Aging Triggers H3K27 Trimethylation Hoarding in the Chromatin of Nothobranchius furzeri Skeletal Muscle
Abstract
:1. Introduction
2. Materials and Methods
2.1. In Vivo Experiments
2.2. Cells and Treatments
2.3. Quantification of Global DNA Methylation
2.4. RNA Extraction and qRT-PCR
2.5. Sequencing and Bioinformatics Analysis
2.6. Western Blot
2.7. Histology and Morphometric Analysis
2.8. Statistical Analysis
2.9. Ethics Statement
3. Results
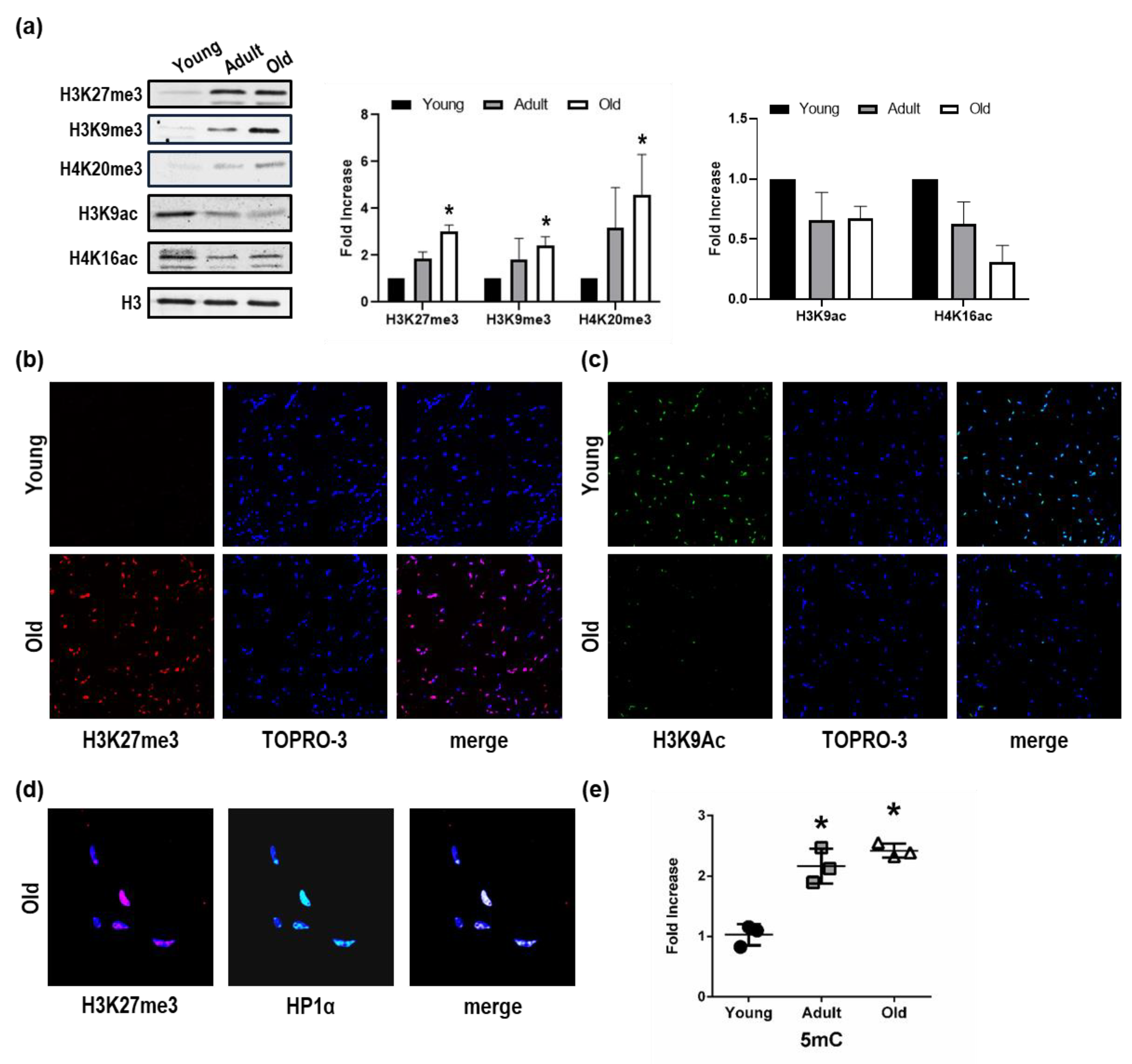
3.1. Chromatin Landscape Discriminates Among Young, Adult, and Old Nfu
3.2. Epigenetic Enzymes Responsible for Chromatin Condensation are Differentially Expressed Among Young, Adult, and Old Nfu
3.3. RNA Sequencing of Nfu Skeletal Muscle Tissue Shows Age-Specific Expression Pattern
3.4. H3K27me3 and H3K9ac Show Opposite Regulation and Role During Aging
3.5. Impairment of Cell Cycle, Differentiation, and DNA Repair Mechanics in Old Nfu Skeletal Muscle Tissue
3.6. Up-Regulation of Inflammation and Senescence in Old Nfu Skeletal Muscle Tissue
4. Discussion
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Cencioni, C.; Spallotta, F.; Martelli, F.; Valente, S.; Mai, A.; Zeiher, A.M.; Gaetano, C. Oxidative stress and epigenetic regulation in ageing and age-related diseases. Int. J. Mol. Sci. 2013, 14, 17643–17663. [Google Scholar] [CrossRef] [PubMed]
- Booth, L.N.; Brunet, A. The Aging Epigenome. Mol. Cell 2016, 62, 728–744. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brunet, A.; Berger, S.L. Epigenetics of aging and aging-related disease. J. Gerontol. A Biol. Sci. Med. Sci. 2014, 69, S17–S20. [Google Scholar] [CrossRef] [PubMed]
- Zenin, A.; Tsepilov, Y.; Sharapov, S.; Getmantsev, E.; Menshikov, L.I.; Fedichev, P.O.; Aulchenko, Y. Identification of 12 genetic loci associated with human healthspan. Commun. Biol. 2019, 2, 41. [Google Scholar] [CrossRef] [PubMed]
- Benayoun, B.A.; Pollina, E.A.; Brunet, A. Epigenetic regulation of ageing: Linking environmental inputs to genomic stability. Nat. Rev. Mol. Cell Biol. 2015, 16, 593–610. [Google Scholar] [CrossRef] [PubMed]
- Tieland, M.; Trouwborst, I.; Clark, B.C. Skeletal muscle performance and ageing. J. Cachexia Sarcopenia Muscle 2018, 9, 3–19. [Google Scholar] [CrossRef] [PubMed]
- Shafiee, G.; Keshtkar, A.; Soltani, A.; Ahadi, Z.; Larijani, B.; Heshmat, R. Prevalence of sarcopenia in the world: A systematic review and meta- analysis of general population studies. J. Diabetes Metab. Disord. 2017, 16, 21. [Google Scholar] [CrossRef]
- Fuggle, N.; Shaw, S.; Dennison, E.; Cooper, C. Sarcopenia. Best Pract. Res. Clin. Rheumatol. 2017, 31, 218–242. [Google Scholar] [CrossRef] [Green Version]
- McCormick, R.; Vasilaki, A. Age-related changes in skeletal muscle: Changes to life-style as a therapy. Biogerontology 2018, 19, 519–536. [Google Scholar] [CrossRef]
- Larsson, L.; Degens, H.; Li, M.; Salviati, L.; Lee, Y.I.; Thompson, W.; Kirkland, J.L.; Sandri, M. Sarcopenia: Aging-Related Loss of Muscle Mass and Function. Physiol. Rev. 2019, 99, 427–511. [Google Scholar] [CrossRef]
- Allis, C.D.; Jenuwein, T. The molecular hallmarks of epigenetic control. Nat. Rev. Genet. 2016, 17, 487–500. [Google Scholar] [CrossRef] [PubMed]
- Jung, M.; Pfeifer, G.P. Aging and DNA methylation. BMC Biol. 2015, 13, 7. [Google Scholar] [CrossRef] [PubMed]
- Horvath, S. DNA methylation age of human tissues and cell types. Genome Biol. 2013, 14, R115. [Google Scholar] [CrossRef] [PubMed]
- Cedar, H.; Bergman, Y. Linking DNA methylation and histone modification: Patterns and paradigms. Nat. Rev. Genet. 2009, 10, 295–304. [Google Scholar] [CrossRef]
- Chandra, T.; Narita, M. High-order chromatin structure and the epigenome in SAHFs. Nucleus 2013, 4, 23–28. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hernandez-Segura, A.; Nehme, J.; Demaria, M. Hallmarks of Cellular Senescence. Trends Cell Biol. 2018, 28, 436–453. [Google Scholar] [CrossRef]
- Aird, K.M.; Zhang, R. Detection of senescence-associated heterochromatin foci (SAHF). Methods Mol. Biol. 2013, 965, 185–196. [Google Scholar] [CrossRef]
- Roth, S.Y.; Denu, J.M.; Allis, C.D. Histone acetyltransferases. Annu. Rev. Biochem. 2001, 70, 81–120. [Google Scholar] [CrossRef]
- Gates, L.A.; Shi, J.; Rohira, A.D.; Feng, Q.; Zhu, B.; Bedford, M.T.; Sagum, C.A.; Jung, S.Y.; Qin, J.; Tsai, M.J.; et al. Acetylation on histone H3 lysine 9 mediates a switch from transcription initiation to elongation. J. Biol. Chem. 2017, 292, 14456–14472. [Google Scholar] [CrossRef] [Green Version]
- Taylor, G.C.; Eskeland, R.; Hekimoglu-Balkan, B.; Pradeepa, M.M.; Bickmore, W.A. H4K16 acetylation marks active genes and enhancers of embryonic stem cells, but does not alter chromatin compaction. Genome Res. 2013, 23, 2053–2065. [Google Scholar] [CrossRef] [Green Version]
- Weidner, C.I.; Wagner, W. The epigenetic tracks of aging. Biol. Chem. 2014, 395, 1307–1314. [Google Scholar] [CrossRef] [PubMed]
- O’Sullivan, R.J.; Karlseder, J. The great unravelling: Chromatin as a modulator of the aging process. Trends Biochem. Sci. 2012, 37, 466–476. [Google Scholar] [CrossRef] [PubMed]
- Aranda, S.; Mas, G.; Di Croce, L. Regulation of gene transcription by Polycomb proteins. Sci. Adv. 2015, 1, e1500737. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cellerino, A.; Valenzano, D.R.; Reichard, M. From the bush to the bench: The annual Nothobranchius fishes as a new model system in biology. Biol. Rev. Camb. Philos. Soc. 2016, 91, 511–533. [Google Scholar] [CrossRef] [PubMed]
- Genade, T.; Benedetti, M.; Terzibasi, E.; Roncaglia, P.; Valenzano, D.R.; Cattaneo, A.; Cellerino, A. Annual fishes of the genus Nothobranchius as a model system for aging research. Aging Cell 2005, 4, 223–233. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.; Nam, H.G.; Valenzano, D.R. The short-lived African turquoise killifish: An emerging experimental model for ageing. Dis. Model. Mech. 2016, 9, 115–129. [Google Scholar] [CrossRef] [PubMed]
- Terzibasi, E.; Valenzano, D.R.; Benedetti, M.; Roncaglia, P.; Cattaneo, A.; Domenici, L.; Cellerino, A. Large differences in aging phenotype between strains of the short-lived annual fish Nothobranchius furzeri. PLoS ONE 2008, 3, e3866. [Google Scholar] [CrossRef] [PubMed]
- Di Cicco, E.; Tozzini, E.T.; Rossi, G.; Cellerino, A. The short-lived annual fish Nothobranchius furzeri shows a typical teleost aging process reinforced by high incidence of age-dependent neoplasias. Exp. Gerontol. 2011, 46, 249–256. [Google Scholar] [CrossRef]
- Hartmann, N.; Reichwald, K.; Lechel, A.; Graf, M.; Kirschner, J.; Dorn, A.; Terzibasi, E.; Wellner, J.; Platzer, M.; Rudolph, K.L.; et al. Telomeres shorten while Tert expression increases during ageing of the short-lived fish Nothobranchius furzeri. Mech. Ageing Dev. 2009, 130, 290–296. [Google Scholar] [CrossRef]
- Harel, I.; Benayoun, B.A.; Machado, B.; Singh, P.P.; Hu, C.K.; Pech, M.F.; Valenzano, D.R.; Zhang, E.; Sharp, S.C.; Artandi, S.E.; et al. A platform for rapid exploration of aging and diseases in a naturally short-lived vertebrate. Cell 2015, 160, 1013–1026. [Google Scholar] [CrossRef]
- Valenzano, D.R.; Terzibasi, E.; Cattaneo, A.; Domenici, L.; Cellerino, A. Temperature affects longevity and age-related locomotor and cognitive decay in the short-lived fish Nothobranchius furzeri. Aging Cell 2006, 5, 275–278. [Google Scholar] [CrossRef] [PubMed]
- Reichwald, K.; Petzold, A.; Koch, P.; Downie, B.R.; Hartmann, N.; Pietsch, S.; Baumgart, M.; Chalopin, D.; Felder, M.; Bens, M.; et al. Insights into Sex Chromosome Evolution and Aging from the Genome of a Short-Lived Fish. Cell 2015, 163, 1527–1538. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Valenzano, D.R.; Benayoun, B.A.; Singh, P.P.; Zhang, E.; Etter, P.D.; Hu, C.K.; Clement-Ziza, M.; Willemsen, D.; Cui, R.; Harel, I.; et al. The African Turquoise Killifish Genome Provides Insights into Evolution and Genetic Architecture of Lifespan. Cell 2015, 163, 1539–1554. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Evangelou, K.; Gorgoulis, V.G. Sudan Black B, The Specific Histochemical Stain for Lipofuscin: A Novel Method to Detect Senescent Cells. Methods Mol. Biol. 2017, 1534, 111–119. [Google Scholar] [CrossRef] [PubMed]
- Garcia, V.; Garcia, J.M.; Pena, C.; Silva, J.; Dominguez, G.; Rodriguez, R.; Maximiano, C.; Espinosa, R.; Espana, P.; Bonilla, F. The GADD45, ZBRK1 and BRCA1 pathway: Quantitative analysis of mRNA expression in colon carcinomas. J. Pathol. 2005, 206, 92–99. [Google Scholar] [CrossRef]
- Higashi, H.; Vallbohmer, D.; Warnecke-Eberz, U.; Hokita, S.; Xi, H.; Brabender, J.; Metzger, R.; Baldus, S.E.; Natsugoe, S.; Aikou, T.; et al. Down-regulation of Gadd45 expression is associated with tumor differentiation in non-small cell lung cancer. Anticancer Res. 2006, 26, 2143–2147. [Google Scholar] [PubMed]
- Kuo, L.J.; Yang, L.X. Gamma-H2AX-a novel biomarker for DNA double-strand breaks. In Vivo 2008, 22, 305–309. [Google Scholar]
- Rahman, S.M.; Janssen, R.C.; Choudhury, M.; Baquero, K.C.; Aikens, R.M.; de la Houssaye, B.A.; Friedman, J.E. CCAAT/enhancer-binding protein beta (C/EBPβ) expression regulates dietary-induced inflammation in macrophages and adipose tissue in mice. J. Biol. Chem. 2012, 287, 34349–34360. [Google Scholar] [CrossRef]
- Wang, H.; Brown, J.; Gao, S.; Liang, S.; Jotwani, R.; Zhou, H.; Suttles, J.; Scott, D.A.; Lamont, R.J. The role of JAK-3 in regulating TLR-mediated inflammatory cytokine production in innate immune cells. J. Immunol. 2013, 191, 1164–1174. [Google Scholar] [CrossRef]
- Bottazzi, B.; Inforzato, A.; Messa, M.; Barbagallo, M.; Magrini, E.; Garlanda, C.; Mantovani, A. The pentraxins PTX3 and SAP in innate immunity, regulation of inflammation and tissue remodelling. J. Hepatol. 2016, 64, 1416–1427. [Google Scholar] [CrossRef] [Green Version]
- Georgakopoulou, E.A.; Tsimaratou, K.; Evangelou, K.; Fernandez Marcos, P.J.; Zoumpourlis, V.; Trougakos, I.P.; Kletsas, D.; Bartek, J.; Serrano, M.; Gorgoulis, V.G. Specific lipofuscin staining as a novel biomarker to detect replicative and stress-induced senescence. A method applicable in cryo-preserved and archival tissues. Aging (Albany N.Y.) 2013, 5, 37–50. [Google Scholar] [CrossRef] [PubMed]
- Graf, M.; Hartmann, N.; Reichwald, K.; Englert, C. Absence of replicative senescence in cultured cells from the short-lived killifish Nothobranchius furzeri. Exp. Gerontol. 2013, 48, 17–28. [Google Scholar] [CrossRef] [PubMed]
- Wendler, S.; Hartmann, N.; Hoppe, B.; Englert, C. Age-dependent decline in fin regenerative capacity in the short-lived fish Nothobranchius furzeri. Aging Cell 2015, 14, 857–866. [Google Scholar] [CrossRef] [PubMed]
- Freund, A.; Laberge, R.M.; Demaria, M.; Campisi, J. Lamin B1 loss is a senescence-associated biomarker. Mol. Biol. Cell 2012, 23, 2066–2075. [Google Scholar] [CrossRef] [PubMed]
- Fang, E.F.; Scheibye-Knudsen, M.; Chua, K.F.; Mattson, M.P.; Croteau, D.L.; Bohr, V.A. Nuclear DNA damage signalling to mitochondria in ageing. Nat. Rev. Mol. Cell Biol. 2016, 17, 308–321. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lopez-Otin, C.; Blasco, M.A.; Partridge, L.; Serrano, M.; Kroemer, G. The hallmarks of aging. Cell 2013, 153, 1194–1217. [Google Scholar] [CrossRef] [PubMed]
- Blazek, R.; Polacik, M.; Reichard, M. Rapid growth, early maturation and short generation time in African annual fishes. EvoDevo 2013, 4, 24. [Google Scholar] [CrossRef]
- Tian, X.; Firsanov, D.; Zhang, Z.; Cheng, Y.; Luo, L.; Tombline, G.; Tan, R.; Simon, M.; Henderson, S.; Steffan, J.; et al. SIRT6 Is Responsible for More Efficient DNA Double-Strand Break Repair in Long-Lived Species. Cell 2019, 177, 622–638 e622. [Google Scholar] [CrossRef]
- Lidzbarsky, G.; Gutman, D.; Shekhidem, H.A.; Sharvit, L.; Atzmon, G. Genomic Instabilities, Cellular Senescence, and Aging: In Vitro, In Vivo and Aging-Like Human Syndromes. Front. Med. (Lausanne) 2018, 5, 104. [Google Scholar] [CrossRef] [Green Version]
- Janssen, A.; Colmenares, S.U.; Karpen, G.H. Heterochromatin: Guardian of the Genome. Annu. Rev. Cell Dev. Biol. 2018, 34, 265–288. [Google Scholar] [CrossRef] [Green Version]
- Larson, K.; Yan, S.J.; Tsurumi, A.; Liu, J.; Zhou, J.; Gaur, K.; Guo, D.; Eickbush, T.H.; Li, W.X. Heterochromatin formation promotes longevity and represses ribosomal RNA synthesis. PLoS Genet. 2012, 8, e1002473. [Google Scholar] [CrossRef] [PubMed]
- Kozak, M.L.; Chavez, A.; Dang, W.; Berger, S.L.; Ashok, A.; Guo, X.; Johnson, F.B. Inactivation of the Sas2 histone acetyltransferase delays senescence driven by telomere dysfunction. EMBO J. 2010, 29, 158–170. [Google Scholar] [CrossRef] [PubMed]
- Han, S.; Brunet, A. Histone methylation makes its mark on longevity. Trends Cell Biol. 2012, 22, 42–49. [Google Scholar] [CrossRef] [Green Version]
- Maures, T.J.; Greer, E.L.; Hauswirth, A.G.; Brunet, A. The H3K27 demethylase UTX-1 regulates C. elegans lifespan in a germline-independent, insulin-dependent manner. Aging Cell 2011, 10, 980–990. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bu, H.; Wedel, S.; Cavinato, M.; Jansen-Durr, P. MicroRNA Regulation of Oxidative Stress-Induced Cellular Senescence. Oxid. Med. Cell. Longev. 2017, 2017, 2398696. [Google Scholar] [CrossRef]
- Xie, Y.; Chen, Y. microRNAs: Emerging Targets Regulating Oxidative Stress in the Models of Parkinson’s Disease. Front. Neurosci. 2016, 10, 298. [Google Scholar] [CrossRef] [PubMed]
- Romanello, V.; Sandri, M. Mitochondrial Quality Control and Muscle Mass Maintenance. Front. Physiol. 2015, 6, 422. [Google Scholar] [CrossRef]
- Powers, S.K. Can antioxidants protect against disuse muscle atrophy? Sports Med. 2014, 44, S155–S165. [Google Scholar] [CrossRef]
- Hartmann, N.; Reichwald, K.; Wittig, I.; Drose, S.; Schmeisser, S.; Luck, C.; Hahn, C.; Graf, M.; Gausmann, U.; Terzibasi, E.; et al. Mitochondrial DNA copy number and function decrease with age in the short-lived fish Nothobranchius furzeri. Aging Cell 2011, 10, 824–831. [Google Scholar] [CrossRef]
- Franceschi, C.; Campisi, J. Chronic inflammation (inflammaging) and its potential contribution to age-associated diseases. J. Gerontol. A Biol. Sci. Med. Sci. 2014, 69, S4–S9. [Google Scholar] [CrossRef]
- Baumgart, M.; Groth, M.; Priebe, S.; Savino, A.; Testa, G.; Dix, A.; Ripa, R.; Spallotta, F.; Gaetano, C.; Ori, M.; et al. RNA-seq of the aging brain in the short-lived fish N. furzeri-conserved pathways and novel genes associated with neurogenesis. Aging Cell 2014, 13, 965–974. [Google Scholar] [CrossRef] [PubMed]
- Cellerino, A.; Ori, A. What have we learned on aging from omics studies? Semin. Cell Dev. Biol. 2017, 70, 177–189. [Google Scholar] [CrossRef] [PubMed]
- Piantanelli, L.; Rossolini, G.; Basso, A.; Piantanelli, A.; Malavolta, M.; Zaia, A. Use of mathematical models of survivorship in the study of biomarkers of aging: The role of heterogeneity. Mech. Ageing Dev. 2001, 122, 1461–1475. [Google Scholar] [CrossRef]
- Aramillo Irizar, P.; Schauble, S.; Esser, D.; Groth, M.; Frahm, C.; Priebe, S.; Baumgart, M.; Hartmann, N.; Marthandan, S.; Menzel, U.; et al. Transcriptomic alterations during ageing reflect the shift from cancer to degenerative diseases in the elderly. Nat. Commun. 2018, 9, 327. [Google Scholar] [CrossRef] [PubMed]





| Gene | Forward 5’−3’ | Reverse 5’−3’ |
|---|---|---|
| cbx2 | TCCCAACGGACAAAAGAAAC | TTGTTGGGTTTGGTGGATTT |
| cbx7.1 | GAGCAAGTGTTTGCTGTGGA | CTTTGGCACCTTTCTTCCTG |
| cbx7.2 | GGAGACAGGCTGGATTTTGA | GCCATGGTAACCGACTGATT |
| cbx8a | CAGTCAATCGGGGTGAAAGT | TTAGACTCCTCCGGGAACCT |
| cbx8b | AGGTGGCGAGTATCTGCTGT | CGGTTCCCAAGTGCTGTATT |
| ccnb | GGTGGGAGACTTTGCCTACA | AGAGGGTCAGCTCCATCAGA |
| ccnd | CTGTGAGCTTTGCTGCTTTG | ACGCTCAGCAAACACATACG |
| eed | AGTCCTGTGAAAACGCCATC | AACGTAAAGCTTCCCCACCT |
| ezh1 | CAAGAGGATTCCCAGCGATA | GGGTTGGAGGAAACAGTCGTA |
| ezh2 | ATTCTGTCAACCCCAACTGC | ATGCCCACGTACTTCAGAGC |
| gadd45γ | ATTGCGCTTCAGATCCACTT | CGCAGAACAGACTCAGCTTG |
| ira | TGCCTCTTCAAACCCTGAGT | AGGATGGCGATCTTATCACG |
| kdm6a | GTCAAACCCTACCCCCTCAT | TGTGGAGAGAGGAGCCAACT |
| kdm6b | CAAAGCCAGCTTTCTGGAAC | TCTGGATGTGAGGAGCACAG |
| myog | GTTCGACCAAGCTGGCTATC | CATGGTCACCGTCTTCCTTT |
| p21 | CCCTGCGTAAAGATCTGGAG | ACCACCACCCTTCCTCTTT |
| pcna | ACCCTCAGAGCAGAGGACAA | CATGGGAAAGGATCTGGAAA |
| polγ | TCCCCGTTAATCAGAACTGG | TCTGCTGCTTTTTGGGAGTT |
| ssbp1 | CTGGAGAGACGGAAACAAGC | CTGACGTTGTCGCTCAGAAA |
| suz12 | AAAGGAGCAAAGGTGGAGGT | GACGGTTGTGACCACTGATG |
| tfam | TACGTGTCCGAGCACTTTCA | CATGTGGTCTTCCCAGGACT |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cencioni, C.; Heid, J.; Krepelova, A.; Rasa, S.M.M.; Kuenne, C.; Guenther, S.; Baumgart, M.; Cellerino, A.; Neri, F.; Spallotta, F.; et al. Aging Triggers H3K27 Trimethylation Hoarding in the Chromatin of Nothobranchius furzeri Skeletal Muscle. Cells 2019, 8, 1169. https://doi.org/10.3390/cells8101169
Cencioni C, Heid J, Krepelova A, Rasa SMM, Kuenne C, Guenther S, Baumgart M, Cellerino A, Neri F, Spallotta F, et al. Aging Triggers H3K27 Trimethylation Hoarding in the Chromatin of Nothobranchius furzeri Skeletal Muscle. Cells. 2019; 8(10):1169. https://doi.org/10.3390/cells8101169
Chicago/Turabian StyleCencioni, Chiara, Johanna Heid, Anna Krepelova, Seyed Mohammad Mahdi Rasa, Carsten Kuenne, Stefan Guenther, Mario Baumgart, Alessandro Cellerino, Francesco Neri, Francesco Spallotta, and et al. 2019. "Aging Triggers H3K27 Trimethylation Hoarding in the Chromatin of Nothobranchius furzeri Skeletal Muscle" Cells 8, no. 10: 1169. https://doi.org/10.3390/cells8101169
APA StyleCencioni, C., Heid, J., Krepelova, A., Rasa, S. M. M., Kuenne, C., Guenther, S., Baumgart, M., Cellerino, A., Neri, F., Spallotta, F., & Gaetano, C. (2019). Aging Triggers H3K27 Trimethylation Hoarding in the Chromatin of Nothobranchius furzeri Skeletal Muscle. Cells, 8(10), 1169. https://doi.org/10.3390/cells8101169






