The miRFIB-Score: A Serological miRNA-Based Scoring Algorithm for the Diagnosis of Significant Liver Fibrosis
Abstract
1. Introduction
2. Materials and Methods
2.1. Animal Studies
2.2. Patient Cohort
2.3. Blood Collection
2.4. Serological Scoring of Fibrosis
2.5. Messenger RNA and microRNA Analysis
2.6. Histological Evaluation
2.7. Immunocytochemistry
2.8. Statistical Analysis
3. Results
3.1. Identification of Candidate HSC-Linked miRNAs
3.2. Identification of Ankrd52, Clcn5 and Peg10 as Potential Target Genes
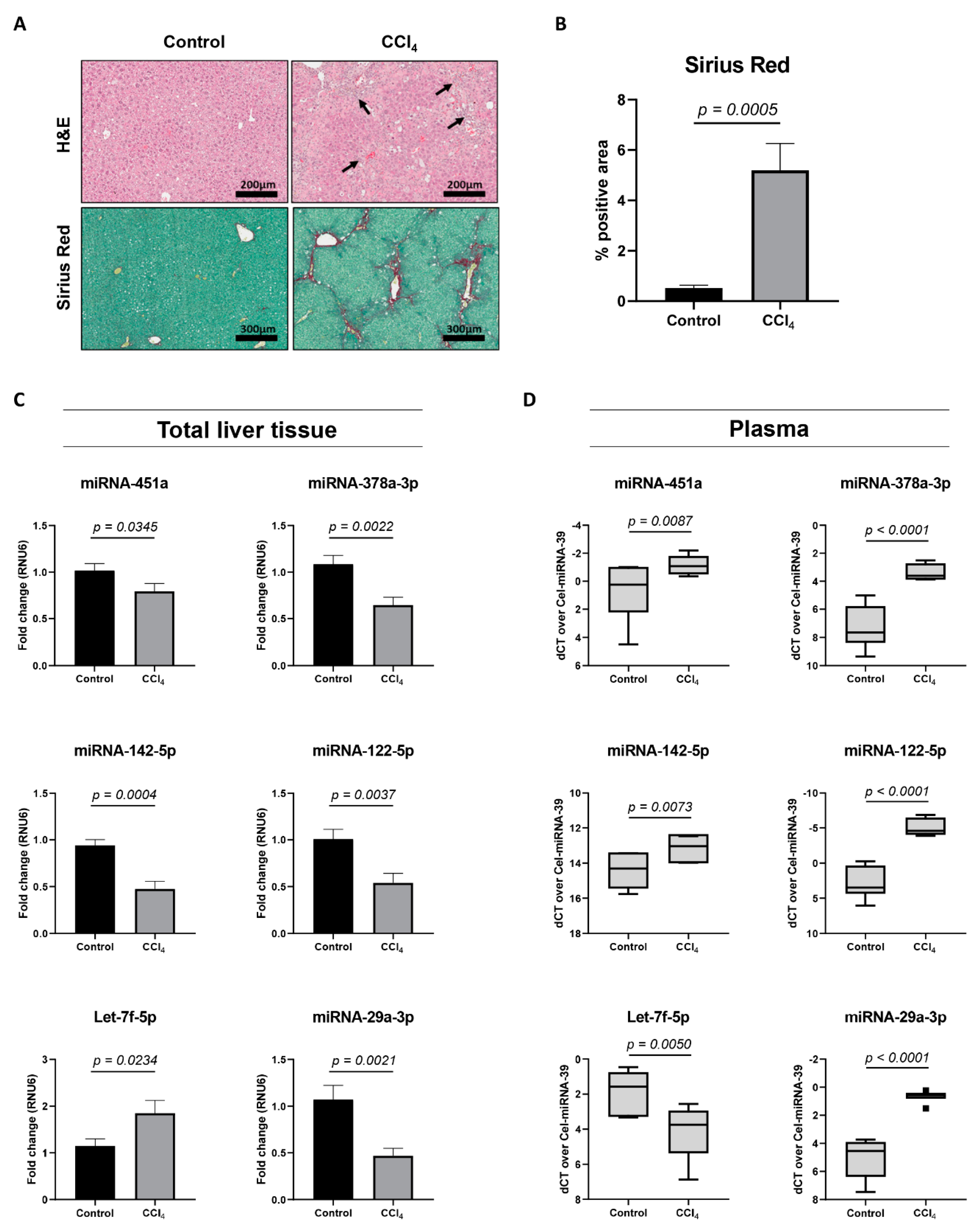
3.3. miRNA Expression Analysis in the CCl4-Mouse Model
3.4. Patient Characteristics and Plasma miRNA Alterations During Liver Fibrosis Progression
3.5. Association of Circulating miRNAs With Clinical Variables
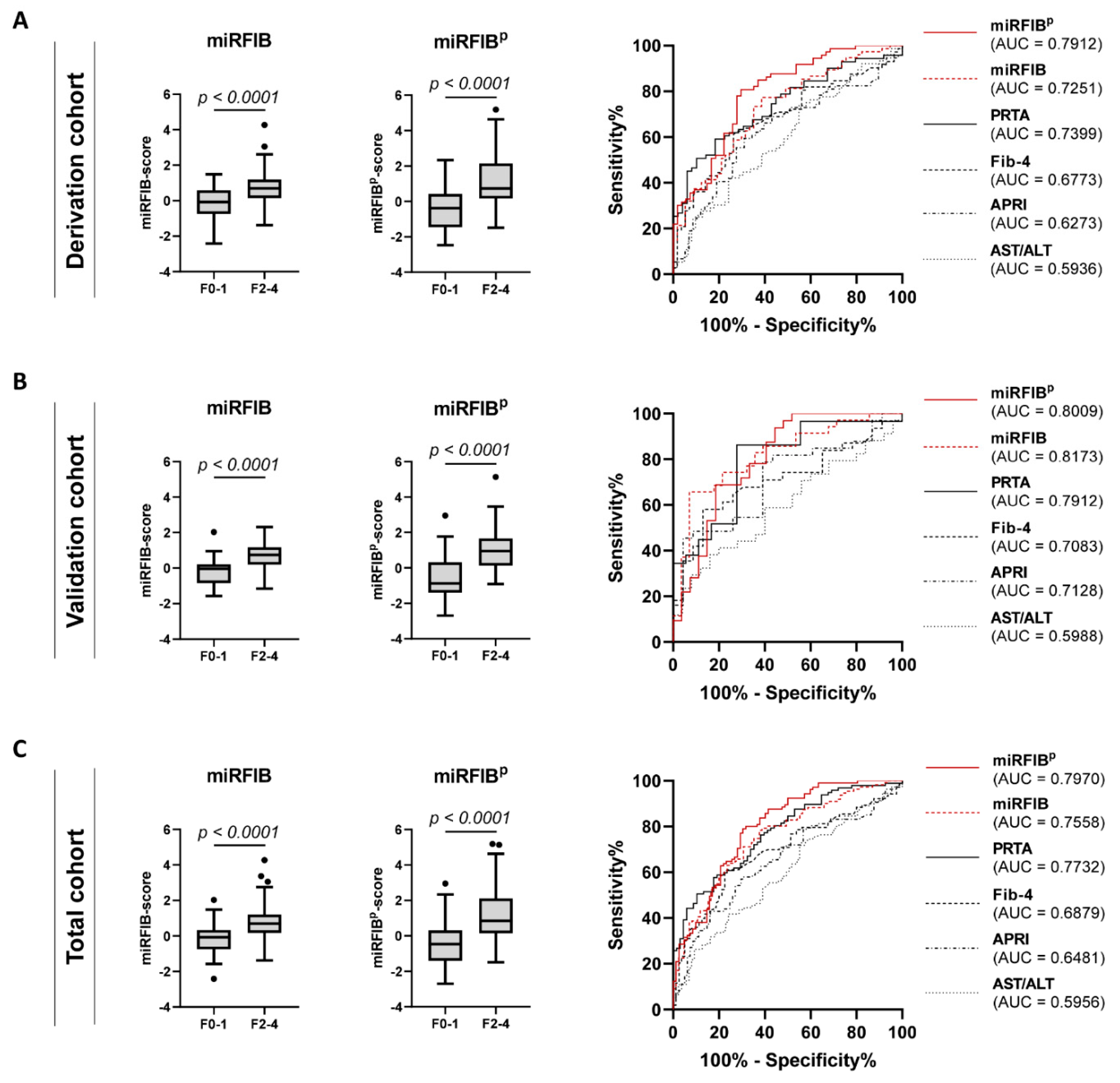
3.6. Discrimination of Significant Liver Fibrosis by the miRFIB-Score
3.7. Inclusion of PDGFRβ Improves the Diagnostic Power of the miRFIB-Score
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- World Health Organization. Global Health Estimates for Cause Specific Mortality. 2016. Available online: https://www.who.int/healthinfo/global_burden_disease/estimates/en/ (accessed on 11 July 2019).
- Tsochatzis, E.A.; Bosch, J.; Burroughs, A.K. Liver cirrhosis. Lancet 2014, 383, 1749–1761. [Google Scholar] [CrossRef]
- Tsuchida, T.; Friedman, S.L. Mechanisms of hepatic stellate cell activation. Nat. Rev. Gastroenterol. Hepatol. 2017, 14, 397–411. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.A.; Wallace, M.C.; Friedman, S.L. Pathobiology of liver fibrosis: A translational success story. Gut 2015, 64, 830–841. [Google Scholar] [CrossRef] [PubMed]
- Brenner, D.A. Reversibility of liver fibrosis. Gastroenterol. Hepatol. 2013, 9, 737–739. [Google Scholar]
- Regev, A.; Berho, M.; Jeffers, L.J.; Milikowski, C.; Molina, E.G.; Pyrsopoulos, N.T.; Feng, Z.Z.; Reddy, K.R.; Schiff, E.R. Sampling error and intraobserver variation in liver biopsy in patients with chronic HCV infection. Am. J. Gastroenterol. 2002, 97, 2614–2618. [Google Scholar] [CrossRef] [PubMed]
- Pasha, T.; Gabriel, S.; Therneau, T.; Dickson, E.R.; Lindor, K.D. Cost-effectiveness of ultrasound-guided liver biopsy. Hepatology 1998, 27, 1220–1226. [Google Scholar] [CrossRef] [PubMed]
- Cadranel, J.F.; Rufat, P.; Degos, F. Practices of liver biopsy in France: Results of a prospective nationwide survey. For the Group of Epidemiology of the French Association for the Study of the Liver (AFEF). Hepatology 2000, 32, 477–481. [Google Scholar] [CrossRef] [PubMed]
- Lambrecht, J.; Verhulst, S.; Mannaerts, I.; Reynaert, H.; van Grunsven, L.A. Prospects in non-invasive assessment of liver fibrosis: Liquid biopsy as the future gold standard? Biochim. Biophys. Acta Mol. Basis Dis. 2018, 1864, 1024–1036. [Google Scholar] [CrossRef] [PubMed]
- Lambrecht, J.; Mannaerts, I.; van Grunsven, L.A. The role of miRNAs in stress-responsive hepatic stellate cells during liver fibrosis. Front. Physiol. 2015, 6, 209. [Google Scholar] [CrossRef] [PubMed]
- Turchinovich, A.; Weiz, L.; Burwinkel, B. Extracellular miRNAs: The mystery of their origin and function. Trends Biochem. Sci. 2012, 37, 460–465. [Google Scholar] [CrossRef] [PubMed]
- Loosen, S.H.; Schueller, F.; Trautwein, C.; Roy, S.; Roderburg, C. Role of circulating microRNAs in liver diseases. World J. Hepatol. 2017, 9, 586–594. [Google Scholar] [CrossRef] [PubMed]
- Hayes, C.N.; Chayama, K. MicroRNAs as Biomarkers for Liver Disease and Hepatocellular Carcinoma. Int. J. Mol. Sci. 2016, 17, 280. [Google Scholar] [CrossRef] [PubMed]
- Lambrecht, J.; Jan Poortmans, P.; Verhulst, S.; Reynaert, H.; Mannaerts, I.; van Grunsven, L.A. Circulating ECV-Associated miRNAs as Potential Clinical Biomarkers in Early Stage HBV and HCV Induced Liver Fibrosis. Front. Pharmacol. 2017, 8, 56. [Google Scholar] [CrossRef] [PubMed]
- Guimaraes, E.L.; Empsen, C.; Geerts, A.; van Grunsven, L.A. Advanced glycation end products induce production of reactive oxygen species via the activation of NADPH oxidase in murine hepatic stellate cells. J. Hepatol. 2010, 52, 389–397. [Google Scholar] [CrossRef] [PubMed]
- Friedrich-Rust, M.; Ong, M.F.; Martens, S.; Sarrazin, C.; Bojunga, J.; Zeuzem, S.; Herrmann, E. Performance of transient elastography for the staging of liver fibrosis: A meta-analysis. Gastroenterology 2008, 134, 960–974. [Google Scholar] [CrossRef] [PubMed]
- Lambrecht, J.; Verhulst, S.; Mannaerts, I.; Sowa, J.P.; Best, J.; Canbay, A.; Reynaert, H.; van Grunsven, L.A. A PDGFRbeta-based score predicts significant liver fibrosis in patients with chronic alcohol abuse, NAFLD and viral liver disease. EBioMedicine 2019, 43, 501–512. [Google Scholar] [CrossRef] [PubMed]
- Seger, S.; Stritt, M.; Vezzali, E.; Nayler, O.; Hess, P.; Groenen, P.M.A.; Stalder, A.K. A fully automated image analysis method to quantify lung fibrosis in the bleomycin-induced rat model. PLoS ONE 2018, 13, e0193057. [Google Scholar] [CrossRef] [PubMed]
- Youden, W.J. Index for rating diagnostic tests. Cancer 1950, 3, 32–35. [Google Scholar] [CrossRef]
- Scholten, D.; Trebicka, J.; Liedtke, C.; Weiskirchen, R. The carbon tetrachloride model in mice. Laboratory Animals 2015, 49, 4–11. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Ghazwani, M.; Zhang, Y.; Lu, J.; Li, J.; Fan, J.; Gandhi, C.R.; Li, S. miR-122 regulates collagen production via targeting hepatic stellate cells and suppressing P4HA1 expression. J. Hepatol. 2013, 58, 522–528. [Google Scholar] [CrossRef]
- Roderburg, C.; Urban, G.W.; Bettermann, K.; Vucur, M.; Zimmermann, H.; Schmidt, S.; Janssen, J.; Koppe, C.; Knolle, P.; Castoldi, M.; et al. Micro-RNA profiling reveals a role for miR-29 in human and murine liver fibrosis. Hepatology 2011, 53, 209–218. [Google Scholar] [CrossRef] [PubMed]
- Ekstedt, M.; Hagstrom, H.; Nasr, P.; Fredrikson, M.; Stal, P.; Kechagias, S.; Hultcrantz, R. Fibrosis stage is the strongest predictor for disease-specific mortality in NAFLD after up to 33 years of follow-up. Hepatology 2015, 61, 1547–1554. [Google Scholar] [CrossRef] [PubMed]
- Angulo, P.; Kleiner, D.E.; Dam-Larsen, S.; Adams, L.A.; Bjornsson, E.S.; Charatcharoenwitthaya, P.; Mills, P.R.; Keach, J.C.; Lafferty, H.D.; Stahler, A.; et al. Liver Fibrosis, but No Other Histologic Features, Is Associated With Long-term Outcomes of Patients With Nonalcoholic Fatty Liver Disease. Gastroenterology 2015, 149, 389–397. [Google Scholar] [CrossRef] [PubMed]
- Mato, J.M.; Lu, S.C. Where are we in the search for noninvasive nonalcoholic steatohepatitis biomarkers? Hepatology 2011, 54, 1115–1117. [Google Scholar] [CrossRef] [PubMed]
- Kim, W.R.; Berg, T.; Asselah, T.; Flisiak, R.; Fung, S.; Gordon, S.C.; Janssen, H.L.; Lampertico, P.; Lau, D.; Bornstein, J.D.; et al. Evaluation of APRI and FIB-4 scoring systems for non-invasive assessment of hepatic fibrosis in chronic hepatitis B patients. J. Hepatol. 2016, 64, 773–780. [Google Scholar] [CrossRef] [PubMed]
- Xiao, G.; Yang, J.; Yan, L. Comparison of diagnostic accuracy of aspartate aminotransferase to platelet ratio index and fibrosis-4 index for detecting liver fibrosis in adult patients with chronic hepatitis B virus infection: A systemic review and meta-analysis. Hepatology 2015, 61, 292–302. [Google Scholar] [CrossRef]
- Ampuero, J.; Romero-Gomez, M. Editorial: Looking for patients at risk of cirrhosis in the general population-many needles in a haystack. Aliment. Pharmacol. Ther. 2018, 47, 692–694. [Google Scholar] [CrossRef]
- Hur, W.; Lee, J.H.; Kim, S.W.; Kim, J.H.; Bae, S.H.; Kim, M.; Hwang, D.; Kim, Y.S.; Park, T.; Um, S.J.; et al. Downregulation of microRNA-451 in non-alcoholic steatohepatitis inhibits fatty acid-induced proinflammatory cytokine production through the AMPK/AKT pathway. Int. J. Biochem. Cell Biol. 2015, 64, 265–276. [Google Scholar] [CrossRef]
- Yamada, H.; Suzuki, K.; Ichino, N.; Ando, Y.; Sawada, A.; Osakabe, K.; Sugimoto, K.; Ohashi, K.; Teradaira, R.; Inoue, T.; et al. Associations between circulating microRNAs (miR-21, miR-34a, miR-122 and miR-451) and non-alcoholic fatty liver. Clin. Chim. Acta 2013, 424, 99–103. [Google Scholar] [CrossRef]
- Jin, B.X.; Zhang, Y.H.; Jin, W.J.; Sun, X.Y.; Qiao, G.F.; Wei, Y.Y.; Sun, L.B.; Zhang, W.H.; Li, N. MicroRNA panels as disease biomarkers distinguishing hepatitis B virus infection caused hepatitis and liver cirrhosis. Sci. Rep. 2015, 5, 15026. [Google Scholar] [CrossRef]
- Su, S.; Zhao, Q.; He, C.; Huang, D.; Liu, J.; Chen, F.; Chen, J.; Liao, J.Y.; Cui, X.; Zeng, Y.; et al. miR-142-5p and miR-130a-3p are regulated by IL-4 and IL-13 and control profibrogenic macrophage program. Nat. Commun. 2015, 6, 8523. [Google Scholar] [CrossRef] [PubMed]
- Tsang, F.H.; Au, S.L.; Wei, L.; Fan, D.N.; Lee, J.M.; Wong, C.C.; Ng, I.O.; Wong, C.M. MicroRNA-142-3p and microRNA-142-5p are downregulated in hepatocellular carcinoma and exhibit synergistic effects on cell motility. Front. Med. 2015, 9, 331–343. [Google Scholar] [CrossRef] [PubMed]
- Matsuura, K.; De Giorgi, V.; Schechterly, C.; Wang, R.Y.; Farci, P.; Tanaka, Y.; Alter, H.J. Circulating let-7 levels in plasma and extracellular vesicles correlate with hepatic fibrosis progression in chronic hepatitis C. Hepatology 2016, 64, 732–745. [Google Scholar] [CrossRef] [PubMed]
- Schueller, F.; Roy, S.; Trautwein, C.; Luedde, T.; Roderburg, C. miR-122 expression is not regulated during activation of hepatic stellate cells. J. Hepatol. 2016, 65, 865–867. [Google Scholar] [CrossRef] [PubMed]
- Esau, C.; Davis, S.; Murray, S.F.; Yu, X.X.; Pandey, S.K.; Pear, M.; Watts, L.; Booten, S.L.; Graham, M.; McKay, R.; et al. miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab. 2006, 3, 87–98. [Google Scholar] [CrossRef] [PubMed]
- Trebicka, J.; Anadol, E.; Elfimova, N.; Strack, I.; Roggendorf, M.; Viazov, S.; Wedemeyer, I.; Drebber, U.; Rockstroh, J.; Sauerbruch, T.; et al. Hepatic and serum levels of miR-122 after chronic HCV-induced fibrosis. J. Hepatol. 2013, 58, 234–239. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.Z.; Lin, D.D.; Jin, B.X.; Sun, X.Y.; Li, N. Plasma microRNA: A novel non-invasive biomarker for HBV-associated liver fibrosis staging. Exp. Ther. Med. 2019, 17, 1919–1929. [Google Scholar] [CrossRef] [PubMed]
- Ye, D.; Zhang, T.; Lou, G.; Xu, W.; Dong, F.; Chen, G.; Liu, Y. Plasma miR-17, miR-20a, miR-20b and miR-122 as potential biomarkers for diagnosis of NAFLD in type 2 diabetes mellitus patients. Life Sci. 2018, 208, 201–207. [Google Scholar] [CrossRef]
- Shaker, O.G.; Senousy, M.A. Serum microRNAs as predictors for liver fibrosis staging in hepatitis C virus-associated chronic liver disease patients. J. Viral. Hepat. 2017, 24, 636–644. [Google Scholar] [CrossRef]
- Miyaaki, H.; Ichikawa, T.; Kamo, Y.; Taura, N.; Honda, T.; Shibata, H.; Milazzo, M.; Fornari, F.; Gramantieri, L.; Bolondi, L.; et al. Significance of serum and hepatic microRNA-122 levels in patients with non-alcoholic fatty liver disease. Liver Int. 2014, 34, e302–e307. [Google Scholar] [CrossRef]
- Waidmann, O.; Koberle, V.; Brunner, F.; Zeuzem, S.; Piiper, A.; Kronenberger, B. Serum microRNA-122 predicts survival in patients with liver cirrhosis. PLoS ONE 2012, 7, e45652. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Jia, Y.; Zheng, R.; Guo, Y.; Wang, Y.; Guo, H.; Fei, M.; Sun, S. Plasma microRNA-122 as a biomarker for viral-, alcohol-, and chemical-related hepatic diseases. Clin. Chem. 2010, 56, 1830–1838. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.L.; Wang, F.S.; Li, S.C.; Tiao, M.M.; Huang, Y.H. MicroRNA-29a Alleviates Bile Duct Ligation Exacerbation of Hepatic Fibrosis in Mice through Epigenetic Control of Methyltransferases. Int. J. Mol. Sci. 2017, 18, 192. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.H.; Kuo, H.C.; Yang, Y.L.; Wang, F.S. MicroRNA-29a is a key regulon that regulates BRD4 and mitigates liver fibrosis in mice by inhibiting hepatic stellate cell activation. Int. J. Med. Sci. 2019, 16, 212–220. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.L.; Kuo, H.C.; Wang, F.S.; Huang, Y.H. MicroRNA-29a Disrupts DNMT3b to Ameliorate Diet-Induced Non-Alcoholic Steatohepatitis in Mice. Int. J. Mol. Sci. 2019, 20, 1499. [Google Scholar] [CrossRef] [PubMed]
- Li, S.C.; Wang, F.S.; Yang, Y.L.; Tiao, M.M.; Chuang, J.H.; Huang, Y.H. Microarray Study of Pathway Analysis Expression Profile Associated with MicroRNA-29a with Regard to Murine Cholestatic Liver Injuries. Int. J. Mol. Sci. 2016, 17, 324. [Google Scholar] [CrossRef] [PubMed]
- Jampoka, K.; Muangpaisarn, P.; Khongnomnan, K.; Treeprasertsuk, S.; Tangkijvanich, P.; Payungporn, S. Serum miR-29a and miR-122 as Potential Biomarkers for Non-Alcoholic Fatty Liver Disease (NAFLD). Microrna 2018, 7, 215–222. [Google Scholar] [CrossRef]
- Bao, S.; Zheng, J.; Li, N.; Huang, C.; Chen, M.; Cheng, Q.; Yu, K.; Chen, S.; Zhu, M.; Shi, G. Serum MicroRNA Levels as a Noninvasive Diagnostic Biomarker for the Early Diagnosis of Hepatitis B Virus-Related Liver Fibrosis. Gut Liver 2017, 11, 860–869. [Google Scholar] [CrossRef]
- Gebert, L.F.R.; MacRae, I.J. Regulation of microRNA function in animals. Nat. Rev. Mol. Cell Biol. 2019, 20, 21–37. [Google Scholar] [CrossRef]
- Dong, H.; Ge, X.; Shen, Y.; Chen, L.; Kong, Y.; Zhang, H.; Man, X.; Tang, L.; Yuan, H.; Wang, H.; et al. Gene expression profile analysis of human hepatocellular carcinoma using SAGE and LongSAGE. BMC Med. Genomics 2009, 2, 5. [Google Scholar] [CrossRef]
- Okabe, H.; Satoh, S.; Furukawa, Y.; Kato, T.; Hasegawa, S.; Nakajima, Y.; Yamaoka, Y.; Nakamura, Y. Involvement of PEG10 in human hepatocellular carcinogenesis through interaction with SIAH1. Cancer Res. 2003, 63, 3043–3048. [Google Scholar] [PubMed]
- Li, C.M.; Margolin, A.A.; Salas, M.; Memeo, L.; Mansukhani, M.; Hibshoosh, H.; Szabolcs, M.; Klinakis, A.; Tycko, B. PEG10 is a c-MYC target gene in cancer cells. Cancer Res. 2006, 66, 665–672. [Google Scholar] [CrossRef] [PubMed]
- Ip, W.K.; Lai, P.B.; Wong, N.L.; Sy, S.M.; Beheshti, B.; Squire, J.A.; Wong, N. Identification of PEG10 as a progression related biomarker for hepatocellular carcinoma. Cancer Lett. 2007, 250, 284–291. [Google Scholar] [CrossRef] [PubMed]
- Bang, H.; Ha, S.Y.; Hwang, S.H.; Park, C.K. Expression of PEG10 Is Associated with Poor Survival and Tumor Recurrence in Hepatocellular Carcinoma. Cancer Res. Treat. 2015, 47, 844–852. [Google Scholar] [CrossRef] [PubMed]
- Zou, C.D.; Zhao, W.M.; Wang, X.N.; Li, Q.; Huang, H.; Cheng, W.P.; Jin, J.F.; Zhang, H.; Wu, M.J.; Tai, S.; et al. MicroRNA-107: A novel promoter of tumor progression that targets the CPEB3/EGFR axis in human hepatocellular carcinoma. Oncotarget 2016, 7, 266–278. [Google Scholar] [CrossRef] [PubMed]
- Tang, H.; Zhang, J.; Yu, Z.; Ye, L.; Li, K.; Ding, F.; Feng, X.; Meng, W. Mir-452-3p: A Potential Tumor Promoter That Targets the CPEB3/EGFR Axis in Human Hepatocellular Carcinoma. Technol. Cancer Res. Treat. 2017, 16, 1136–1149. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Yao, J.; Huan, L.; Lian, J.; Bao, C.; Li, Y.; Ge, C.; Li, J.; Yao, M.; Liang, L.; et al. GNAI3 inhibits tumor cell migration and invasion and is post-transcriptionally regulated by miR-222 in hepatocellular carcinoma. Cancer Lett. 2015, 356, 978–984. [Google Scholar] [CrossRef] [PubMed]
- Pritchard, C.C.; Kroh, E.; Wood, B.; Arroyo, J.D.; Dougherty, K.J.; Miyaji, M.M.; Tait, J.F.; Tewari, M. Blood cell origin of circulating microRNAs: A cautionary note for cancer biomarker studies. Cancer Prev. Res. 2012, 5, 492–497. [Google Scholar] [CrossRef] [PubMed]
- Kirschner, M.B.; Kao, S.C.; Edelman, J.J.; Armstrong, N.J.; Vallely, M.P.; van Zandwijk, N.; Reid, G. Haemolysis during sample preparation alters microRNA content of plasma. PLoS ONE 2011, 6, e24145. [Google Scholar] [CrossRef] [PubMed]
- Koberle, V.; Pleli, T.; Schmithals, C.; Augusto Alonso, E.; Haupenthal, J.; Bonig, H.; Peveling-Oberhag, J.; Biondi, R.M.; Zeuzem, S.; Kronenberger, B.; et al. Differential stability of cell-free circulating microRNAs: Implications for their utilization as biomarkers. PLoS ONE 2013, 8, e75184. [Google Scholar] [CrossRef] [PubMed]
- Appourchaux, K.; Dokmak, S.; Resche-Rigon, M.; Treton, X.; Lapalus, M.; Gattolliat, C.H.; Porchet, E.; Martinot-Peignoux, M.; Boyer, N.; Vidaud, M.; et al. MicroRNA-based diagnostic tools for advanced fibrosis and cirrhosis in patients with chronic hepatitis B and C. Sci. Rep. 2016, 6, 34935. [Google Scholar] [CrossRef] [PubMed]



| Patient Cohorts | F0–1 | F2–4 | p Value |
|---|---|---|---|
| Individuals, n | 92 | 116 | |
| Disease aetiology: n (%) | |||
| Alcoholic liver disease | 6 (7%) | 27 (23%) | |
| Viral liver disease | 46 (50%) | 28 (24%) | |
| NAFLD | 40 (43%) | 61 (53%) | |
| Characteristics | |||
| Age (years): median (IQR) | 52 (42–63) | 57 (51–65) | 0.0003 |
| Male, n (%) | 52 (57%) | 84 (72%) | 0.0267 |
| BMI (kg/m2): median (IQR) | 27.44 (24.16–32.35) | 29.96 (25.18–34.23) | ns |
| Laboratory parameters: median (IQR) | |||
| AST (IU/L) | 34 (24–48) | 40 (28–67) | 0.0048 |
| ALT (IU/L) | 43 (34–62) | 48 (32–71) | ns |
| Alk Phos (IU/L) | 68 (54–87) | 87 (66–130) | <0.0001 |
| GGT (IU/L) | 39 (23–83) | 71 (40–154) | <0.0001 |
| Total bilirubin (mg/dL) | 0.60 (0.47–0.79) | 0.76 (0.57–1.20) | 0.0005 |
| Albumin (g/L) | 43 (41–46) | 42 (39–45) | 0.0035 |
| Thrombocytes (×103/mm3) | 231 (202–277) | 202 (149–255) | 0.0006 |
| Creatinine (mg/dL) | 0.85 (0.70–1.02) | 0.87 (0.74–1.04) | ns |
| Fibrosis scoring: median (IQR) | |||
| AST/ALT ratio | 0.76 (0.62–0.87) | 0.82 (0.67–1.18) | 0.0211 |
| APRI | 0.35 (0.24–0.57) | 0.56 (0.32–0.88) | 0.0005 |
| Fib-4 | 1.13 (0.83–1.49) | 1.67 (1.08–2.80) | <0.0001 |
| PRTA-score | 7.18 (4.49–9.87) | 11.63 (7.95–20.40) | <0.0001 |
| miRNA-451a | miRNA-142-5p | Let-7f-5p | miRNA-378a-3p | miRNA-122-5p | miRNA-29a-3p | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| r | p | r | p | r | p | r | p | r | p | r | p | |
| Age | −0.0409 | ns | −0.1602 | 0.0234 | 0.1416 | 0.0413 | 0.0905 | ns | 0.2628 | 0.0001 | 0.0096 | ns |
| BMI | 0.0280 | ns | −0.0397 | ns | −0.1563 | 0.0287 | −0.1037 | ns | −0.0667 | ns | −0.2161 | 0.0025 |
| AST | −0.1554 | 0.0318 | 0.0076 | ns | 0.1371 | ns | −0.1038 | ns | −0.2625 | 0.0002 | 0.1150 | ns |
| ALT | −0.0727 | ns | 0.0207 | ns | 0.0534 | ns | −0.1412 | 0.0484 | −0.4093 | <0.0001 | −0.0166 | ns |
| Alk Phos | −0.0022 | ns | −0.0804 | ns | 0.2027 | 0.0050 | −0.0047 | ns | 0.2059 | 0.0046 | 0.1600 | 0.0287 |
| GGT | −0.1372 | ns | −0.1756 | 0.0174 | 0.1730 | 0.0163 | −0.0621 | ns | 0.0060 | ns | 0.0899 | ns |
| Bilirubin | −0.0609 | ns | −0.0237 | ns | 0.3194 | <0.0001 | 0.1295 | ns | 0.1213 | ns | 0.2790 | 0.0001 |
| Albumin | −0.1064 | ns | 0.0365 | ns | −0.2031 | 0.0067 | −0.0148 | ns | −0.2394 | 0.0014 | −0.1747 | 0.0207 |
| Platelet count | 0.1243 | ns | −0.1139 | ns | −0.3778 | <0.0001 | −0.1629 | 0.0255 | −0.1050 | ns | −0.3514 | <0.0001 |
| Creatinine | −0.0419 | ns | −0.0399 | ns | 0.0054 | ns | −0.0094 | ns | 0.0652 | ns | −0.0257 | ns |
| AST/ALT | −0.1026 | ns | −0.0126 | ns | 0.0756 | ns | 0.0311 | ns | 0.1920 | 0.0072 | 0.1627 | 0.0234 |
| Fib-4 | −0.1223 | ns | 0.0439 | ns | 0.3215 | <0.0001 | 0.1400 | ns | 0.0607 | ns | 0.2852 | 0.0001 |
| APRI | −0.1551 | 0.0365 | 0.1039 | ns | 0.2820 | <0.0001 | 0.0321 | ns | −0.1573 | 0.0321 | 0.2551 | 0.0005 |
| PRTA-score | −0.0559 | ns | 0.0266 | ns | 0.4332 | <0.0001 | 0.1479 | ns | 0.2401 | 0.0020 | 0.3447 | <0.0001 |
| AUC | 95% CI | Optimal Cut-Off | Sensitivity (%) | Specificity (%) | PPV | NPV | |
|---|---|---|---|---|---|---|---|
| AST/ALT | |||||||
| Derivation | 0.5936 | 0.4986–0.6886 | 0.6948 | 73.68 | 45.16 | 62.87 | 58.65 |
| Validation | 0.5988 | 0.4546–0.7430 | 1.025 | 38.24 | 84.00 | 75.08 | 51.90 |
| Total | 0.5956 | 0.5166–0.6747 | 0.8725 | 41.82 | 75.86 | 68.59 | 50.85 |
| APRI | |||||||
| Derivation | 0.6273 | 0.5313–0.7234 | 0.4928 | 59.46 | 68.97 | 70.72 | 57.44 |
| Validation | 0.7128 | 0.5773–0.8482 | 0.7531 | 45.45 | 95.65 | 92.94 | 58.18 |
| Total | 0.6481 | 0.5696–0.7267 | 0.4928 | 57.01 | 70.37 | 70.80 | 56.49 |
| Fib-4 | |||||||
| Derivation | 0.6773 | 0.5847–0.7698 | 1.505 | 61.11 | 73.68 | 74.53 | 60.05 |
| Validation | 0.7083 | 0.5692–0.8474 | 1.520 | 58.06 | 86.96 | 84.88 | 62.19 |
| Total | 0.6879 | 0.6112–0.7647 | 1.505 | 60.19 | 77.50 | 77.13 | 60.70 |
| PRTA-score | |||||||
| Derivation | 0.7399 | 0.6525–0.8272 | 10.36 | 59.15 | 81.63 | 80.23 | 61.32 |
| Validation | 0.7912 | 0.6566–0.9258 | 7.842 | 86.21 | 72.22 | 79.64 | 80.60 |
| Total | 0.7732 | 0.7033–0.8431 | 11.59 | 50.52 | 89.71 | 86.09 | 58.99 |
| miRFIB | |||||||
| Derivation | 0.7251 | 0.6393–0.8110 | 0.1109 | 77.33 | 61.40 | 71.63 | 68.24 |
| Validation | 0.8173 | 0.7112–0.9235 | 0.3412 | 65.71 | 92.86 | 92.06 | 68.24 |
| Total | 0.7558 | 0.6887–0.8229 | 0.1404 | 78.38 | 62.35 | 72.41 | 69.59 |
| miRFIBp | |||||||
| Derivation | 0.7912 | 0.7120–0.8704 | 0.0673 | 80.82 | 70.37 | 77.47 | 74.43 |
| Validation | 0.8009 | 0.6844–0.9175 | 0.3840 | 68.75 | 81.48 | 82.39 | 67.41 |
| Total | 0.7970 | 0.7329–0.8611 | 0.1043 | 79.05 | 69.51 | 76.57 | 72.47 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lambrecht, J.; Verhulst, S.; Reynaert, H.; van Grunsven, L.A. The miRFIB-Score: A Serological miRNA-Based Scoring Algorithm for the Diagnosis of Significant Liver Fibrosis. Cells 2019, 8, 1003. https://doi.org/10.3390/cells8091003
Lambrecht J, Verhulst S, Reynaert H, van Grunsven LA. The miRFIB-Score: A Serological miRNA-Based Scoring Algorithm for the Diagnosis of Significant Liver Fibrosis. Cells. 2019; 8(9):1003. https://doi.org/10.3390/cells8091003
Chicago/Turabian StyleLambrecht, Joeri, Stefaan Verhulst, Hendrik Reynaert, and Leo A. van Grunsven. 2019. "The miRFIB-Score: A Serological miRNA-Based Scoring Algorithm for the Diagnosis of Significant Liver Fibrosis" Cells 8, no. 9: 1003. https://doi.org/10.3390/cells8091003
APA StyleLambrecht, J., Verhulst, S., Reynaert, H., & van Grunsven, L. A. (2019). The miRFIB-Score: A Serological miRNA-Based Scoring Algorithm for the Diagnosis of Significant Liver Fibrosis. Cells, 8(9), 1003. https://doi.org/10.3390/cells8091003






