N-Glycomic and Transcriptomic Changes Associated with CDX1 mRNA Expression in Colorectal Cancer Cell Lines
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Cells and Cell Culture
2.3. N-glycan Release, Derivatization, and Purification
2.4. MALDI-TOF-MS Analysis
2.5. Data Processing and Analysis of MALDI-TOF-MS Spectra
2.6. Gene Expression Microarrays and Data Analysis
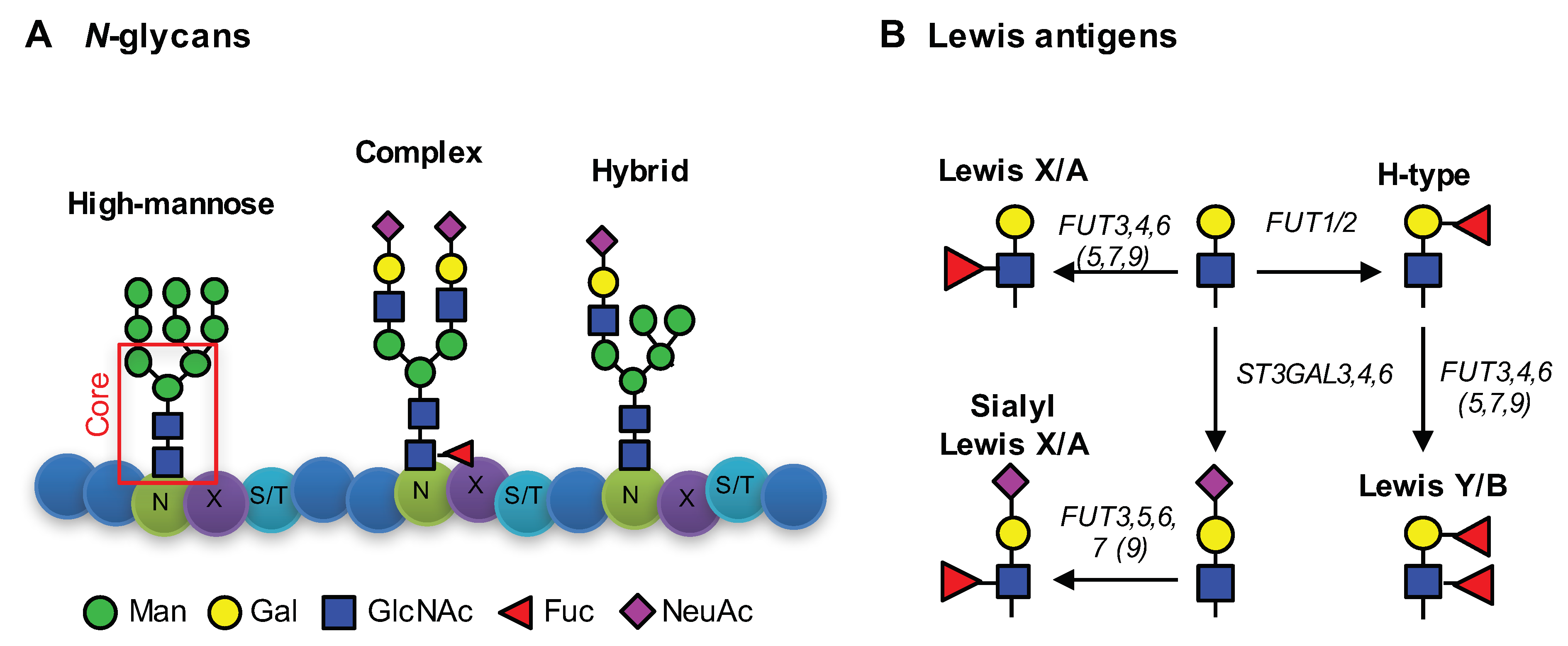
3. Results
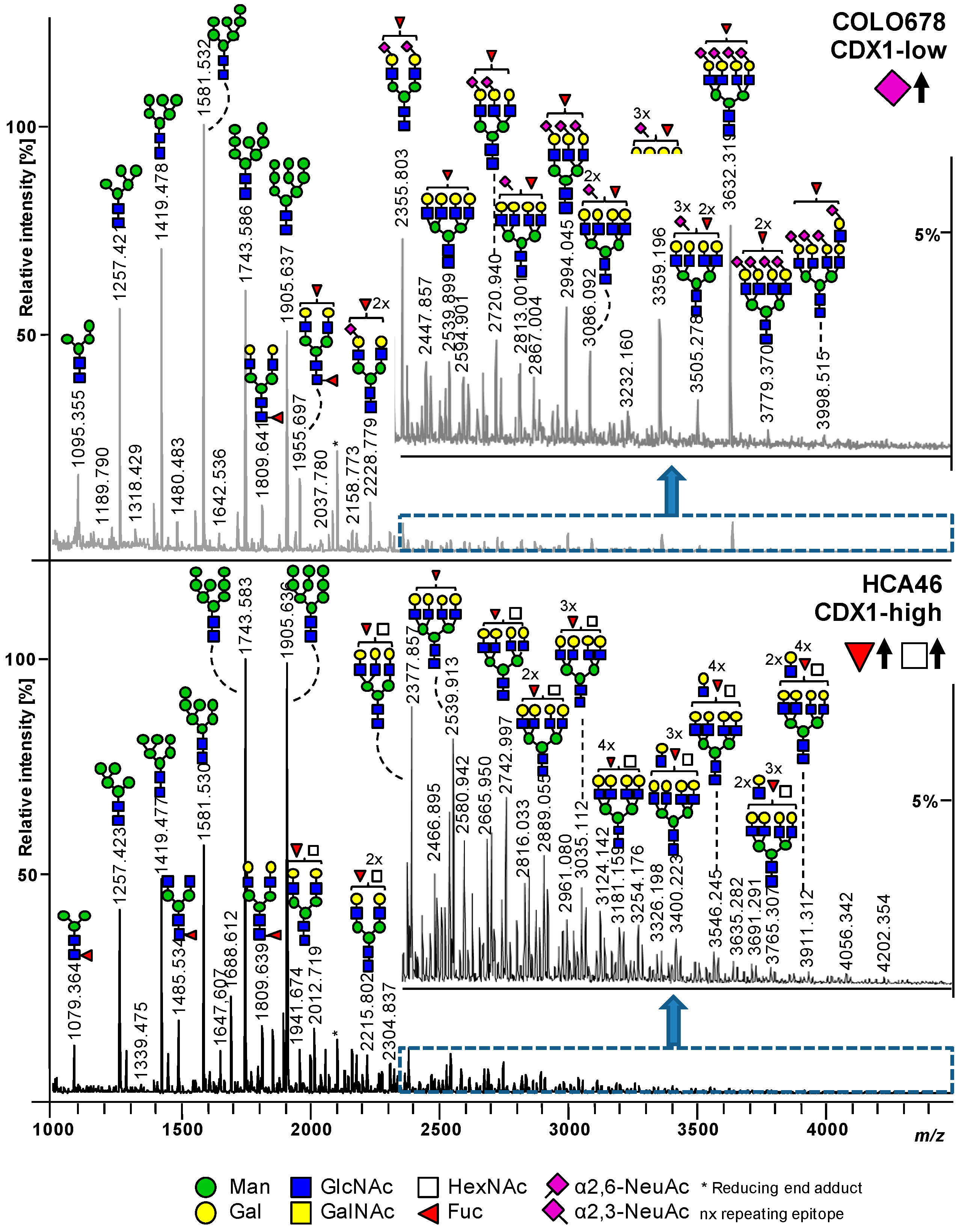
3.1. CDX1high and CDX1low CRC Cells Exhibit Different N-Glycan Profiles
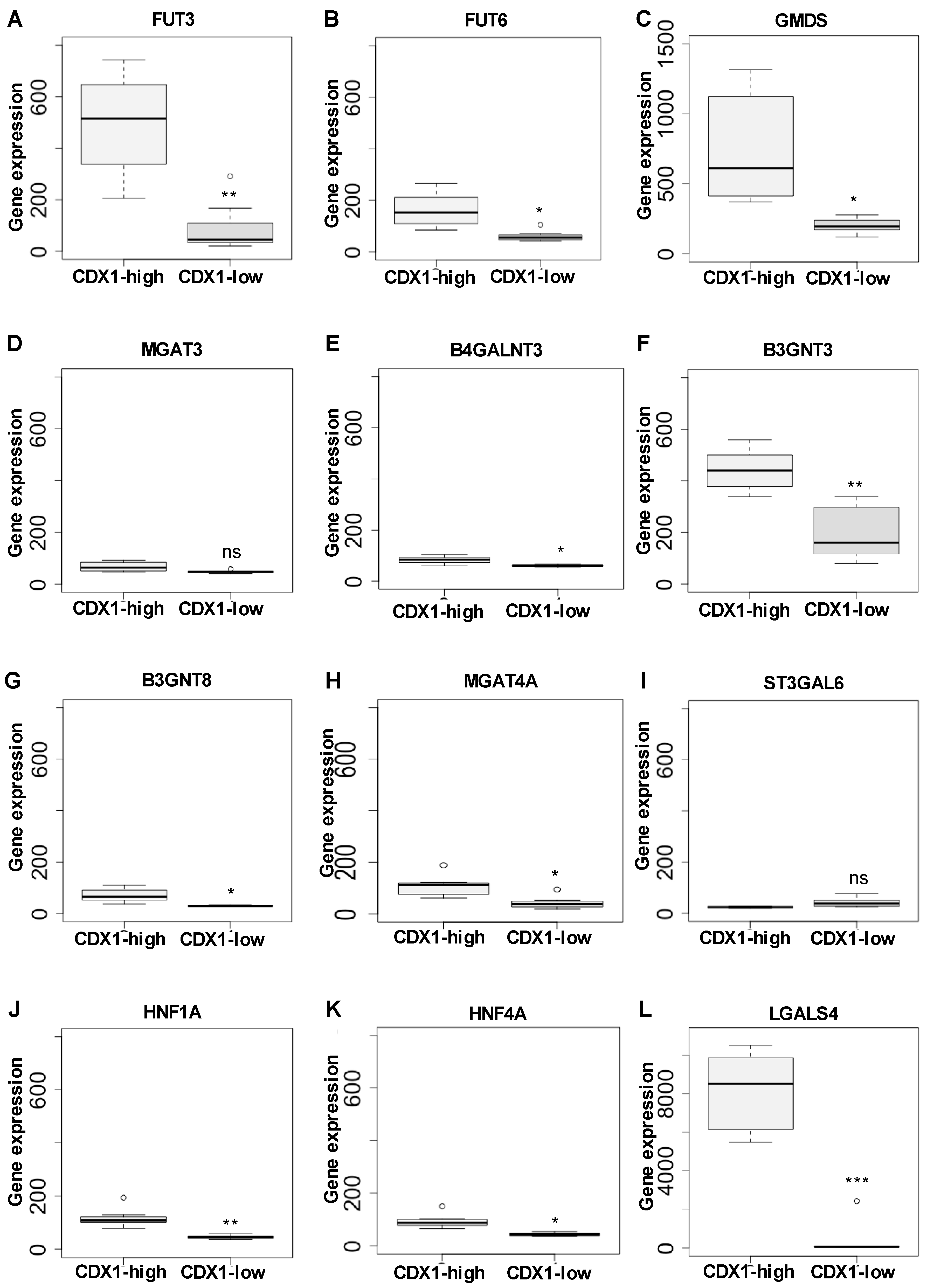
3.2. Higher Antenna Fucosylation on N-Glycans Characterized CDX1-high Expressing Colorectal Cancer Cell Lines
3.3. CDX1high Expressing CRC Cell Lines Exhibit a Lower Level of Sialylated N-Glycans as Compared to CDX1-Low Expressing Cells
3.4. CDX1 Expression in CRC Cell Lines Associated with N-Glycans CARRYING Additional N-Acetylhexosamine
3.5. CDX1 Expression in CRC Cell Lines Associated with Higher Branched N-Glycan-Derived Traits
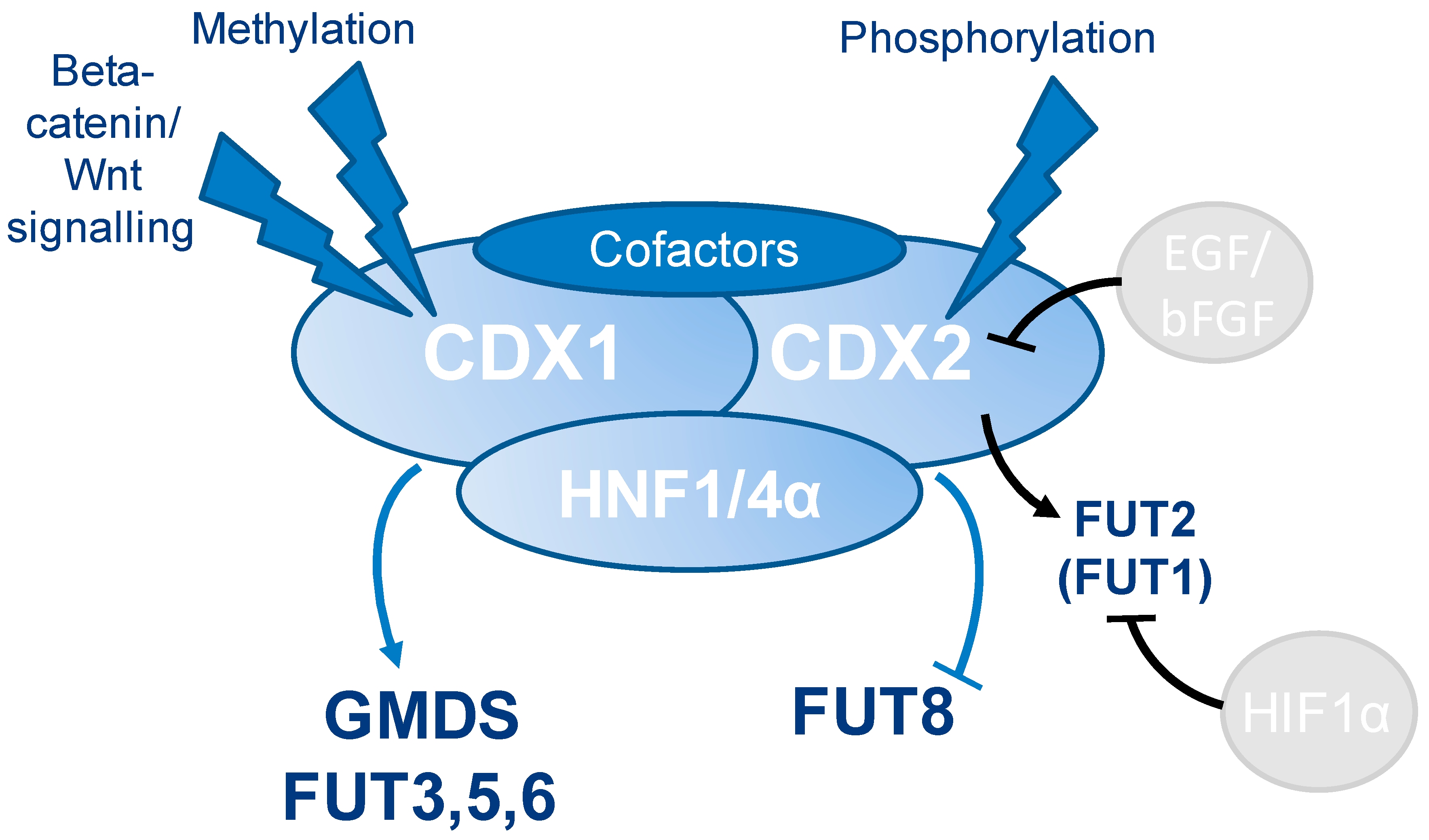
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Schnaar, R.L. Glycobiology simplified: Diverse roles of glycan recognition in inflammation. J. Leukoc. Biol. 2016, 99, 825–838. [Google Scholar] [CrossRef]
- Cummings, R.D.; Pierce, J.M. The Challenge and Promise of Glycomics. Chem. Biol. 2014, 21, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Taniguchi, N.; Kizuka, Y. Glycans and cancer: Role of N-glycans in cancer biomarker, progression and metastasis, and therapeutics. Adv. Cancer Res. 2015, 126, 11–51. [Google Scholar]
- Stanley, P.; Schachter, H.; Taniguchi, N. N-Glycans. In Essentials of Glycobiology, 2nd ed.; Varki, A., Cummings, R.D., Esko, J.D., Freeze, H.H., Stanley, P., Bertozzi, C.R., Hart, G.W., Etzler, M.E., Eds.; CSHL Press: Cold Spring Harbor, NY, USA, 2009. [Google Scholar]
- Pinho, S.S.; Reis, C.A. Glycosylation in cancer: Mechanisms and clinical implications. Nat. Rev. Cancer 2015, 15, 540–555. [Google Scholar] [CrossRef] [PubMed]
- de-Freitas-Junior, J.C.; Morgado-Diaz, J.A. The role of N-glycans in colorectal cancer progression: Potential biomarkers and therapeutic applications. Oncotarget 2015, 12, 19395–19413. [Google Scholar]
- Glavey, S.V.; Huynh, D.; Reagan, M.R.; Manier, S.; Moschetta, M.; Kawano, Y.; Roccaro, A.M.; Ghobrial, I.M.; Joshi, L.; O’Dwyer, M.E. The cancer glycome: Carbohydrates as mediators of metastasis. Blood Rev. 2015, 29, 269–279. [Google Scholar] [CrossRef] [PubMed]
- Holst, S.; Wuhrer, M.; Rombouts, Y. Chapter Six—Glycosylation Characteristics of Colorectal Cancer. In Advances in Cancer Research; Richard, R.D., Lauren, E.B., Eds.; Academic Press: London, UK, 2015; Volume 126, pp. 203–256. [Google Scholar]
- Holst, S.; Deuss, A.J.; van Pelt, G.W.; van Vliet, S.J.; Garcia-Vallejo, J.J.; Koeleman, C.A.; Deelder, A.M.; Mesker, W.E.; Tollenaar, R.A.; Rombouts, Y.; et al. N-glycosylation Profiling of Colorectal Cancer Cell Lines Reveals Association of Fucosylation with Differentiation and Caudal Type Homebox 1 (CDX1)/Villin mRNA Expression. Mol. Cell. Proteomics 2016, 15, 124–140. [Google Scholar] [CrossRef] [PubMed]
- Dotz, V.; Wuhrer, M. Histo-blood group glycans in the context of personalized medicine. Biochim. Biophys. Acta 2016, 1860, 1596–1607. [Google Scholar] [CrossRef] [PubMed]
- Le Pendu, J.; Marionneau, S.; Cailleau-Thomas, A.; Rocher, J.; Le Moullac-Vaidye, B.; Clement, M. ABH and Lewis histo-blood group antigens in cancer. APMIS 2001, 109, 9–31. [Google Scholar] [CrossRef] [PubMed]
- Mollicone, R.; Cailleau, A.; Oriol, R. Molecular genetics of H, Se, Lewis and other fucosyltransferase genes. Transfus. Clin. Biol. 1995, 2, 235–242. [Google Scholar] [CrossRef]
- Le Pendu, J. Histo-blood group antigen and human milk oligosaccharides: Genetic polymorphism and risk of infectious diseases. Adv. Exp. Med. Biol. 2004, 554, 135–143. [Google Scholar] [PubMed]
- Becker, D.J.; Lowe, J.B. Fucose: Biosynthesis and biological function in mammals. Glycobiology 2003, 13, 41R–53R. [Google Scholar] [CrossRef] [PubMed]
- Vajaria, B.N.; Patel, P.S. Glycosylation: A hallmark of cancer? Glycoconj. J. 2017, 34, 147–156. [Google Scholar] [CrossRef] [PubMed]
- Miyoshi, E.; Moriwaki, K.; Nakagawa, T. Biological Function of Fucosylation in Cancer Biology. J. Biochem. 2008, 143, 725–729. [Google Scholar] [CrossRef] [PubMed]
- Moriwaki, K.; Miyoshi, E. Fucosylation and gastrointestinal cancer. World J. Hepatol. 2010, 2, 151–161. [Google Scholar] [CrossRef] [PubMed]
- Sethi, M.K.; Hancock, W.S.; Fanayan, S. Identifying N-Glycan Biomarkers in Colorectal Cancer by Mass Spectrometry. Acc. Chem. Res. 2016, 49, 2099–2106. [Google Scholar] [CrossRef] [PubMed]
- Kaprio, T.; Satomaa, T.; Heiskanen, A.; Hokke, C.H.; Deelder, A.M.; Mustonen, H.; Hagstrom, J.; Carpen, O.; Saarinen, J.; Haglund, C. N-glycomic profiling as a tool to separate rectal adenomas from carcinomas. Mol. Cell. Proteomics 2015, 14, 277–288. [Google Scholar] [CrossRef] [PubMed]
- Miyoshi, E.; Moriwaki, K.; Terao, N.; Tan, C.C.; Terao, M.; Nakagawa, T.; Matsumoto, H.; Shinzaki, S.; Kamada, Y. Fucosylation is a promising target for cancer diagnosis and therapy. Biomolecules 2012, 2, 34–45. [Google Scholar] [CrossRef] [PubMed]
- Moriwaki, K.; Noda, K.; Furukawa, Y.; Ohshima, K.; Uchiyama, A.; Nakagawa, T.; Taniguchi, N.; Daigo, Y.; Nakamura, Y.; Hayashi, N.; et al. Deficiency of GMDS leads to escape from NK cell-mediated tumor surveillance through modulation of TRAIL signaling. Gastroenterology 2009, 137, 188–198. [Google Scholar] [CrossRef] [PubMed]
- Nakagoe, T.; Fukushima, K.; Nanashima, A.; Sawai, T.; Tsuji, T.; Jibiki, M.; Yamaguchi, H.; Yasutake, T.; Ayabe, H.; Matuo, T.; et al. Expression of Lewisa, sialyl Lewisa, Lewisx, sialyl Lewisx, antigens as prognostic factors in patients with colorectal cancer. Can. J. Gastroenterol. Hepatol. 2000, 14, 753–760. [Google Scholar] [CrossRef]
- Konno, A.; Hoshino, Y.; Terashima, S.; Motoki, R.; Kawaguchi, T. Carbohydrate expression profile of colorectal cancer cells is relevant to metastatic pattern and prognosis. Clin. Exp. Metastasis 2002, 19, 61–70. [Google Scholar] [CrossRef] [PubMed]
- Guo, R.J.; Suh, E.R.; Lynch, J.P. The role of Cdx proteins in intestinal development and cancer. Cancer Biol. Ther. 2004, 3, 593–601. [Google Scholar] [CrossRef] [PubMed]
- Freund, J.N.; Domon-Dell, C.; Kedinger, M.; Duluc, I. The Cdx-1 and Cdx-2 homeobox genes in the intestine. Biochem. Cell Biol. 1998, 76, 957–969. [Google Scholar] [CrossRef]
- Domon-Dell, C.; Schneider, A.; Moucadel, V.; Guerin, E.; Guenot, D.; Aguillon, S.; Duluc, I.; Martin, E.; Iovanna, J.; Launay, J.F.; et al. Cdx1 homeobox gene during human colon cancer progression. Oncogene 2003, 22, 7913–7921. [Google Scholar] [CrossRef] [PubMed]
- Arango, D.; Al-Obaidi, S.; Williams, D.S.; Dopeso, H.; Mazzolini, R.; Corner, G.; Byun, D.S.; Carr, A.A.; Murone, C.; Togel, L.; et al. Villin expression is frequently lost in poorly differentiated colon cancer. Am. J. Pathol. 2012, 180, 1509–1521. [Google Scholar] [CrossRef] [PubMed]
- Chan, C.W.; Wong, N.A.; Liu, Y.; Bicknell, D.; Turley, H.; Hollins, L.; Miller, C.J.; Wilding, J.L.; Bodmer, W.F. Gastrointestinal differentiation marker Cytokeratin 20 is regulated by homeobox gene CDX1. Proc. Natl. Acad. Sci. USA 2009, 106, 1936–1941. [Google Scholar] [CrossRef] [PubMed]
- Al-Maghrabi, J.; Gomaa, W.; Buhmeida, A.; Al-Qahtani, M.; Al-Ahwal, M. Loss of villin immunoexpression in colorectal carcinoma is associated with poor differentiation and survival. ISRN Gastroenterol. 2013, 2013, 679724. [Google Scholar] [CrossRef] [PubMed]
- Silberg, D.G.; Furth, E.E.; Taylor, J.K.; Schuck, T.; Chiou, T.; Traber, P.G. CDX1 protein expression in normal, metaplastic, and neoplastic human alimentary tract epithelium. Gastroenterology 1997, 113, 478–486. [Google Scholar] [CrossRef] [PubMed]
- Vider, B.Z.; Zimber, A.; Hirsch, D.; Estlein, D.; Chastre, E.; Prevot, S.; Gespach, C.; Yaniv, A.; Gazit, A. Human colorectal carcinogenesis is associated with deregulation of homeobox gene expression. Biochem. Biophys. Res. Commun. 1997, 232, 742–748. [Google Scholar] [CrossRef] [PubMed]
- Mallo, G.V.; Soubeyran, P.; Lissitzky, J.C.; Andre, F.; Farnarier, C.; Marvaldi, J.; Dagorn, J.C.; Iovanna, J.L. Expression of the Cdx1 and Cdx2 homeotic genes leads to reduced malignancy in colon cancer-derived cells. J. Biol. Chem. 1998, 273, 14030–14036. [Google Scholar] [CrossRef] [PubMed]
- Wong, N.A.; Britton, M.P.; Choi, G.S.; Stanton, T.K.; Bicknell, D.C.; Wilding, J.L.; Bodmer, W.F. Loss of CDX1 expression in colorectal carcinoma: Promoter methylation, mutation, and loss of heterozygosity analyses of 37 cell lines. Proc. Natl. Acad. Sci. USA 2004, 101, 574–579. [Google Scholar] [CrossRef] [PubMed]
- Lauc, G.; Essafi, A.; Huffman, J.E.; Hayward, C.; Knezevic, A.; Kattla, J.J.; Polasek, O.; Gornik, O.; Vitart, V.; Abrahams, J.L.; et al. Genomics meets glycomics-the first GWAS study of human N-Glycome identifies HNF1alpha as a master regulator of plasma protein fucosylation. PLoS Genet. 2010, 6, e1001256. [Google Scholar] [CrossRef] [PubMed]
- Cantarino, N.; Fernandez-Figueras, M.T.; Valero, V.; Musulen, E.; Malinverni, R.; Granada, I.; Goldie, S.J.; Martin-Caballero, J.; Douet, J.; Forcales, S.V.; et al. A cellular model reflecting the phenotypic heterogeneity of mutant HRAS driven squamous cell carcinoma. Int. J. Cancer. 2016, 139, 1106–1116. [Google Scholar] [CrossRef] [PubMed]
- Lussier, C.R.; Babeu, J.P.; Auclair, B.A.; Perreault, N.; Boudreau, F. Hepatocyte nuclear factor-4alpha promotes differentiation of intestinal epithelial cells in a coculture system. Am. J. Physiol. Gastrointest. Liver Physiol. 2008, 294, G418–G428. [Google Scholar] [CrossRef] [PubMed]
- Reiding, K.R.; Blank, D.; Kuijper, D.M.; Deelder, A.M.; Wuhrer, M. High-throughput profiling of protein N-glycosylation by MALDI-TOF-MS employing linkage-specific sialic acid esterification. Anal. Chem. 2014, 86, 5784–5793. [Google Scholar] [CrossRef] [PubMed]
- Strohalm, M.; Hassman, M.; Kosata, B.; Kodicek, M. mMass data miner: An open source alternative for mass spectrometric data analysis. Rapid Commun. Mass Spectrom. 2008, 22, 905–908. [Google Scholar] [CrossRef] [PubMed]
- Jansen, B.C.; Reiding, K.R.; Bondt, A.; Hipgrave Ederveen, A.L.; Palmblad, M.; Falck, D.; Wuhrer, M. MassyTools: A High-Throughput Targeted Data Processing Tool for Relative Quantitation and Quality Control Developed for Glycomic and Glycoproteomic MALDI-MS. J. Proteome Res. 2015, 14, 5088–5098. [Google Scholar] [CrossRef]
- Domon, B.; Costello, C.E. A Systematic Nomenclature for Carbohydrate Fragmentations in Fab-Ms Ms Spectra of Glycoconjugates. Glycoconj. J. 1988, 5, 397–409. [Google Scholar] [CrossRef]
- Boudreau, F.; Rings, E.H.; van Wering, H.M.; Kim, R.K.; Swain, G.P.; Krasinski, S.D.; Moffett, J.; Grand, R.J.; Suh, E.R.; Traber, P.G. Hepatocyte nuclear factor-1 alpha, GATA-4, and caudal related homeodomain protein Cdx2 interact functionally to modulate intestinal gene transcription. Implication for the developmental regulation of the sucrase-isomaltase gene. J. Biol. Chem. 2002, 277, 31909–31917. [Google Scholar] [CrossRef]
- Krasinski, S.D.; Van Wering, H.M.; Tannemaat, M.R.; Grand, R.J. Differential activation of intestinal gene promoters: Functional interactions between GATA-5 and HNF-1 alpha. Am. J. Physiol. Gastrointest. Liver Physiol. 2001, 281, G69–G84. [Google Scholar] [CrossRef]
- Boyd, M.; Hansen, M.; Jensen, T.G.; Perearnau, A.; Olsen, A.K.; Bram, L.L.; Bak, M.; Tommerup, N.; Olsen, J.; Troelsen, J.T. Genome-wide analysis of CDX2 binding in intestinal epithelial cells (Caco-2). J. Biol. Chem. 2010, 285, 25115–25125. [Google Scholar] [CrossRef] [PubMed]
- Huflejt, M.E.; Leffler, H. Galectin-4 in normal tissues and cancer. Glycoconj. J. 2004, 20, 247–255. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.Y.; Takahashi, M.; Gu, J.G.; Miyoshi, E.; Matsumoto, A.; Kitazume, S.; Taniguchi, N. Functional roles of N-glycans in cell signaling and cell adhesion in cancer. Cancer Sci. 2008, 99, 1304–1310. [Google Scholar] [CrossRef] [PubMed]
- Christiansen, M.N.; Chik, J.; Lee, L.; Anugraham, M.; Abrahams, J.L.; Packer, N.H. Cell surface protein glycosylation in cancer. Proteomics 2014, 14, 525–546. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.; Pierce, J.M. Transcriptional Regulation of Glycan Expression. In Glycoscience: Biology and Medicine; Endo, T., Seeberger, H.P., Hart, W.G., Wong, C.-H., Taniguchi, N., Eds.; Springer: Tokyo, Japan, 2014; pp. 1–7. [Google Scholar]
- Trinchera, M.; Aronica, A.; Dall’Olio, F. Selectin Ligands Sialyl-Lewis a and Sialyl-Lewis x in Gastrointestinal Cancers. Biology 2017, 6, 16. [Google Scholar] [CrossRef] [PubMed]
- Yeung, T.M.; Gandhi, S.C.; Wilding, J.L.; Muschel, R.; Bodmer, W.F. Cancer stem cells from colorectal cancer-derived cell lines. Proc. Natl. Acad. Sci. USA 2010, 107, 3722–3727. [Google Scholar] [CrossRef]
- Breiman, A.; Lopez Robles, M.D.; de Carne Trecesson, S.; Echasserieau, K.; Bernardeau, K.; Drickamer, K.; Imberty, A.; Barille-Nion, S.; Altare, F.; Le Pendu, J. Carcinoma-associated fucosylated antigens are markers of the epithelial state and can contribute to cell adhesion through CLEC17A (Prolectin). Oncotarget 2016, 7, 14064–14082. [Google Scholar] [CrossRef]
- Vellinga, T.T.; den Uil, S.; Rinkes, I.H.; Marvin, D.; Ponsioen, B.; Alvarez-Varela, A.; Fatrai, S.; Scheele, C.; Zwijnenburg, D.A.; Snippert, H.; et al. Collagen-rich stroma in aggressive colon tumors induces mesenchymal gene expression and tumor cell invasion. Oncogene 2016, 35, 5263–5271. [Google Scholar] [CrossRef]
- Kim, S.W.; Park, K.C.; Jeon, S.M.; Ohn, T.B.; Kim, T.I.; Kim, W.H.; Cheon, J.H. Abrogation of galectin-4 expression promotes tumorigenesis in colorectal cancer. Cell. Oncol. 2013, 36, 169–178. [Google Scholar] [CrossRef] [PubMed]
- Satelli, A.; Rao, P.S.; Thirumala, S.; Rao, U.S. Galectin-4 functions as a tumor suppressor of human colorectal cancer. Int. J. Cancer. 2011, 129, 799–809. [Google Scholar] [CrossRef]
- Belo, A.I.; van der Sar, A.M.; Tefsen, B.; van Die, I. Galectin-4 Reduces Migration and Metastasis Formation of Pancreatic Cancer Cells. PLoS ONE 2013, 8, e65957. [Google Scholar] [CrossRef] [PubMed]
- Sakuma, K.; Aoki, M.; Kannagi, R. Transcription factors c-Myc and CDX2 mediate E-selectin ligand expression in colon cancer cells undergoing EGF/bFGF-induced epithelial-mesenchymal transition. Proc. Natl. Acad. Sci. USA 2012, 109, 7776–7781. [Google Scholar] [CrossRef] [PubMed]
- Hanamatsu, H.; Nishikaze, T.; Miura, N.; Piao, J.; Okada, K.; Sekiya, S.; Iwamoto, S.; Sakamoto, N.; Tanaka, K.; Furukawa, J.I. Sialic Acid Linkage Specific Derivatization of Glycosphingolipid Glycans by Ring-Opening Aminolysis of Lactones. Anal. Chem. 2018, 90, 13193–13199. [Google Scholar] [CrossRef] [PubMed]
- Varki, A. Sialic acids in human health and disease. Trends Mol. Med. 2008, 14, 351–360. [Google Scholar] [CrossRef] [PubMed]
- Corfield, A.P.; Myerscough, N.; Warren, B.F.; Durdey, P.; Paraskeva, C.; Schauer, R. Reduction of sialic acid O-acetylation in human colonic mucins in the adenoma-carcinoma sequence. Glycoconj. J. 1999, 16, 307–317. [Google Scholar] [CrossRef] [PubMed]
- Holst, S.; Stavenhagen, K.; Balog, C.I.; Koeleman, C.A.; McDonnell, L.M.; Mayboroda, O.A.; Verhoeven, A.; Mesker, W.E.; Tollenaar, R.A.; Deelder, A.M.; et al. Investigations on aberrant glycosylation of glycosphingolipids in colorectal cancer tissues using liquid chromatography and matrix-assisted laser desorption time-of-flight mass spectrometry (MALDI-TOF-MS). Mol. Cell. Proteomics 2013, 12, 3081–3093. [Google Scholar] [CrossRef] [PubMed]
- Falck, D.; Haberger, M.; Plomp, R.; Hook, M.; Bulau, P.; Wuhrer, M.; Reusch, D. Affinity purification of erythropoietin from cell culture supernatant combined with MALDI-TOF-MS analysis of erythropoietin N-glycosylation. Sci. Rep. 2017, 7, 5324. [Google Scholar] [CrossRef] [PubMed]
- Isshiki, S.; Kudo, T.; Nishihara, S.; Ikehara, Y.; Togayachi, A.; Furuya, A.; Shitara, K.; Kubota, T.; Watanabe, M.; Kitajima, M.; et al. Lewis type 1 antigen synthase (beta3Gal-T5) is transcriptionally regulated by homeoproteins. J. Biol. Chem. 2003, 278, 36611–36620. [Google Scholar] [CrossRef] [PubMed]
- Groux-Degroote, S.; Wavelet, C.; Krzewinski-Recchi, M.A.; Portier, L.; Mortuaire, M.; Mihalache, A.; Trinchera, M.; Delannoy, P.; Malagolini, N.; Chiricolo, M.; et al. B4GALNT2 gene expression controls the biosynthesis of Sda and sialyl Lewis X antigens in healthy and cancer human gastrointestinal tract. Int. J. Biochem. Cell Biol. 2014, 53, 442–449. [Google Scholar] [CrossRef] [PubMed]
- Hirano, K.; Matsuda, A.; Shirai, T.; Furukawa, K. Expression of LacdiNAc groups on N-glycans among human tumors is complex. Biomed. Res. Int. 2014, 2014, 981627. [Google Scholar] [CrossRef] [PubMed]
- Gu, J.; Sato, Y.; Kariya, Y.; Isaji, T.; Taniguchi, N.; Fukuda, T. A mutual regulation between cell-cell adhesion and N-glycosylation: Implication of the bisecting GlcNAc for biological functions. J. Proteome Res. 2009, 8, 431–435. [Google Scholar] [CrossRef] [PubMed]
- Yoshimura, M.; Ihara, Y.; Matsuzawa, Y.; Taniguchi, N. Aberrant glycosylation of E-cadherin enhances cell-cell binding to suppress metastasis. J. Biol. Chem. 1996, 271, 13811–13815. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Isaji, T.; Im, S.; Fukuda, T.; Kameyama, A.; Gu, J. Expression of N-acetylglucosaminyltransferase III suppresses alpha2,3 sialylation and its distinctive functions in cell migration are attributed to alpha2,6 sialylation levels. J. Biol. Chem. 2016, 291, 5708. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Wang, X.; Tan, Z.; Chen, S.; Guan, F. Role of Glycans in Cancer Cells Undergoing Epithelial-Mesenchymal Transition. Front. Oncol. 2016, 6, 33. [Google Scholar] [CrossRef] [PubMed]
- Tan, Z.; Lu, W.; Li, X.; Yang, G.; Guo, J.; Yu, H.; Li, Z.; Guan, F. Altered N-Glycan expression profile in epithelial-to-mesenchymal transition of NMuMG cells revealed by an integrated strategy using mass spectrometry and glycogene and lectin microarray analysis. J. Proteome Res. 2014, 13, 2783–2795. [Google Scholar] [CrossRef] [PubMed]
- Hirakawa, M.; Takimoto, R.; Tamura, F.; Yoshida, M.; Ono, M.; Murase, K.; Sato, Y.; Osuga, T.; Sato, T.; Iyama, S.; et al. Fucosylated TGF-[beta] receptors transduces a signal for epithelial-mesenchymal transition in colorectal cancer cells. Br. J. Cancer 2014, 110, 156–163. [Google Scholar] [CrossRef] [PubMed]
- Hryniuk, A.; Grainger, S.; Savory, J.G.; Lohnes, D. Cdx1 and Cdx2 function as tumor suppressors. J. Biol. Chem. 2014, 289, 33343–33354. [Google Scholar] [CrossRef] [PubMed]
- Ishikawa, F.; Nose, K.; Shibanuma, M. Downregulation of hepatocyte nuclear factor-4alpha and its role in regulation of gene expression by TGF-beta in mammary epithelial cells. Exp. Cell Res. 2008, 314, 2131–2140. [Google Scholar] [CrossRef]
- Pelletier, L.; Rebouissou, S.; Vignjevic, D.; Bioulac-Sage, P.; Zucman-Rossi, J. HNF1alpha inhibition triggers epithelial-mesenchymal transition in human liver cancer cell lines. BMC Cancer 2011, 11, 427. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; Li, S.N.; Anjum, K.M.; Gui, L.X.; Zhu, S.S.; Liu, J.; Chen, J.K.; Liu, Q.F.; Ye, G.D.; Wang, W.J.; et al. A double-negative feedback loop between Wnt-beta-catenin signaling and HNF4alpha regulates epithelial-mesenchymal transition in hepatocellular carcinoma. J. Cell Sci. 2013, 126, 5692–5703. [Google Scholar] [CrossRef]


| Cell Lines | CDX1 mRNA [a.u.] | Tumor | Duke’s/grade | Differentiation | Lumen Formation |
|---|---|---|---|---|---|
| HCA46 | 3308.59 | sigmoid colon adenocarcinoma | C | poorly | Lumen |
| HCC56 | 4115.35 | colon adenocarcinoma/liver? | n.a. | moderately | Dense |
| GP2D | 3058.48 | colon adenocarcinoma | B | poorly | Intermediate |
| PCJW | 2598.75 | colon adenocarcinoma | C | poorly | Intermediate |
| LS174T | 1147.36 | colon adenocarcinoma | B/grade I | well | Intermediate |
| LIM1863 | 2862.38 | colon adenocarcinoma | C/grade III | poorly | Lumen |
| SW403 | 2964.14 | colon adenocarcinoma | C/grade III | n.a. | Intermediate |
| RCM1 | 1152.22 | colon adenocarcinoma | n.a. | n.a. | Intermediate |
| ISRECO1 | 32.10 | colon adenocarcinoma | n.a. | n.a. | n.a. |
| VACO429 | 28.69 | colon adenocarcinoma | n.a. | n.a. | Dense |
| HCT116 | 59.94 | colon adenocarcinoma | grade IV | poorly | Dense |
| CC20 | 59.51 | sigmoid colon adenocarcinoma | B | well | Network/stellate |
| CAR1 | 49.35 | colon adenocarcinoma | n.a. | n.a. | Dense |
| COLO678 | 16.21 | colon adenocarcinoma, metastatic lymph node | n.a. | n.a. | Dense |
| HDC8 | 29.31 | colon adenocarcinoma | C/grade III | n.a. | Intermediate |
| OXCO1 | 49.69 | colon adenocarcinoma | n.a. | n.a. | n.a. |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Holst, S.; Wilding, J.L.; Koprowska, K.; Rombouts, Y.; Wuhrer, M. N-Glycomic and Transcriptomic Changes Associated with CDX1 mRNA Expression in Colorectal Cancer Cell Lines. Cells 2019, 8, 273. https://doi.org/10.3390/cells8030273
Holst S, Wilding JL, Koprowska K, Rombouts Y, Wuhrer M. N-Glycomic and Transcriptomic Changes Associated with CDX1 mRNA Expression in Colorectal Cancer Cell Lines. Cells. 2019; 8(3):273. https://doi.org/10.3390/cells8030273
Chicago/Turabian StyleHolst, Stephanie, Jennifer L. Wilding, Kamila Koprowska, Yoann Rombouts, and Manfred Wuhrer. 2019. "N-Glycomic and Transcriptomic Changes Associated with CDX1 mRNA Expression in Colorectal Cancer Cell Lines" Cells 8, no. 3: 273. https://doi.org/10.3390/cells8030273
APA StyleHolst, S., Wilding, J. L., Koprowska, K., Rombouts, Y., & Wuhrer, M. (2019). N-Glycomic and Transcriptomic Changes Associated with CDX1 mRNA Expression in Colorectal Cancer Cell Lines. Cells, 8(3), 273. https://doi.org/10.3390/cells8030273





