Novel Mechanistic Insight into the Anticancer Activity of Cucurbitacin D against Pancreatic Cancer (Cuc D Attenuates Pancreatic Cancer)
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Culture
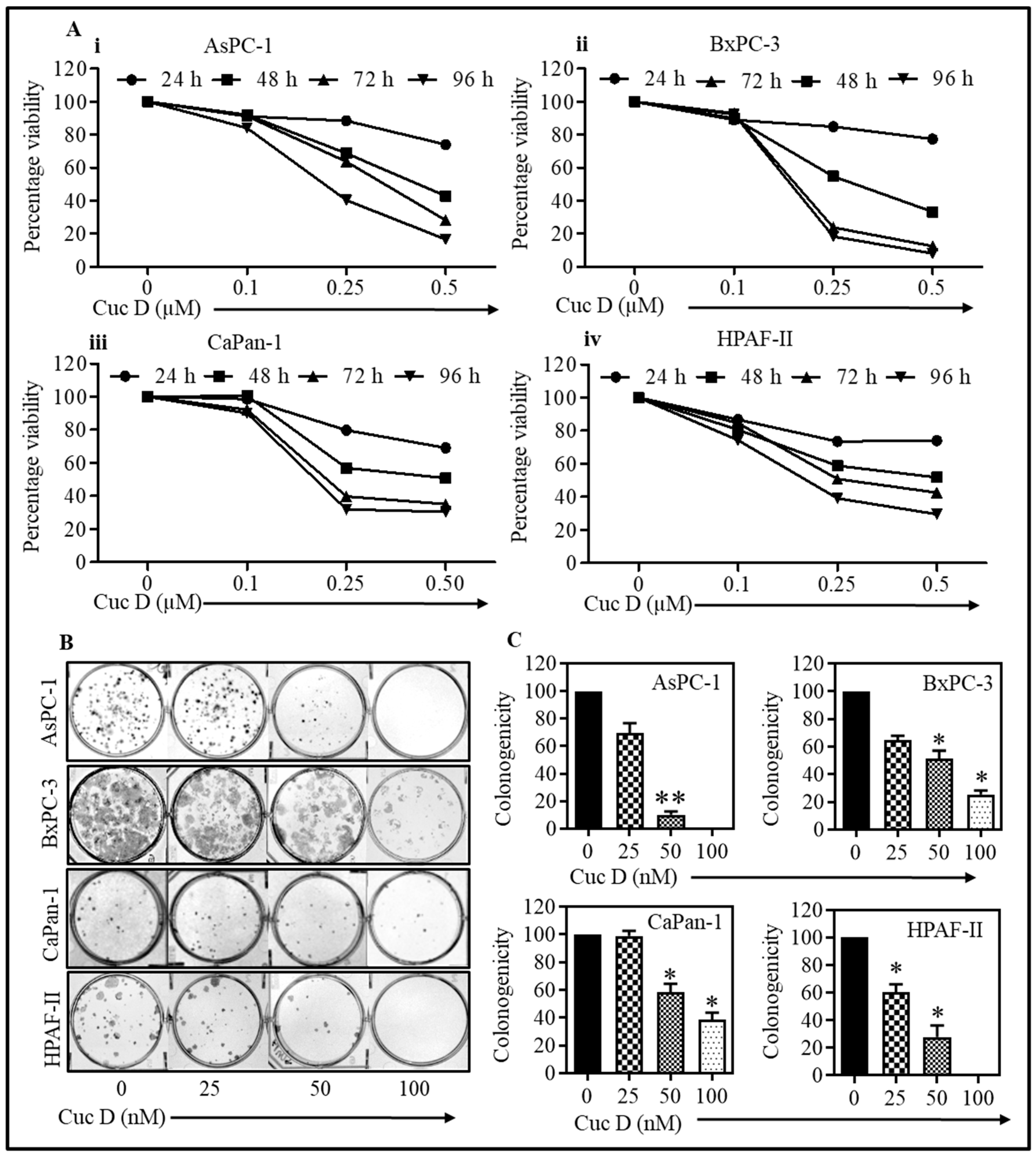
2.2. Cell Proliferation Assay
2.3. Colony Formation Assay
2.4. Cell Cycle Analysis
2.5. Migration Assay
2.6. Agarose Bead Assay
2.7. Wound Healing Assay
2.8. Invasion Assay
2.9. Western Blotting
2.10. Transfection and Quantitative Real-Time Polymerase
2.11. Molecular Docking
2.12. Xenograft Study
2.13. Immunohistochemistry (IHC)
2.14. In Situ Hybridization
2.15. Bioavailability Studies of Cuc D
2.16. Statistical Analysis
3. Results
3.1. Cuc D Treatment Suppresses Proliferation and Clonogenicity of Pancreatic Cancer Cells
3.2. Cuc D Treatment Effectively Arrests Cell Cycle in G2/M Phase
3.3. Cuc D Treatment Inhibited Migratory Potential of PanCa Cells
3.4. Cuc D Treatment Suppresses Invasiveness of PanCa Cells
3.5. Cuc D Treatment Inhibits MUC13 Expression in PanCa Cells
3.6. Molecular Docking Analysis
3.7. Cuc D Treatment Induced Gemcitabine Sensitivity in Gemcitabine Resistant PanCa Cells
3.8. Cuc D Effectively Inhibits Tumor Growth in NOD SCID Gamma (NSG) Xenograft Mouse Model
3.9. Bioavailability of Cuc D in Mice
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Availability of Data
References
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2018. CA Cancer J. Clin. 2018, 68, 7–30. [Google Scholar] [CrossRef] [PubMed]
- Rahib, L.; Smith, B.D.; Aizenberg, R.; Rosenzweig, A.B.; Fleshman, J.M.; Matrisian, L.M. Projecting cancer incidence and deaths to 2030: The unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer Res. 2014, 74, 2913–2921. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morris, J.P.T.; Wang, S.C.; Hebrok, M. KRAS, Hedgehog, Wnt and the twisted developmental biology of pancreatic ductal adenocarcinoma. Nat. Rev. Cancer 2010, 10, 683–695. [Google Scholar] [CrossRef] [PubMed]
- Conroy, T.; Desseigne, F.; Ychou, M.; Bouche, O.; Guimbaud, R.; Becouarn, Y.; Adenis, A.; Raoul, J.L.; Gourgou-Bourgade, S.; de la Fouchardiere, C.; et al. FOLFIRINOX versus gemcitabine for metastatic pancreatic cancer. N. Engl. J. Med. 2011, 364, 1817–1825. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lopez-Casas, P.P.; Lopez-Fernandez, L.A. Gene-expression profiling in pancreatic cancer. Expert Rev. Mol. Diagn. 2010, 10, 591–601. [Google Scholar] [CrossRef]
- Luo, Y.; Cobb, R.E.; Zhao, H. Recent advances in natural product discovery. Curr. Opin. Biotechnol. 2014, 30, 230–237. [Google Scholar] [CrossRef] [Green Version]
- Hu, M. Commentary: Bioavailability of Flavonoids and Polyphenols: Call to Arms. Mol. Pharm. 2007, 4, 803–806. [Google Scholar] [CrossRef] [Green Version]
- Sikander, M.; Malik, S.; Parveen, K.; Ahmad, M.; Yadav, D.; Hafeez, Z.B.; Bansal, M. Hepatoprotective effect of Origanum vulgare in Wistar rats against carbon tetrachloride-induced hepatotoxicity. Protoplasma 2013, 250, 483–493. [Google Scholar] [CrossRef]
- Sikander, M.; Malik, S.; Yadav, D.; Biswas, S.; Katare, D.P.; Jain, S.K. Cytoprotective activity of a trans-chalcone against hydrogen peroxide induced toxicity in hepatocellular carcinoma (HepG2) cells. Asian Pac. J. Cancer Prev. 2011, 12, 2513–2516. [Google Scholar]
- Zaman, M.S.; Chauhan, N.; Yallapu, M.M.; Gara, R.K.; Maher, D.M.; Kumari, S.; Sikander, M.; Khan, S.; Zafar, N.; Jaggi, M.; et al. Curcumin Nanoformulation for Cervical Cancer Treatment. Sci. Rep. 2016, 6, 20051. [Google Scholar] [CrossRef] [Green Version]
- Jayaprakasam, B.; Seeram, N.P.; Nair, M.G. Anticancer and antiinflammatory activities of cucurbitacins from Cucurbita andreana. Cancer Lett. 2003, 189, 11–16. [Google Scholar] [CrossRef]
- Chen, J.C.; Chiu, M.H.; Nie, R.L.; Cordell, G.A.; Qiu, S.X. Cucurbitacins and cucurbitane glycosides: Structures and biological activities. Nat. Prod. Rep. 2005, 22, 386–399. [Google Scholar] [CrossRef] [PubMed]
- Ding, N.; Yamashita, U.; Matsuoka, H.; Sugiura, T.; Tsukada, J.; Noguchi, J.; Yoshida, Y. Apoptosis induction through proteasome inhibitory activity of cucurbitacin D in human T-cell leukemia. Cancer 2011, 117, 2735–2746. [Google Scholar] [CrossRef] [PubMed]
- Nakanishi, T.; Song, Y.; He, C.; Wang, D.; Morita, K.; Tsukada, J.; Kanazawa, T.; Yoshida, Y. Autophagy is associated with cucurbitacin D-induced apoptosis in human T cell leukemia cells. Med. Oncol. 2016, 33, 30. [Google Scholar] [CrossRef] [PubMed]
- Ku, J.M.; Kim, S.R.; Hong, S.H.; Choi, H.S.; Seo, H.S.; Shin, Y.C.; Ko, S.G. Cucurbitacin D induces cell cycle arrest and apoptosis by inhibiting STAT3 and NF-kappaB signaling in doxorubicin-resistant human breast carcinoma (MCF7/ADR) cells. Mol. Cell Biochem. 2015, 409, 33–43. [Google Scholar] [CrossRef] [Green Version]
- Sikander, M.; Hafeez, B.B.; Malik, S.; Alsayari, A.; Halaweish, F.T.; Yallapu, M.M.; Chauhan, S.C.; Jaggi, M. Cucurbitacin D exhibits potent anti-cancer activity in cervical cancer. Sci. Rep. 2016, 6, 36594. [Google Scholar] [CrossRef]
- Sikander, M.; Malik, S.; Chauhan, N.; Khan, P.; Kumari, S.; Kashyap, V.K.; Khan, S.; Ganju, A.; Halaweish, F.T.; Yallapu, M.M.; et al. Cucurbitacin D Reprograms Glucose Metabolic Network in Prostate Cancer. Cancers 2019, 11, 364. [Google Scholar] [CrossRef] [Green Version]
- Hall, J.A.; Seedarala, S.; Rice, N.; Kopel, L.; Halaweish, F.; Blagg, B.S. Cucurbitacin D Is a Disruptor of the HSP90 Chaperone Machinery. J. Nat. Prod. 2015, 78, 873–879. [Google Scholar] [CrossRef] [Green Version]
- Chauhan, S.C.; Ebeling, M.C.; Maher, D.M.; Koch, M.D.; Watanabe, A.; Aburatani, H.; Lio, Y.; Jaggi, M. MUC13 mucin augments pancreatic tumorigenesis. Mol. Cancer Ther. 2012, 11, 24–33. [Google Scholar] [CrossRef] [Green Version]
- Khan, S.; Ebeling, M.C.; Zaman, M.S.; Sikander, M.; Yallapu, M.M.; Chauhan, N.; Yacoubian, A.M.; Behrman, S.W.; Zafar, N.; Kumar, D.; et al. MicroRNA-145 targets MUC13 and suppresses growth and invasion of pancreatic cancer. Oncotarget 2014, 5, 7599–7609. [Google Scholar] [CrossRef]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Voura, M.; Khan, P.; Thysiadis, S.; Katsamakas, S.; Queen, A.; Hasan, G.M.; Ali, S.; Sarli, V.; Hassan, M.I. Probing the Inhibition of Microtubule Affinity Regulating Kinase 4 by N-Substituted Acridones. Sci. Rep. 2019, 9, 1676. [Google Scholar] [CrossRef] [PubMed]
- Webb, B.; Sali, A. Protein Structure Modeling with MODELLER. Methods Mol. Biol. 2017, 1654, 39–54. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Skolnick, J.; Zhang, Y. Ab initio modeling of small proteins by iterative TASSER simulations. BMC Biol. 2007, 5, 17. [Google Scholar] [CrossRef] [Green Version]
- Ramachandran, G.N.; Ramakrishnan, C.; Sasisekharan, V. Stereochemistry of polypeptide chain configurations. J. Mol. Biol. 1963, 7, 95–99. [Google Scholar] [CrossRef]
- Vermeulen, K.; Van Bockstaele, D.R.; Berneman, Z.N. The cell cycle: A review of regulation, deregulation and therapeutic targets in cancer. Cell Prolif. 2003, 36, 131–149. [Google Scholar] [CrossRef]
- Leber, M.F.; Efferth, T. Molecular principles of cancer invasion and metastasis. Int. J. Oncol. 2009, 34, 881–895. [Google Scholar] [CrossRef] [Green Version]
- Dallakyan, S.; Olson, A.J. Small-molecule library screening by docking with PyRx. Methods Mol. Biol. 2015, 1263, 243–250. [Google Scholar] [CrossRef]
- Long, J.; Zhang, Y.; Yu, X.; Yang, J.; LeBrun, D.G.; Chen, C.; Yao, Q.; Li, M. Overcoming drug resistance in pancreatic cancer. Expert Opin. Ther. Targets 2011, 15, 817–828. [Google Scholar] [CrossRef] [Green Version]
- Wang, C.; Zhang, W.; Fu, M.; Yang, A.; Huang, H.; Xie, J. Establishment of human pancreatic cancer gemcitabineresistant cell line with ribonucleotide reductase overexpression. Oncol. Rep. 2015, 33, 383–390. [Google Scholar] [CrossRef] [Green Version]
- Wong, H.H.; Lemoine, N.R. Pancreatic cancer: Molecular pathogenesis and new therapeutic targets. Nat. Rev. Gastroenterol. Hepatol. 2009, 6, 412–422. [Google Scholar] [CrossRef] [PubMed]
- Lui, V.W.; Yau, D.M.; Wong, E.Y.; Ng, Y.K.; Lau, C.P.; Ho, Y.; Chan, J.P.; Hong, B.; Ho, K.; Cheung, C.S.; et al. Cucurbitacin I elicits anoikis sensitization, inhibits cellular invasion and in vivo tumor formation ability of nasopharyngeal carcinoma cells. Carcinogenesis 2009, 30, 2085–2094. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ishii, T.; Kira, N.; Yoshida, T.; Narahara, H. Cucurbitacin D induces growth inhibition, cell cycle arrest, and apoptosis in human endometrial and ovarian cancer cells. Tumor Biol. 2013, 34, 285–291. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.R.; Seo, H.S.; Choi, H.S.; Cho, S.G.; Kim, Y.K.; Hong, E.H.; Shin, Y.C.; Ko, S.G. Trichosanthes kirilowii Ethanol Extract and Cucurbitacin D Inhibit Cell Growth and Induce Apoptosis through Inhibition of STAT3 Activity in Breast Cancer Cells. Evid. Based Complement. Altern. Med. 2013, 2013, 975350. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Song, Y.; Ding, N.; Kanazawa, T.; Yamashita, U.; Yoshida, Y. Cucurbitacin D is a new inflammasome activator in macrophages. Int. Immunopharmacol. 2013, 17, 1044–1050. [Google Scholar] [CrossRef] [PubMed]
- Spear, S.A.; Burns, S.S.; Oblinger, J.L.; Ren, Y.; Pan, L.; Kinghorn, A.D.; Welling, D.B.; Chang, L.S. Natural compounds as potential treatments of NF2-deficient schwannoma and meningioma: Cucurbitacin D and goyazensolide. Otol. Neurotol. 2013, 34, 1519–1527. [Google Scholar] [CrossRef] [Green Version]
- Takahashi, N.; Yoshida, Y.; Sugiura, T.; Matsuno, K.; Fujino, A.; Yamashita, U. Cucurbitacin D isolated from Trichosanthes kirilowii induces apoptosis in human hepatocellular carcinoma cells in vitro. Int. Immunopharmacol. 2009, 9, 508–513. [Google Scholar] [CrossRef]
- Zhang, Y.Z.; Wang, C.F.; Zhang, L.F. Cucurbitacin D impedes gastric cancer cell survival via activation of the iNOS/NO and inhibition of the Akt signalling pathway. Oncol. Rep. 2018, 39, 2595–2603. [Google Scholar] [CrossRef]
- Kufe, D.W. Mucins in cancer: Function, prognosis and therapy. Nat. Rev. Cancer 2009, 9, 874–885. [Google Scholar] [CrossRef] [Green Version]
- Rubinstein, D.B.; Karmely, M.; Pichinuk, E.; Ziv, R.; Benhar, I.; Feng, N.; Smorodinsky, N.I.; Wreschner, D.H. The MUC1 oncoprotein as a functional target: Immunotoxin binding to alpha/beta junction mediates cell killing. Int. J. Cancer 2009, 124, 46–54. [Google Scholar] [CrossRef]
- Singh, R.; Bandyopadhyay, D. MUC1: A target molecule for cancer therapy. Cancer Biol. Ther. 2007, 6, 481–486. [Google Scholar] [CrossRef] [PubMed]
- Torres, M.P.; Chakraborty, S.; Souchek, J.; Batra, S.K. Mucin-based targeted pancreatic cancer therapy. Curr. Pharm. Des. 2012, 18, 2472–2481. [Google Scholar] [CrossRef] [PubMed]
- Rachagani, S.; Torres, M.P.; Moniaux, N.; Batra, S.K. Current status of mucins in the diagnosis and therapy of cancer. Biofactors 2009, 35, 509–527. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Macha, M.A.; Rachagani, S.; Gupta, S.; Pai, P.; Ponnusamy, M.P.; Batra, S.K.; Jain, M. Guggulsterone decreases proliferation and metastatic behavior of pancreatic cancer cells by modulating JAK/STAT and Src/FAK signaling. Cancer Lett. 2013, 341, 166–177. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Torres, M.P.; Ponnusamy, M.P.; Chakraborty, S.; Smith, L.M.; Das, S.; Arafat, H.A.; Batra, S.K. Effects of thymoquinone in the expression of mucin 4 in pancreatic cancer cells: Implications for the development of novel cancer therapies. Mol. Cancer Ther. 2010, 9, 1419–1431. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Torres, M.P.; Rachagani, S.; Purohit, V.; Pandey, P.; Joshi, S.; Moore, E.D.; Johansson, S.L.; Singh, P.K.; Ganti, A.K.; Batra, S.K. Graviola: A novel promising natural-derived drug that inhibits tumorigenicity and metastasis of pancreatic cancer cells in vitro and in vivo through altering cell metabolism. Cancer Lett. 2012, 323, 29–40. [Google Scholar] [CrossRef] [PubMed] [Green Version]






© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sikander, M.; Malik, S.; Khan, S.; Kumari, S.; Chauhan, N.; Khan, P.; Halaweish, F.T.; Chauhan, B.; Yallapu, M.M.; Jaggi, M.; et al. Novel Mechanistic Insight into the Anticancer Activity of Cucurbitacin D against Pancreatic Cancer (Cuc D Attenuates Pancreatic Cancer). Cells 2020, 9, 103. https://doi.org/10.3390/cells9010103
Sikander M, Malik S, Khan S, Kumari S, Chauhan N, Khan P, Halaweish FT, Chauhan B, Yallapu MM, Jaggi M, et al. Novel Mechanistic Insight into the Anticancer Activity of Cucurbitacin D against Pancreatic Cancer (Cuc D Attenuates Pancreatic Cancer). Cells. 2020; 9(1):103. https://doi.org/10.3390/cells9010103
Chicago/Turabian StyleSikander, Mohammed, Shabnam Malik, Sheema Khan, Sonam Kumari, Neeraj Chauhan, Parvez Khan, Fathi T. Halaweish, Bhavin Chauhan, Murali M. Yallapu, Meena Jaggi, and et al. 2020. "Novel Mechanistic Insight into the Anticancer Activity of Cucurbitacin D against Pancreatic Cancer (Cuc D Attenuates Pancreatic Cancer)" Cells 9, no. 1: 103. https://doi.org/10.3390/cells9010103
APA StyleSikander, M., Malik, S., Khan, S., Kumari, S., Chauhan, N., Khan, P., Halaweish, F. T., Chauhan, B., Yallapu, M. M., Jaggi, M., & Chauhan, S. C. (2020). Novel Mechanistic Insight into the Anticancer Activity of Cucurbitacin D against Pancreatic Cancer (Cuc D Attenuates Pancreatic Cancer). Cells, 9(1), 103. https://doi.org/10.3390/cells9010103









