Scholar] [CrossRef]
- Winckler, B.; Faundez, V.; Maday, S.; Cai, Q.; Guimas Almeida, C.; Zhang, H. The Endolysosomal System and Proteostasis: From Development to Degeneration. J. Neurosci. 2018, 38, 9364–9374. [Google Scholar] [CrossRef] [PubMed]
- Rowland, A.M.; Richmond, J.E.; Olsen, J.G.; Hall, D.H.; Bamber, B.A. Presynaptic terminals independently regulate synaptic clustering and autophagy of GABAA receptors in Caenorhabditis elegans. J. Neurosci. 2006, 26, 1711–1720. [Google Scholar] [CrossRef] [PubMed]
- Shehata, M.; Matsumura, H.; Okubo-Suzuki, R.; Ohkawa, N.; Inokuchi, K. Neuronal stimulation induces autophagy in hippocampal neurons that is involved in AMPA receptor degradation after chemical long-term depression. J. Neurosci. 2012, 32, 10413–10422. [Google Scholar] [CrossRef]
- Hernandez, D.; Torres, C.A.; Setlik, W.; Cebrián, C.; Mosharov, E.V.; Tang, G.; Cheng, H.-C.; Kholodilov, N.; Yarygina, O.; Burke, R.E.; et al. Regulation of presynaptic neurotransmission by macroautophagy. Neuron 2012, 74, 277–284. [Google Scholar] [CrossRef]
- Binotti, B.; Pavlos, N.J.; Riedel, D.; Wenzel, D.; Vorbrüggen, G.; Schalk, A.M.; Kühnel, K.; Boyken, J.; Erck, C.; Martens, H.; et al. The GTPase Rab26 links synaptic vesicles to the autophagy pathway. Elife 2015, 4, e05597. [Google Scholar] [CrossRef]
- Harris, J.J.; Jolivet, R.; Attwell, D. Synaptic energy use and supply. Neuron 2012, 75, 762–777. [Google Scholar] [CrossRef]
- Devine, M.J.; Kittler, J.T. Mitochondria at the neuronal presynapse in health and disease. Nat. Rev. Neurosci. 2018, 19, 63–80. [Google Scholar] [CrossRef]
- Cai, Q.; Zakaria, H.M.; Simone, A.; Sheng, Z.-H. Spatial parkin translocation and degradation of damaged mitochondria via mitophagy in live cortical neurons. Curr. Biol. 2012, 22, 545–552. [Google Scholar] [CrossRef] [PubMed]
- Wong, Y.C.; Holzbaur, E.L.F. Optineurin is an autophagy receptor for damaged mitochondria in parkin-mediated mitophagy that is disrupted by an ALS-linked mutation. Proc. Natl. Acad. Sci. USA 2014, 111, E4439–E4448. [Google Scholar] [CrossRef] [PubMed]
- Ashrafi, G.; Schlehe, J.S.; LaVoie, M.J.; Schwarz, T.L. Mitophagy of damaged mitochondria occurs locally in distal neuronal axons and requires PINK1 and Parkin. J. Cell Biol. 2014, 206, 655–670. [Google Scholar] [CrossRef]
- Devireddy, S.; Liu, A.; Lampe, T.; Hollenbeck, P.J. The Organization of Mitochondrial Quality Control and Life Cycle in the Nervous System In Vivo in the Absence of PINK1. J. Neurosci. 2015, 35, 9391–9401. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.; Zhou, B.; Lin, M.-Y.; Wang, S.; Foust, K.D.; Sheng, Z.-H. Endolysosomal Deficits Augment Mitochondria Pathology in Spinal Motor Neurons of Asymptomatic fALS Mice. Neuron 2015, 87, 355–370. [Google Scholar] [CrossRef]
- Ebrahimi-Fakhari, D.; Saffari, A.; Wahlster, L.; Di Nardo, A.; Turner, D.; Lewis, T.L.; Conrad, C.; Rothberg, J.M.; Lipton, J.O.; Kölker, S.; et al. Impaired Mitochondrial Dynamics and Mitophagy in Neuronal Models of Tuberous Sclerosis Complex. Cell Rep. 2016, 17, 1053–1070. [Google Scholar] [CrossRef] [PubMed]
- Ploumi, C.; Daskalaki, I.; Tavernarakis, N. Mitochondrial biogenesis and clearance: A balancing act. FEBS J. 2017, 284, 183–195. [Google Scholar] [CrossRef]
- Nikoletopoulou, V.; Sidiropoulou, K.; Kallergi, E.; Dalezios, Y.; Tavernarakis, N. Modulation of Autophagy by BDNF Underlies Synaptic Plasticity. Cell Metab. 2017, 26, 230–242.e5. [Google Scholar] [CrossRef]
- Soukup, S.-F.; Kuenen, S.; Vanhauwaert, R.; Manetsberger, J.; Hernández-Díaz, S.; Swerts, J.; Schoovaerts, N.; Vilain, S.; Gounko, N.V.; Vints, K.; et al. A LRRK2-Dependent EndophilinA Phosphoswitch Is Critical for Macroautophagy at Presynaptic Terminals. Neuron 2016, 92, 829–844. [Google Scholar] [CrossRef]
- Murdoch, J.D.; Rostosky, C.M.; Gowrisankaran, S.; Arora, A.S.; Soukup, S.-F.; Vidal, R.; Capece, V.; Freytag, S.; Fischer, A.; Verstreken, P.; et al. Endophilin-A Deficiency Induces the Foxo3a-Fbxo32 Network in the Brain and Causes Dysregulation of Autophagy and the Ubiquitin-Proteasome System. Cell Rep. 2016, 17, 1071–1086. [Google Scholar] [CrossRef]
- Vanhauwaert, R.; Kuenen, S.; Masius, R.; Bademosi, A.; Manetsberger, J.; Schoovaerts, N.; Bounti, L.; Gontcharenko, S.; Swerts, J.; Vilain, S.; et al. The SAC1 domain in synaptojanin is required for autophagosome maturation at presynaptic terminals. EMBO J. 2017, 36, 1392–1411. [Google Scholar] [CrossRef] [PubMed]
- Song, W.; Zinsmaier, K.E. Endophilin and synaptojanin hook up to promote synaptic vesicle endocytosis. Neuron 2003, 40, 665–667. [Google Scholar] [CrossRef]
- Azarnia Tehran, D.; Kuijpers, M.; Haucke, V. Presynaptic endocytic factors in autophagy and neurodegeneration. Curr. Opin. Neurobiol. 2018, 48, 153–159. [Google Scholar] [CrossRef] [PubMed]
- Okerlund, N.D.; Schneider, K.; Leal-Ortiz, S.; Montenegro-Venegas, C.; Kim, S.A.; Garner, L.C.; Waites, C.L.; Gundelfinger, E.D.; Reimer, R.J.; Garner, C.C. Bassoon Controls Presynaptic Autophagy through Atg5. Neuron 2017, 93, 897–913.e7. [Google Scholar] [CrossRef] [PubMed]
- Klionsky, D.J.; Abdelmohsen, K.; Abe, A.; Abedin, M.J.; Abeliovich, H.; Acevedo Arozena, A.; Adachi, H.; Adams, C.M.; Adams, P.D.; Adeli, K.; et al. Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition). Autophagy 2016, 12, 1–222. [Google Scholar] [CrossRef]
- Mathai, B.; Meijer, A.; Simonsen, A. Studying Autophagy in Zebrafish. Cells 2017, 6, 21. [Google Scholar] [CrossRef] [PubMed]
- Lőrincz, P.; Mauvezin, C.; Juhász, G. Exploring Autophagy in Drosophila. Cells 2017, 6, 22. [Google Scholar] [CrossRef]
- Jiménez-Moreno, N.; Stathakos, P.; Caldwell, M.; Lane, J. Induced Pluripotent Stem Cell Neuronal Models for the Study of Autophagy Pathways in Human Neurodegenerative Disease. Cells 2017, 6, 24. [Google Scholar] [CrossRef]
- Benito-Cuesta, I.; Diez, H.; Ordoñez, L.; Wandosell, F. Assessment of Autophagy in Neurons and Brain Tissue. Cells 2017, 6, 25. [Google Scholar] [CrossRef]
- Richter, J.D.; Klann, E. Making synaptic plasticity and memory last: Mechanisms of translational regulation. Genes Dev. 2009, 23, 1–11. [Google Scholar] [CrossRef]
- Bliss, T.V.P.; Collingridge, G.L.; Morris, R.G.M. Synaptic plasticity in health and disease: Introduction and overview. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2014, 369, 20130129. [Google Scholar] [CrossRef] [PubMed]
- Takeuchi, T.; Duszkiewicz, A.J.; Morris, R.G.M. The synaptic plasticity and memory hypothesis: Encoding, storage and persistence. Philos. Trans. R. Soc. B Boil. Sci. 2013, 369, 20130288. [Google Scholar] [CrossRef] [PubMed]
- O’Rourke, N.A.; Weiler, N.C.; Micheva, K.D.; Smith, S.J. Deep molecular diversity of mammalian synapses: Why it matters and how to measure it. Nat. Rev. Neurosci. 2012, 13, 365–379. [Google Scholar] [CrossRef] [PubMed]
- Van Vactor, D.; Sigrist, S.J. Presynaptic morphogenesis, active zone organization and structural plasticity in Drosophila. Curr. Opin. Neurobiol. 2017, 43, 119–129. [Google Scholar] [CrossRef] [PubMed]
- Martin, K.C.; Casadio, A.; Zhu, H.; Yaping, E.; Rose, J.C.; Chen, M.; Bailey, C.H.; Kandel, E.R. Synapse-specific, long-term facilitation of aplysia sensory to motor synapses: A function for local protein synthesis in memory storage. Cell 1997, 91, 927–938. [Google Scholar] [CrossRef]
- Sutton, M.A.; Schuman, E.M. Dendritic protein synthesis, synaptic plasticity, and memory. Cell 2006, 127, 49–58. [Google Scholar] [CrossRef]
- Rangaraju, V.; tom Dieck, S.; Schuman, E.M. Local translation in neuronal compartments: How local is local? EMBO Rep. 2017, 18, 693–711. [Google Scholar] [CrossRef]
- Karpova, A.; Mikhaylova, M.; Thomas, U.; Knöpfel, T.; Behnisch, T. Involvement of protein synthesis and degradation in long-term potentiation of Schaffer collateral CA1 synapses. J. Neurosci. 2006, 26, 4949–4955. [Google Scholar] [CrossRef]
- Fonseca, R.; Vabulas, R.M.; Hartl, F.U.; Bonhoeffer, T.; Nägerl, U.V. A balance of protein synthesis and proteasome-dependent degradation determines the maintenance of LTP. Neuron 2006, 52, 239–245. [Google Scholar] [CrossRef]
- Cajigas, I.J.; Will, T.; Schuman, E.M. Protein homeostasis and synaptic plasticity. EMBO J. 2010, 29, 2746–2752. [Google Scholar] [CrossRef]
- Ho, V.M.; Lee, J.-A.; Martin, K.C. The Cell Biology of Synaptic Plasticity. Science 2011, 334, 623–628. [Google Scholar] [CrossRef] [PubMed]
- Alvarez-Castelao, B.; Schuman, E.M. The Regulation of Synaptic Protein Turnover. J. Biol. Chem. 2015, 290, 28623–28630. [Google Scholar] [CrossRef] [PubMed]
- Cohen, L.D.; Ziv, N.E. Recent insights on principles of synaptic protein degradation. F1000Research 2017, 6, 675. [Google Scholar] [CrossRef] [PubMed]
- Hegde, A.N. Ubiquitin-proteasome-mediated local protein degradation and synaptic plasticity. Prog. Neurobiol. 2004, 73, 311–357. [Google Scholar] [CrossRef] [PubMed]
- Bingol, B.; Sheng, M. Deconstruction for reconstruction: The role of proteolysis in neural plasticity and disease. Neuron 2011, 69, 22–32. [Google Scholar] [CrossRef] [PubMed]
- Shehata, M.; Inokuchi, K. Does autophagy work in synaptic plasticity and memory? Rev. Neurosci. 2014, 25, 543–557. [Google Scholar] [CrossRef]
- Hegde, A.N. Proteolysis, synaptic plasticity and memory. Neurobiol. Learn. Mem. 2017, 138, 98–110. [Google Scholar] [CrossRef]
- Ding, M.; Shen, K. The role of the ubiquitin proteasome system in synapse remodeling and neurodegenerative diseases. BioEssays 2008, 30, 1075–1083. [Google Scholar] [CrossRef]
- Tsai, N.-P. Ubiquitin proteasome system-mediated degradation of synaptic proteins: An update from the postsynaptic side. Biochim. Biophys. Acta 2014, 1843, 2838–2842. [Google Scholar] [CrossRef]
- Lip, P.Z.Y.; Demasi, M.; Bonatto, D. The role of the ubiquitin proteasome system in the memory process. Neurochem. Int. 2017, 102, 57–65. [Google Scholar] [CrossRef]
- Ehlers, M.D. Reinsertion or degradation of AMPA receptors determined by activity-dependent endocytic sorting. Neuron 2000, 28, 511–525. [Google Scholar] [CrossRef]
- Südhof, T.C. Molecular Neuroscience in the 21st Century: A Personal Perspective. Neuron 2017, 96, 536–541. [Google Scholar] [CrossRef] [PubMed]
- Cowansage, K.K.; LeDoux, J.E.; Monfils, M.-H. Brain-derived neurotrophic factor: A dynamic gatekeeper of neural plasticity. Curr. Mol. Pharmacol. 2010, 3, 12–29. [Google Scholar] [CrossRef]
- Leal, G.; Comprido, D.; Duarte, C.B. BDNF-induced local protein synthesis and synaptic plasticity. Neuropharmacology 2014, 76 Pt C, 639–656. [Google Scholar] [CrossRef]
- George, A.A.; Hayden, S.; Holzhausen, L.C.; Ma, E.Y.; Suzuki, S.C.; Brockerhoff, S.E. Synaptojanin 1 Is Required for Endolysosomal Trafficking of Synaptic Proteins in Cone Photoreceptor Inner Segments. PLoS ONE 2014, 9, e84394. [Google Scholar] [CrossRef] [PubMed]
- George, A.A.; Hayden, S.; Stanton, G.R.; Brockerhoff, S.E. Arf6 and the 5’phosphatase of synaptojanin 1 regulate autophagy in cone photoreceptors: PIP2 effectors, SynJ1 and Arf6, regulate autophagy. BioEssays 2016, 38, S119–S135. [Google Scholar] [CrossRef] [PubMed]
- Eisenberg, T.; Knauer, H.; Schauer, A.; Büttner, S.; Ruckenstuhl, C.; Carmona-Gutierrez, D.; Ring, J.; Schroeder, S.; Magnes, C.; Antonacci, L.; et al. Induction of autophagy by spermidine promotes longevity. Nat. Cell Biol. 2009, 11, 1305–1314. [Google Scholar] [CrossRef]
- Gupta, V.K.; Scheunemann, L.; Eisenberg, T.; Mertel, S.; Bhukel, A.; Koemans, T.S.; Kramer, J.M.; Liu, K.S.Y.; Schroeder, S.; Stunnenberg, H.G.; et al. Restoring polyamines protects from age-induced memory impairment in an autophagy-dependent manner. Nat. Neurosci. 2013, 16, 1453–1460. [Google Scholar] [CrossRef]
- Petzoldt, A.G.; Lützkendorf, J.; Sigrist, S.J. Mechanisms controlling assembly and plasticity of presynaptic active zone scaffolds. Curr. Opin. Neurobiol. 2016, 39, 69–76. [Google Scholar] [CrossRef]
- Gupta, V.K.; Pech, U.; Bhukel, A.; Fulterer, A.; Ender, A.; Mauermann, S.F.; Andlauer, T.F.M.; Antwi-Adjei, E.; Beuschel, C.; Thriene, K.; et al. Spermidine Suppresses Age-Associated Memory Impairment by Preventing Adverse Increase of Presynaptic Active Zone Size and Release. PLoS Biol. 2016, 14, e1002563. [Google Scholar] [CrossRef]
- Bhukel, A.; Madeo, F.; Sigrist, S.J. Spermidine boosts autophagy to protect from synapse aging. Autophagy 2017, 13, 444–445. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y. Emerging Connections: Synaptic Autophagy in Brain Aging and Disease. In Autophagy in Health and Disease; Turksen, K., Ed.; Springer International Publishing: Cham, Switzerland, 2018; pp. 135–152. ISBN 978-3-319-98145-1. [Google Scholar]
- Haack, T.B.; Hogarth, P.; Kruer, M.C.; Gregory, A.; Wieland, T.; Schwarzmayr, T.; Graf, E.; Sanford, L.; Meyer, E.; Kara, E.; et al. Exome sequencing reveals de novo WDR45 mutations causing a phenotypically distinct, X-linked dominant form of NBIA. Am. J. Hum. Genet. 2012, 91, 1144–1149. [Google Scholar] [CrossRef] [PubMed]
- Hayflick, S.J.; Kruer, M.C.; Gregory, A.; Haack, T.B.; Kurian, M.A.; Houlden, H.H.; Anderson, J.; Boddaert, N.; Sanford, L.; Harik, S.I.; et al. β-Propeller protein-associated neurodegeneration: A new X-linked dominant disorder with brain iron accumulation. Brain 2013, 136, 1708–1717. [Google Scholar] [CrossRef] [PubMed]
- Saitsu, H.; Nishimura, T.; Muramatsu, K.; Kodera, H.; Kumada, S.; Sugai, K.; Kasai-Yoshida, E.; Sawaura, N.; Nishida, H.; Hoshino, A.; et al. De novo mutations in the autophagy gene WDR45 cause static encephalopathy of childhood with neurodegeneration in adulthood. Nat. Genet. 2013, 45, 445–449. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.G.; Sun, L.; Miao, G.; Ji, C.; Zhao, H.; Sun, H.; Miao, L.; Yoshii, S.R.; Mizushima, N.; Wang, X.; et al. The autophagy gene Wdr45/Wipi4 regulates learning and memory function and axonal homeostasis. Autophagy 2015, 11, 881–890. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Hao, S.; Yin, H.; Gao, J.; Yang, Z. Autophagy ameliorates cognitive impairment through activation of PVT1 and apoptosis in diabetes mice. Behav. Brain Res. 2016, 305, 265–277. [Google Scholar] [CrossRef]
- Fu, J.; Wang, H.; Gao, J.; Yu, M.; Wang, R.; Yang, Z.; Zhang, T. Rapamycin Effectively Impedes Melamine-Induced Impairments of Cognition and Synaptic Plasticity in Wistar Rats. Mol. Neurobiol. 2017, 54, 819–832. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Shang, Y.; Xiao, X.; Yu, M.; Zhang, T. Prenatal stress-induced impairments of cognitive flexibility and bidirectional synaptic plasticity are possibly associated with autophagy in adolescent male-offspring. Exp. Neurol. 2017, 298, 68–78. [Google Scholar] [CrossRef]
- Zhai, B.; Shang, X.; Fu, J.; Li, F.; Zhang, T. Rapamycin relieves anxious emotion and synaptic plasticity deficits induced by hindlimb unloading in mice. Neurosci. Lett. 2018, 677, 44–48. [Google Scholar] [CrossRef]
- Wang, F.; Zhang, C.; Hou, S.; Geng, X. Synergistic Effects of Mesenchymal Stem Cell Transplantation and Repetitive Transcranial Magnetic Stimulation on Promoting Autophagy and Synaptic Plasticity in Vascular Dementia. J. Gerontol. Ser. A 2018. [Google Scholar] [CrossRef]
- Yan, J.; Porch, M.W.; Court-Vazquez, B.; Bennett, M.V.L.; Zukin, R.S. Activation of autophagy rescues synaptic and cognitive deficits in fragile X mice. Proc. Natl. Acad. Sci. USA 2018, 115, E9707–E9716. [Google Scholar] [CrossRef] [PubMed]
- Shehata, M.; Abdou, K.; Choko, K.; Matsuo, M.; Nishizono, H.; Inokuchi, K. Autophagy Enhances Memory Erasure through Synaptic Destabilization. J. Neurosci. 2018, 38, 3809–3822. [Google Scholar] [CrossRef] [PubMed]
- Scrivo, A.; Bourdenx, M.; Pampliega, O.; Cuervo, A.M. Selective autophagy as a potential therapeutic target for neurodegenerative disorders. Lancet Neurol. 2018, 17, 802–815. [Google Scholar] [CrossRef]
© 2019 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liang, Y. Emerging Concepts and Functions of Autophagy as a Regulator of Synaptic Components and Plasticity. Cells 2019, 8, 34. https://doi.org/10.3390/cells8010034
Liang Y. Emerging Concepts and Functions of Autophagy as a Regulator of Synaptic Components and Plasticity. Cells. 2019; 8(1):34. https://doi.org/10.3390/cells8010034
Chicago/Turabian StyleLiang, YongTian. 2019. "Emerging Concepts and Functions of Autophagy as a Regulator of Synaptic Components and Plasticity" Cells 8, no. 1: 34. https://doi.org/10.3390/cells8010034
APA StyleLiang, Y. (2019). Emerging Concepts and Functions of Autophagy as a Regulator of Synaptic Components and Plasticity. Cells, 8(1), 34. https://doi.org/10.3390/cells8010034




