pH-Dependent Aggregation in Intrinsically Disordered Proteins Is Determined by Charge and Lipophilicity
Abstract
:1. Introduction
2. Materials and Methods
2.1. Generation of Lipophilicity Profiles
2.2. Solubility Modelling
2.3. Data Analysis and Fitting
3. Results
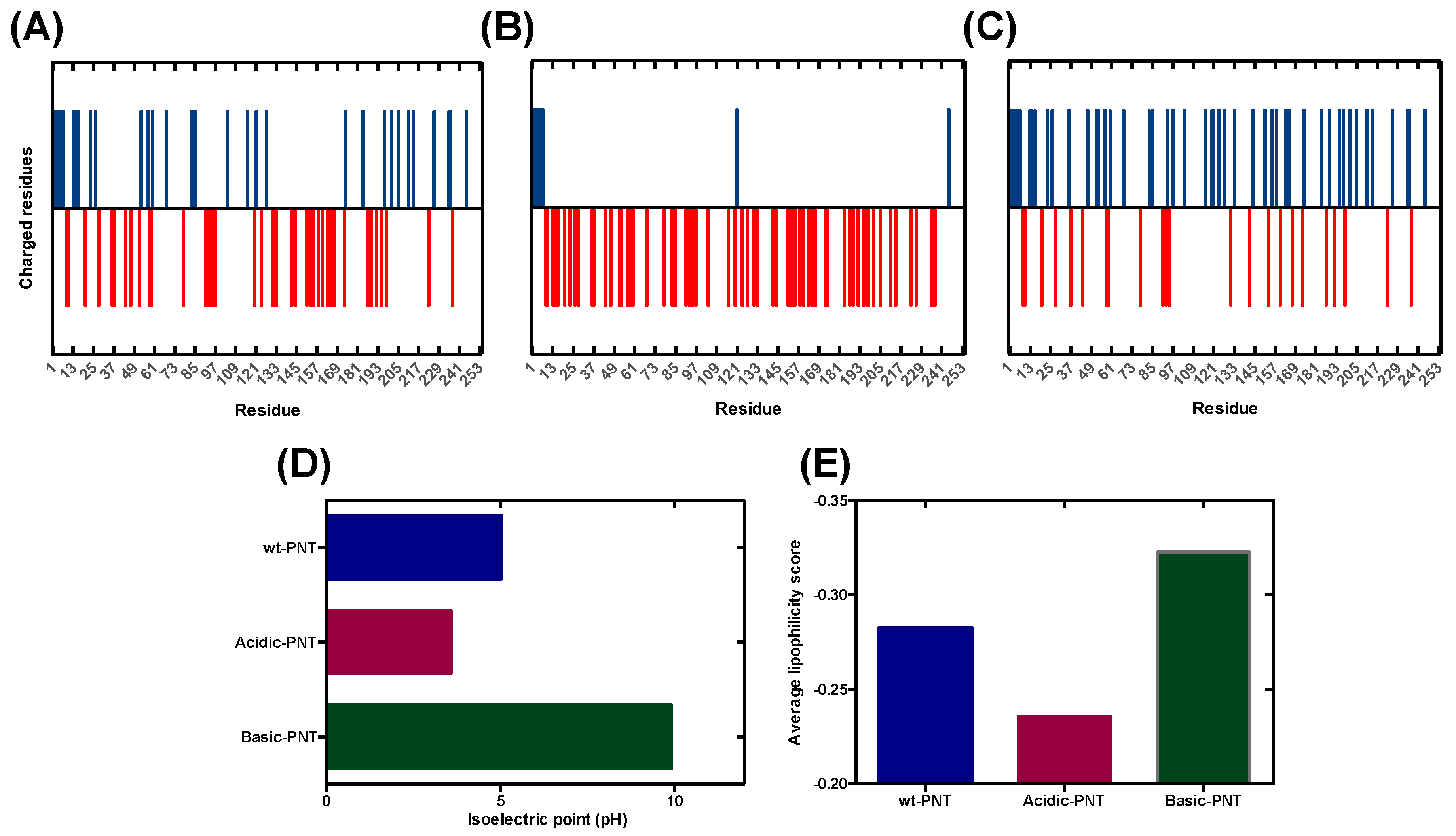
3.1. Rational Analysis of the Molecular Determinants behind pH-Associated Aggregation
3.2. Analysis and Validation of the Lipophilicity Scale as a Proxy for Aggregation Prediction
3.3. Modelling pH-Dependent Solubility usIng Lipophilicity and Net Charge
3.4. pH-Dependent Aggregation Prediction in Disease-Linked Proteins
3.4.1. α-Synuclein (α-S)
3.4.2. Islet Amyloid Polypeptide (IAPP)
3.4.3. Alzheimer’s Disease Related Proteins: Amyloid-Beta Peptides and Tau Protein
3.4.4. Use of a Lipophilicity Term Improves Accuracy in the Prediction of the pH-Dependent Aggregation of Disease-Linked Proteins
3.5. Predicting the Impact of pH on the Aggregation of Functional Amyloids: Context-Dependent Aggregation to Confine Functional Self-Assembly
3.5.1. Pigment Cell-Specific Melanosome Protein
3.5.2. Corticotropin-Releasing Hormone
3.5.3. B Domain of the Bap Protein
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Chiti, F.; Dobson, C.M. Protein misfolding, functional amyloid, and human disease. Annu. Rev. Biochem. 2006, 75, 333–366. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chiti, F.; Dobson, C.M. Protein Misfolding, Amyloid Formation, and Human Disease: A Summary of Progress over the Last Decade. Annu. Rev. Biochem. 2017, 86, 27–68. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.J.; Meyer, J.D.; Carpenter, J.F.; Manning, M.C. Stability of human serum albumin during bioprocessing: Denaturation and aggregation during processing of albumin paste. Pharm. Res. 2000, 17, 391–396. [Google Scholar] [CrossRef] [PubMed]
- Cromwell, M.E.; Hilario, E.; Jacobson, F. Protein aggregation and bioprocessing. AAPS J. 2006, 8, 572–579. [Google Scholar] [CrossRef] [Green Version]
- Loquet, A.; Saupe, S.J.; Romero, D. Functional Amyloids in Health and Disease. J. Mol. Biol. 2018, 430, 3629–3630. [Google Scholar] [CrossRef]
- Camara-Almiron, J.; Caro-Astorga, J.; de Vicente, A.; Romero, D. Beyond the expected: The structural and functional diversity of bacterial amyloids. Crit. Rev. Microbiol. 2018, 44, 653–666. [Google Scholar] [CrossRef]
- McGlinchey, R.P.; Lee, J.C. Why Study Functional Amyloids? Lessons from the Repeat Domain of Pmel17. J. Mol. Biol. 2018, 430, 3696–3706. [Google Scholar] [CrossRef]
- Diaz-Caballero, M.; Navarro, S.; Fuentes, I.; Teixidor, F.; Ventura, S. Minimalist Prion-Inspired Polar Self-Assembling Peptides. ACS Nano 2018, 12, 5394–5407. [Google Scholar] [CrossRef]
- Diaz-Caballero, M.; Fernandez, M.R.; Navarro, S.; Ventura, S. Prion-based nanomaterials and their emerging applications. Prion 2018, 12, 266–272. [Google Scholar] [CrossRef]
- Knowles, T.P.; Mezzenga, R. Amyloid Fibrils as Building Blocks for Natural and Artificial Functional Materials. Adv. Mater. 2016, 28, 6546–6561. [Google Scholar] [CrossRef]
- Wei, G.; Su, Z.; Reynolds, N.P.; Arosio, P.; Hamley, I.W.; Gazit, E.; Mezzenga, R. Self-assembling peptide and protein amyloids: From structure to tailored function in nanotechnology. Chem. Soc. Rev. 2017, 46, 4661–4708. [Google Scholar] [CrossRef] [PubMed]
- Pallares, I.; Ventura, S. Advances in the prediction of protein aggregation propensity. Curr. Med. Chem. 2017, 26, 3911–3920. [Google Scholar] [CrossRef] [PubMed]
- Graña-Montes, R.; Pujol, J.P.; Gómez-Picanyol, C.; Ventura, S. Prediction of Protein Aggregation and Amyloid Formation. In From Protein Structure to Function with Bioinformatics; Rigden, D.J., Ed.; Springer: Berlin/Heidelberg, Germany, 2017; pp. 205–263. [Google Scholar] [CrossRef]
- Conchillo-Sole, O.; de Groot, N.S.; Aviles, F.X.; Vendrell, J.; Daura, X.; Ventura, S. AGGRESCAN: A server for the prediction and evaluation of “hot spots” of aggregation in polypeptides. BMC Bioinform. 2007, 8, 65. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sanchez de Groot, N.; Pallares, I.; Aviles, F.X.; Vendrell, J.; Ventura, S. Prediction of “hot spots” of aggregation in disease-linked polypeptides. BMC Struct. Biol. 2005, 5, 18. [Google Scholar] [CrossRef] [Green Version]
- Garbuzynskiy, S.O.; Lobanov, M.Y.; Galzitskaya, O.V. FoldAmyloid: A method of prediction of amyloidogenic regions from protein sequence. Bioinformatics 2010, 26, 326–332. [Google Scholar] [CrossRef]
- Tsolis, A.C.; Papandreou, N.C.; Iconomidou, V.A.; Hamodrakas, S.J. A consensus method for the prediction of ‘aggregation-prone’ peptides in globular proteins. PLoS ONE 2013, 8, e54175. [Google Scholar] [CrossRef] [Green Version]
- O’Donnell, C.W.; Waldispuhl, J.; Lis, M.; Halfmann, R.; Devadas, S.; Lindquist, S.; Berger, B. A method for probing the mutational landscape of amyloid structure. Bioinformatics 2011, 27, 34–42. [Google Scholar] [CrossRef] [Green Version]
- Walsh, I.; Seno, F.; Tosatto, S.C.; Trovato, A. PASTA 2.0: An improved server for protein aggregation prediction. Nucleic Acids Res. 2014, 42, 301–307. [Google Scholar] [CrossRef]
- Fernandez-Escamilla, A.M.; Rousseau, F.; Schymkowitz, J.; Serrano, L. Prediction of sequence-dependent and mutational effects on the aggregation of peptides and proteins. Nat. Biotechnol. 2004, 22, 1302–1306. [Google Scholar] [CrossRef]
- Rousseau, F.; Schymkowitz, J.; Serrano, L. Protein aggregation and amyloidosis: Confusion of the kinds? Curr. Opin. Struct. Biol. 2006, 16, 118–126. [Google Scholar] [CrossRef]
- Maurer-Stroh, S.; Debulpaep, M.; Kuemmerer, N.; Lopez de la Paz, M.; Martins, I.C.; Reumers, J.; Morris, K.L.; Copland, A.; Serpell, L.; Serrano, L.; et al. Exploring the sequence determinants of amyloid structure using position-specific scoring matrices. Nat. Methods 2010, 7, 237–242. [Google Scholar] [CrossRef] [PubMed]
- Tartaglia, G.G.; Pawar, A.P.; Campioni, S.; Dobson, C.M.; Chiti, F.; Vendruscolo, M. Prediction of aggregation-prone regions in structured proteins. J. Mol. Biol. 2008, 380, 425–436. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Singh, S.; Zeng, D.L.; King, K.; Nema, S. Antibody structure, instability, and formulation. J. Pharm. Sci. 2007, 96, 1–26. [Google Scholar] [CrossRef] [PubMed]
- Roberts, C.J. Therapeutic protein aggregation: Mechanisms, design, and control. Trends Biotechnol. 2014, 32, 372–380. [Google Scholar] [CrossRef] [Green Version]
- Jha, R.K.; Leaver-Fay, A.; Yin, S.; Wu, Y.; Butterfoss, G.L.; Szyperski, T.; Dokholyan, N.V.; Kuhlman, B. Computational design of a PAK1 binding protein. J. Mol. Biol. 2010, 400, 257–270. [Google Scholar] [CrossRef] [Green Version]
- Ventura, S.; Zurdo, J.; Narayanan, S.; Parreno, M.; Mangues, R.; Reif, B.; Chiti, F.; Giannoni, E.; Dobson, C.M.; Aviles, F.X.; et al. Short amino acid stretches can mediate amyloid formation in globular proteins: The Src homology 3 (SH3) case. Proc. Natl. Acad. Sci. USA 2004, 101, 7258–7263. [Google Scholar] [CrossRef] [Green Version]
- Riek, R.; Eisenberg, D.S. The activities of amyloids from a structural perspective. Nature 2016, 539, 227–235. [Google Scholar] [CrossRef]
- Jahn, T.R.; Radford, S.E. Folding versus aggregation: Polypeptide conformations on competing pathways. Arch. Biochem. Biophys. 2008, 469, 100–117. [Google Scholar] [CrossRef] [Green Version]
- Castillo, V.; Grana-Montes, R.; Sabate, R.; Ventura, S. Prediction of the aggregation propensity of proteins from the primary sequence: Aggregation properties of proteomes. Biotechnol. J. 2011, 6, 674–685. [Google Scholar] [CrossRef]
- Simm, S.; Einloft, J.; Mirus, O.; Schleiff, E. 50 years of amino acid hydrophobicity scales: Revisiting the capacity for peptide classification. Biol. Res. 2016, 49, 31. [Google Scholar] [CrossRef] [Green Version]
- MacCallum, J.L.; Tieleman, D.P. Hydrophobicity scales: A thermodynamic looking glass into lipid-protein interactions. Trends Biochem. Sci. 2011, 36, 653–662. [Google Scholar] [CrossRef] [PubMed]
- Tedeschi, G.; Mangiagalli, M.; Chmielewska, S.; Lotti, M.; Natalello, A.; Brocca, S. Aggregation properties of a disordered protein are tunable by pH and depend on its net charge per residue. Biochim. Biophys. Acta Gen. Subj. 2017, 1861, 2543–2550. [Google Scholar] [CrossRef] [PubMed]
- Shaw, K.L.; Grimsley, G.R.; Yakovlev, G.I.; Makarov, A.A.; Pace, C.N. The effect of net charge on the solubility, activity, and stability of ribonuclease Sa. Protein Sci. 2001, 10, 1206–1215. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zamora, W.J.; Campanera, J.M.; Luque, F.J. Development of a Structure-Based, pH-Dependent Lipophilicity Scale of Amino Acids from Continuum Solvation Calculations. J. Phys. Chem. Lett. 2019, 10, 883–889. [Google Scholar] [CrossRef] [PubMed]
- Putnam, C. Protein Calculator. Available online: http://protcalc.sourceforge.net/ (accessed on 19 June 2019).
- Morris, A.M.; Watzky, M.A.; Finke, R.G. Protein aggregation kinetics, mechanism, and curve-fitting: A review of the literature. Biochim. Biophys. Acta 2009, 1794, 375–397. [Google Scholar] [CrossRef] [PubMed]
- Uversky, V.N.; Li, J.; Fink, A.L. Evidence for a partially folded intermediate in alpha-synuclein fibril formation. J. Biol. Chem. 2001, 276, 10737–10744. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jha, S.; Snell, J.M.; Sheftic, S.R.; Patil, S.M.; Daniels, S.B.; Kolling, F.W.; Alexandrescu, A.T. pH dependence of amylin fibrillization. Biochemistry 2014, 53, 300–310. [Google Scholar] [CrossRef]
- Jeganathan, S.; von Bergen, M.; Mandelkow, E.M.; Mandelkow, E. The natively unfolded character of tau and its aggregation to Alzheimer-like paired helical filaments. Biochemistry 2008, 47, 10526–10539. [Google Scholar] [CrossRef]
- Hortschansky, P.; Schroeckh, V.; Christopeit, T.; Zandomeneghi, G.; Fandrich, M. The aggregation kinetics of Alzheimer’s beta-amyloid peptide is controlled by stochastic nucleation. Protein Sci. 2005, 14, 1753–1759. [Google Scholar] [CrossRef] [Green Version]
- Pfefferkorn, C.M.; McGlinchey, R.P.; Lee, J.C. Effects of pH on aggregation kinetics of the repeat domain of a functional amyloid, Pmel17. Proc. Natl. Acad. Sci. USA 2010, 107, 21447–21452. [Google Scholar] [CrossRef] [Green Version]
- Maji, S.K.; Perrin, M.H.; Sawaya, M.R.; Jessberger, S.; Vadodaria, K.; Rissman, R.A.; Singru, P.S.; Nilsson, K.P.; Simon, R.; Schubert, D.; et al. Functional amyloids as natural storage of peptide hormones in pituitary secretory granules. Science 2009, 325, 328–332. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Taglialegna, A.; Navarro, S.; Ventura, S.; Garnett, J.A.; Matthews, S.; Penades, J.R.; Lasa, I.; Valle, J. Staphylococcal Bap Proteins Build Amyloid Scaffold Biofilm Matrices in Response to Environmental Signals. PLoS Pathog. 2016, 12, e1005711. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Soper, D.S. p-Value Calculator for Correlation Coefficients. Available online: http://www.danielsoper.com/statcalc (accessed on 20 June 2018).
- De Groot, N.S.; Aviles, F.X.; Vendrell, J.; Ventura, S. Mutagenesis of the central hydrophobic cluster in Abeta42 Alzheimer’s peptide. Side-chain properties correlate with aggregation propensities. FEBS J. 2006, 273, 658–668. [Google Scholar] [CrossRef] [PubMed]
- Fink, A.L. Protein aggregation: Folding aggregates, inclusion bodies and amyloid. Fold. Des. 1998, 3, 9–23. [Google Scholar] [CrossRef] [Green Version]
- Belli, M.; Ramazzotti, M.; Chiti, F. Prediction of amyloid aggregation in vivo. EMBO Rep. 2011, 12, 657–663. [Google Scholar] [CrossRef] [Green Version]
- Emamzadeh, F.N. Alpha-synuclein structure, functions, and interactions. J. Res. Med. Sci. 2016, 21, 29. [Google Scholar] [CrossRef]
- Goedert, M.; Spillantini, M.G.; Del Tredici, K.; Braak, H. 100 years of Lewy pathology. Nat. Rev. Neurol. 2013, 9, 13–24. [Google Scholar] [CrossRef]
- Spillantini, M.G.; Schmidt, M.L.; Lee, V.M.; Trojanowski, J.Q.; Jakes, R.; Goedert, M. Alpha-synuclein in Lewy bodies. Nature 1997, 388, 839–840. [Google Scholar] [CrossRef]
- Villar-Pique, A.; Lopes da Fonseca, T.; Outeiro, T.F. Structure, function and toxicity of alpha-synuclein: The Bermuda triangle in synucleinopathies. J. Neurochem. 2016, 139, 240–255. [Google Scholar] [CrossRef]
- Lashuel, H.A.; Overk, C.R.; Oueslati, A.; Masliah, E. The many faces of alpha-synuclein: From structure and toxicity to therapeutic target. Nat. Rev. Neurosci. 2013, 14, 38–48. [Google Scholar] [CrossRef] [Green Version]
- Lassen, L.B.; Reimer, L.; Ferreira, N.; Betzer, C.; Jensen, P.H. Protein Partners of alpha-Synuclein in Health and Disease. Brain Pathol. 2016, 26, 389–397. [Google Scholar] [CrossRef] [PubMed]
- Westermark, G.T.; Westermark, P.; Berne, C.; Korsgren, O.; Nordic Network for Clinical Islet Transplantation. Widespread amyloid deposition in transplanted human pancreatic islets. N Engl. J. Med. 2008, 359, 977–979. [Google Scholar] [CrossRef] [PubMed]
- Denroche, H.C.; Verchere, C.B. IAPP and type 1 diabetes: Implications for immunity, metabolism and islet transplants. J. Mol. Endocrinol. 2018, 60, 57–75. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mukherjee, A.; Morales-Scheihing, D.; Salvadores, N.; Moreno-Gonzalez, I.; Gonzalez, C.; Taylor-Presse, K.; Mendez, N.; Shahnawaz, M.; Gaber, A.O.; Sabek, O.M.; et al. Induction of IAPP amyloid deposition and associated diabetic abnormalities by a prion-like mechanism. J. Exp. Med. 2017, 214, 2591–2610. [Google Scholar] [CrossRef]
- Khemtemourian, L.; Domenech, E.; Doux, J.P.; Koorengevel, M.C.; Killian, J.A. Low pH acts as inhibitor of membrane damage induced by human islet amyloid polypeptide. J. Am. Chem. Soc. 2011, 133, 15598–15604. [Google Scholar] [CrossRef]
- Akter, R.; Cao, P.; Noor, H.; Ridgway, Z.; Tu, L.H.; Wang, H.; Wong, A.G.; Zhang, X.; Abedini, A.; Schmidt, A.M.; et al. Islet Amyloid Polypeptide: Structure, Function, and Pathophysiology. J. Diabetes Res. 2016, 2016, 2798269. [Google Scholar] [CrossRef] [Green Version]
- Lane, C.A.; Hardy, J.; Schott, J.M. Alzheimer’s disease. Eur. J. Neurol. 2018, 25, 59–70. [Google Scholar] [CrossRef]
- Meng, F.; Bellaiche, M.M.J.; Kim, J.Y.; Zerze, G.H.; Best, R.B.; Chung, H.S. Highly Disordered Amyloid-beta Monomer Probed by Single-Molecule FRET and MD Simulation. Biophys. J. 2018, 114, 870–884. [Google Scholar] [CrossRef] [Green Version]
- Eliezer, D.; Barre, P.; Kobaslija, M.; Chan, D.; Li, X.; Heend, L. Residual structure in the repeat domain of tau: Echoes of microtubule binding and paired helical filament formation. Biochemistry 2005, 44, 1026–1036. [Google Scholar] [CrossRef]
- Schweers, O.; Schonbrunn-Hanebeck, E.; Marx, A.; Mandelkow, E. Structural studies of tau protein and Alzheimer paired helical filaments show no evidence for beta-structure. J. Biol. Chem. 1994, 269, 24290–24297. [Google Scholar]
- Siddiqua, A.; Margittai, M. Three-and four-repeat Tau coassemble into heterogeneous filaments: An implication for Alzheimer disease. J. Biol. Chem. 2010, 285, 37920–37926. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dinkel, P.D.; Siddiqua, A.; Huynh, H.; Shah, M.; Margittai, M. Variations in filament conformation dictate seeding barrier between three-and four-repeat tau. Biochemistry 2011, 50, 4330–4336. [Google Scholar] [CrossRef] [PubMed]
- Pham, C.L.; Kwan, A.H.; Sunde, M. Functional amyloid: Widespread in Nature, diverse in purpose. Essays Biochem. 2014, 56, 207–219. [Google Scholar] [CrossRef] [PubMed]
- Otzen, D. Functional amyloid: Turning swords into plowshares. Prion 2010, 4, 256–264. [Google Scholar] [CrossRef] [Green Version]
- Jackson, M.P.; Hewitt, E.W. Why are Functional Amyloids Non-Toxic in Humans? Biomolecules 2017, 7, 71. [Google Scholar] [CrossRef] [Green Version]
- Franzmann, T.M.; Jahnel, M.; Pozniakovsky, A.; Mahamid, J.; Holehouse, A.S.; Nuske, E.; Richter, D.; Baumeister, W.; Grill, S.W.; Pappu, R.V.; et al. Phase separation of a yeast prion protein promotes cellular fitness. Science 2018, 359, 5654. [Google Scholar] [CrossRef] [Green Version]
- Zambrano, R.; Jamroz, M.; Szczasiuk, A.; Pujols, J.; Kmiecik, S.; Ventura, S. AGGRESCAN3D (A3D): Server for prediction of aggregation properties of protein structures. Nucleic Acids Res. 2015, 43, 306–313. [Google Scholar] [CrossRef]
- Kuriata, A.; Iglesias, V.; Pujols, J.; Kurcinski, M.; Kmiecik, S.; Ventura, S. Aggrescan3D (A3D) 2.0: Prediction and engineering of protein solubility. Nucleic Acids Res. 2019, 47, 300–307. [Google Scholar] [CrossRef] [Green Version]
- Kuriata, A.; Iglesias, V.; Kurcinski, M.; Ventura, S.; Kmiecik, S. Aggrescan3D standalone package for structure-based prediction of protein aggregation properties. Bioinformatics 2019, 35, 3834–3835. [Google Scholar] [CrossRef]






| Parameter | α | β | γ | δ |
|---|---|---|---|---|
| Values | −97.82 | −0.00747 | 0.8770 | 38.24 |
| Protein | PNTs | α-S (4.67) * | IAPP | Aβ40 | Tau K19 | ||
|---|---|---|---|---|---|---|---|
| Kapp | Tlag | (8.90) * | (5.31) * | (9.68) * | |||
| Charge | R2 | 0.20 | 0.47 | 0.50 | 0.86 | 0.93 | 0.80 |
| p-value | 0.048 | 0.13 | 0.12 | 0.000041 | 0.0019 | 0.000037 | |
| Charge and Lipophilicity | R2 | 0.70 | 0.82 | 0.87 | 0.95 | 0.99 | 0.80 |
| p-value | <0.00001 | 0.013 | 0.0066 | <0.00001 | 0.000039 | 0.000037 | |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Santos, J.; Iglesias, V.; Santos-Suárez, J.; Mangiagalli, M.; Brocca, S.; Pallarès, I.; Ventura, S. pH-Dependent Aggregation in Intrinsically Disordered Proteins Is Determined by Charge and Lipophilicity. Cells 2020, 9, 145. https://doi.org/10.3390/cells9010145
Santos J, Iglesias V, Santos-Suárez J, Mangiagalli M, Brocca S, Pallarès I, Ventura S. pH-Dependent Aggregation in Intrinsically Disordered Proteins Is Determined by Charge and Lipophilicity. Cells. 2020; 9(1):145. https://doi.org/10.3390/cells9010145
Chicago/Turabian StyleSantos, Jaime, Valentín Iglesias, Juan Santos-Suárez, Marco Mangiagalli, Stefania Brocca, Irantzu Pallarès, and Salvador Ventura. 2020. "pH-Dependent Aggregation in Intrinsically Disordered Proteins Is Determined by Charge and Lipophilicity" Cells 9, no. 1: 145. https://doi.org/10.3390/cells9010145
APA StyleSantos, J., Iglesias, V., Santos-Suárez, J., Mangiagalli, M., Brocca, S., Pallarès, I., & Ventura, S. (2020). pH-Dependent Aggregation in Intrinsically Disordered Proteins Is Determined by Charge and Lipophilicity. Cells, 9(1), 145. https://doi.org/10.3390/cells9010145







