Integrated Transcriptome and Proteome Analyses Reveal the Regulatory Role of miR-146a in Human Limbal Epithelium via Notch Signaling
Abstract
1. Introduction
2. Materials and Methods
2.1. Human Corneas
2.2. Primary Limbal Epithelial Cell Isolation and Cell Culture Maintenance
2.3. miRNA Transfection of Primary LECs and HCECs
2.4. Total RNA Isolation
2.5. Quantitative Real-Time RT-PCR (qRT-PCR)
2.6. Library Preparation and RNA Sequencing
2.7. Next-Generation Sequencing Data Analysis
2.8. Protein Extraction and Liquid Chromatography-Mass Spectrometry (LC-MS/MS) Analysis
2.9. Proteomics Data Analysis
2.10. Western Blot Analysis
2.11. Immunostaining
2.12. Statistical Analysis
3. Results
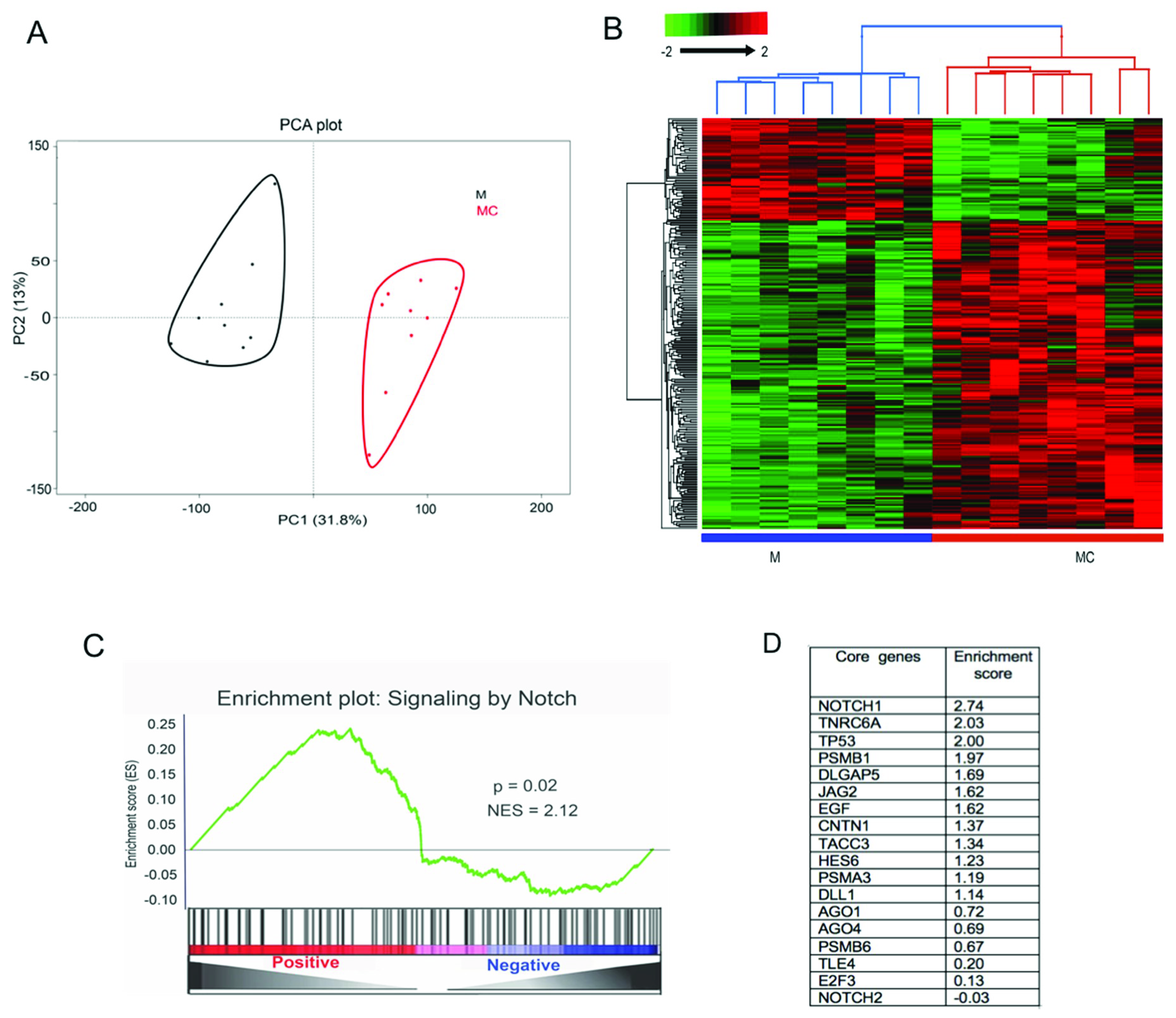
3.1. Transcriptomics Analysis of miR-146a-Transfected LECs
3.2. Proteomics Analysis of miR-146a-Transfected LECs
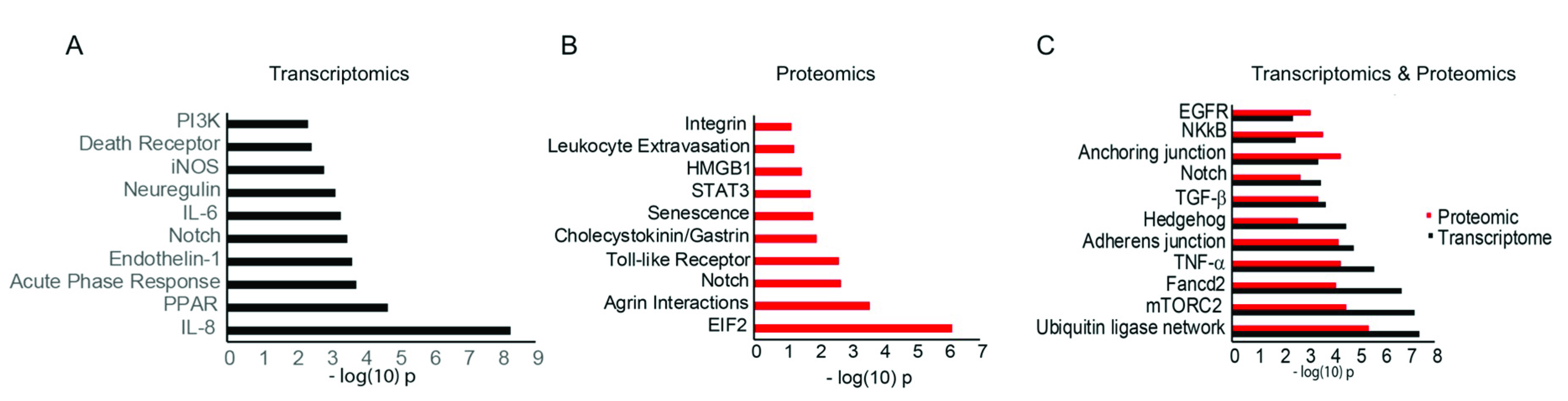
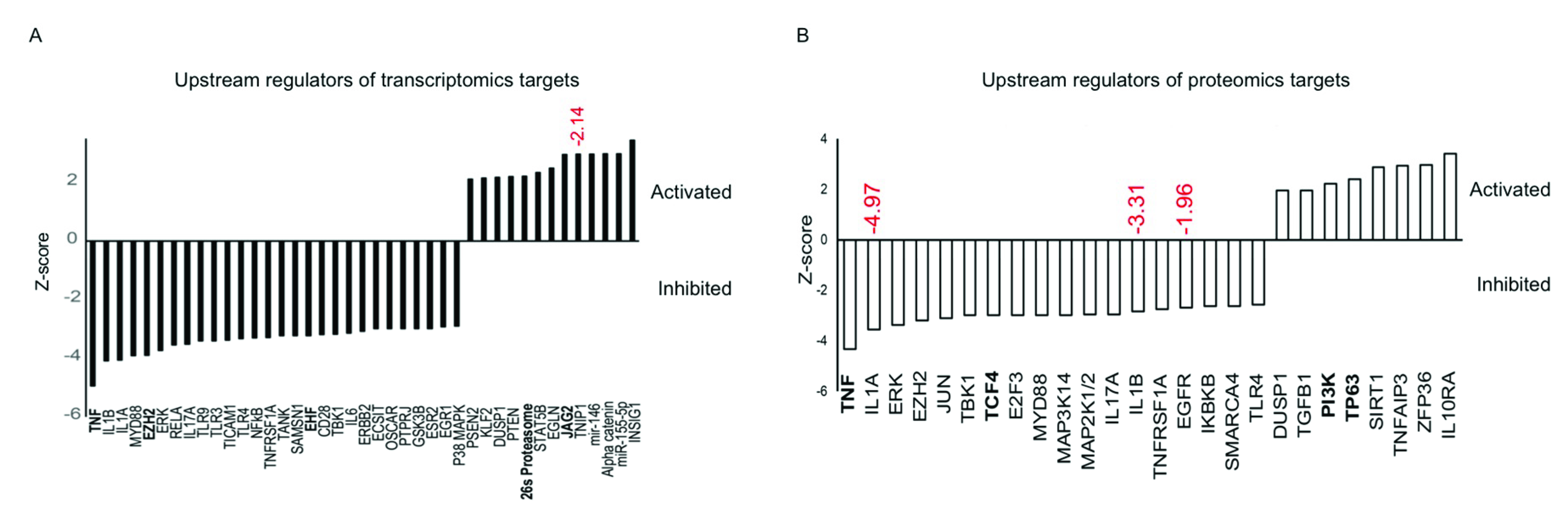
3.3. Functional Analysis of Differentially Expressed mRNAs/Proteins
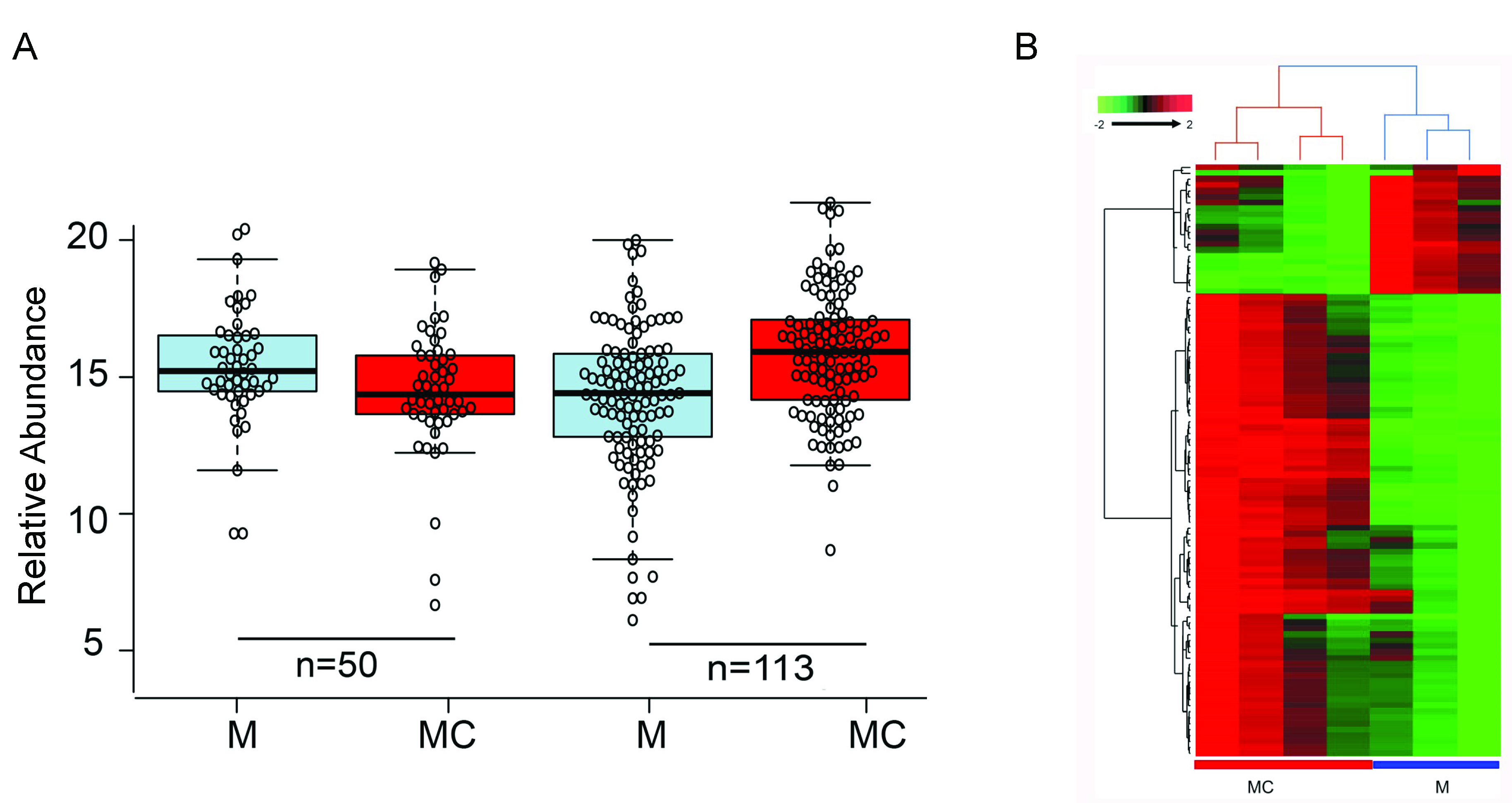
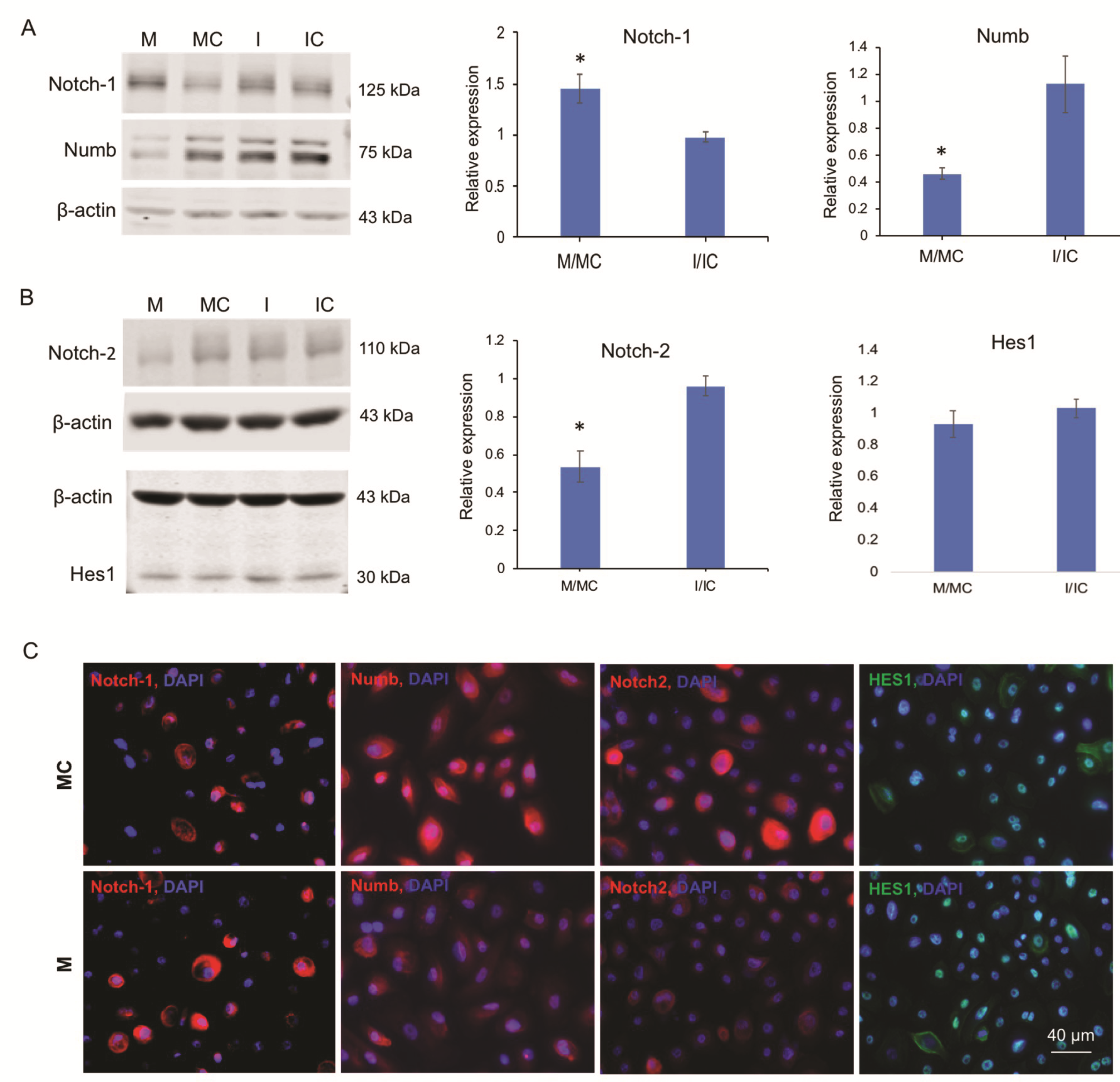
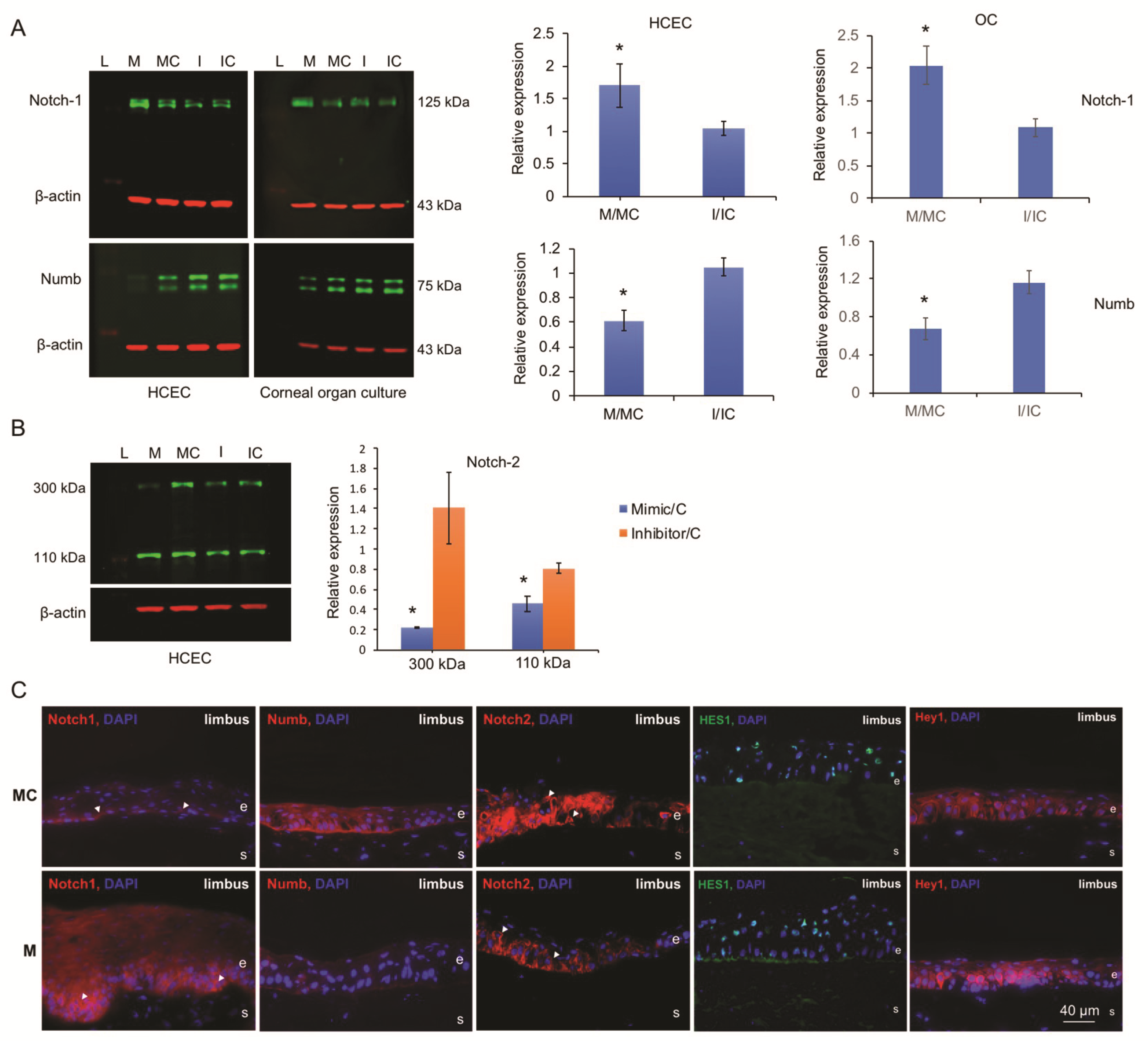
3.4. miR-146a Alters Notch Signaling in Human Primary LECs, HCECs, and Organ-Cultured Corneas
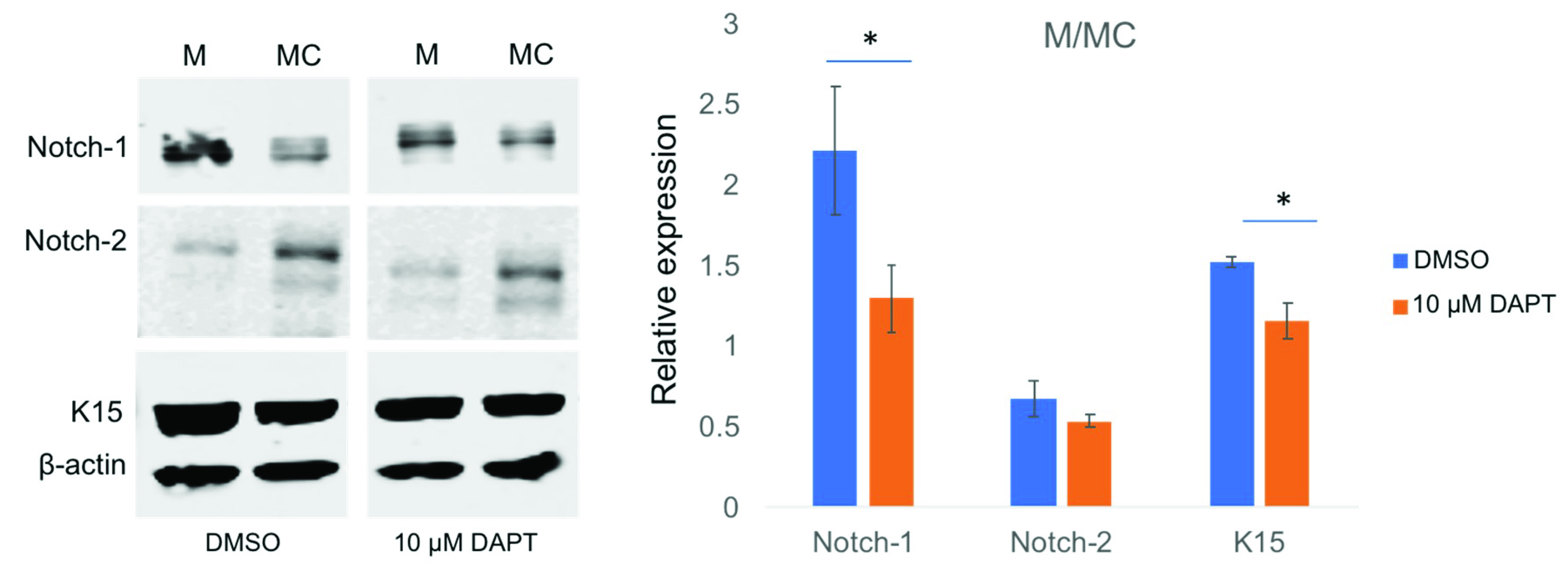
3.5. miR-146a in Limbal Epithelial Stem Cell Maintenance
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Rama, P.; Matuska, S.; Paganoni, G.; Spinelli, A.; De Luca, M.; Pellegrini, G. Limbal stem-cell therapy and long-term corneal regeneration. N. Engl. J. Med. 2010, 147–155. [Google Scholar] [CrossRef]
- Ordonez, P.; Di Girolamo, N. Limbal epithelial stem cells: Role of the niche microenvironment. Stem Cells 2012, 30, 100–107. [Google Scholar] [CrossRef]
- Lehrer, M.S.; Sun, T.T.; Lavker, R.M. Strategies of epithelial repair: Modulation of stem cell and transit amplifying cell proliferation. J. Cell Sci. 1998, 111, 2867–2875. [Google Scholar] [PubMed]
- Ljubimov, A.V.; Saghizadeh, M. Progress in corneal wound healing. Prog. Retin. Eye Res. 2015, 49, 17–45. [Google Scholar] [CrossRef] [PubMed]
- Decembrini, S.; Bressan, D.; Vignali, R.; Pitto, L.; Mariotti, S.; Rainaldi, G.; Wang, X.; Evangelista, M.; Barsacchi, G.; Cremisi, F. MicroRNAs couple cell fate and developmental timing in retina. Proc. Natl. Acad. Sci. USA 2009, 106, 21179–21184. [Google Scholar] [CrossRef] [PubMed]
- Park, J.K.; Yang, W.; Katsnelson, J.; Lavker, R.M.; Peng, H. MicroRNAs Enhance Keratinocyte Proliferative Capacity in a Stem Cell-Enriched Epithelium. PLoS ONE 2015, 10, e0134853. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Humphreys, D.T.; Westman, B.J.; Martin, D.I.; Preiss, T. MicroRNAs control translation initiation by inhibiting eukaryotic initiation factor 4E/cap and poly(A) tail function. Proc. Natl. Acad. Sci. USA 2005, 102, 16961–16966. [Google Scholar] [CrossRef]
- Wu, L.; Fan, J.; Belasco, J.G. MicroRNAs direct rapid deadenylation of mRNA. Proc. Natl. Acad. Sci. USA 2006, 103, 4034–4039. [Google Scholar] [CrossRef]
- Funari, V.A.; Winkler, M.; Brown, J.; Dimitrijevich, S.D.; Ljubimov, A.V.; Saghizadeh, M. Differentially expressed wound healing-related microRNAs in the human diabetic cornea. PLoS ONE 2013, 8, e84425. [Google Scholar] [CrossRef]
- Winkler, M.A.; Dib, C.; Ljubimov, A.V.; Saghizadeh, M. Targeting miR-146a to treat delayed wound healing in human diabetic organ-cultured corneas. PLoS ONE 2014, 9, e114692. [Google Scholar] [CrossRef]
- Starczynowski, D.T.; Kuchenbauer, F.; Wegrzyn, J.; Rouhi, A.; Petriv, O.; Hansen, C.L.; Humphries, R.K.; Karsan, A. MicroRNA-146a disrupts hematopoietic differentiation and survival. Exp. Hematol. 2011, 39, 167–178. [Google Scholar] [CrossRef] [PubMed]
- Taganov, K.D.; Boldin, M.P.; Chang, K.J.; Baltimore, D. NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc. Natl. Acad. Sci. USA 2006, 103, 12481–12486. [Google Scholar] [CrossRef] [PubMed]
- Thomas, P.B.; Liu, Y.H.; Zhuang, F.F.; Selvam, S.; Song, S.W.; Smith, R.E.; Trousdale, M.D.; Yiu, S.C. Identification of Notch-1 expression in the limbal basal epithelium. Mol. Vis. 2007, 13, 337–344. [Google Scholar] [PubMed]
- Ma, A.; Boulton, M.; Zhao, B.; Connon, C.; Cai, J.; Albon, J. A role for notch signaling in human corneal epithelial cell differentiation and proliferation. Investig. Ophthalmol. Vis. Sci. 2007, 48, 3576–3585. [Google Scholar] [CrossRef]
- Nakamura, T.; Ohtsuka, T.; Sekiyama, E.; Cooper, L.J.; Kokubu, H.; Fullwood, N.J.; Barrandon, Y.; Kageyama, R.; Kinoshita, S. Hes1 regulates corneal development and the function of corneal epithelial stem/progenitor cells. Stem Cells 2008, 26, 1265–1274. [Google Scholar] [CrossRef]
- Djalilian, A.R.; Namavari, A.; Ito, A.; Balali, S.; Afshar, A.; Lavker, R.M.; Yue, B.Y. Down-regulation of Notch signaling during corneal epithelial proliferation. Mol. Vis. 2008, 14, 1041–1049. [Google Scholar]
- Ma, A.; Zhao, B.; Boulton, M.; Albon, J. A role for Notch signaling in corneal wound healing. Wound Repair Regen. 2011, 19, 98–106. [Google Scholar] [CrossRef]
- Movahedan, A.; Majdi, M.; Afsharkhamseh, N.; Sagha, H.M.; Saadat, N.S.; Shalileh, K.; Milani, B.Y.; Ying, H.; Djalilian, A.R. Notch inhibition during corneal epithelial wound healing promotes migration. Investig. Ophthalmol. Vis. Sci. 2012, 53, 7476–7483. [Google Scholar] [CrossRef]
- Grisanti, L.; Revenkova, E.; Gordon, R.E.; Iomini, C. Primary cilia maintain corneal epithelial homeostasis by regulation of the Notch signaling pathway. Development 2016, 143, 2160–2171. [Google Scholar] [CrossRef]
- Lu, H.; Lu, Q.; Zheng, Y.; Li, Q. Notch signaling promotes the corneal epithelium wound healing. Mol. Vis. 2012, 18, 403–411. [Google Scholar]
- Saghizadeh, M.; Winkler, M.A.; Kramerov, A.A.; Hemmati, D.M.; Ghiam, C.A.; Dimitrijevich, S.D.; Sareen, D.; Ornelas, L.; Ghiasi, H.; Brunken, W.J.; et al. A simple alkaline method for decellularizing human amniotic membrane for cell culture. PLoS ONE 2013, 8, e79632. [Google Scholar] [CrossRef]
- Sareen, D.; Saghizadeh, M.; Ornelas, L.; Winkler, M.A.; Narwani, K.; Sahabian, A.; Funari, V.A.; Tang, J.; Spurka, L.; Punj, V.; et al. Differentiation of human limbal-derived induced pluripotent stem cells into limbal-like epithelium. Stem Cells Transl. Med. 2014, 3, 1002–1012. [Google Scholar] [CrossRef]
- Kramerov, A.A.; Saghizadeh, M.; Maguen, E.; Rabinowitz, Y.S.; Ljubimov, A.V. Persistence of reduced expression of putative stem cell markers and slow wound healing in cultured diabetic limbal epithelial cells. Mol. Vis. 2015, 21, 1357–1367. [Google Scholar]
- Blazejewska, E.A.; Schlotzer-Schrehardt, U.; Zenkel, M.; Bachmann, B.; Chankiewitz, E.; Jacobi, C.; Kruse, F.E. Corneal limbal microenvironment can induce transdifferentiation of hair follicle stem cells into corneal epithelial-like cells. Stem Cells 2009, 27, 642–652. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 2011, 12, 323. [Google Scholar] [CrossRef]
- Anders, S.; Huber, W. Differential expression analysis for sequence count data. Genome Biol. 2010, 11, R106. [Google Scholar] [CrossRef]
- Kani, K.; Garri, C.; Tiemann, K.; Malihi, P.D.; Punj, V.; Nguyen, A.L.; Lee, J.; Hughes, L.D.; Alvarez, R.M.; Wood, D.M.; et al. JUN-Mediated Downregulation of EGFR Signaling Is Associated with Resistance to Gefitinib in EGFR-mutant NSCLC Cell Lines. Mol. Cancer Ther. 2017, 16, 1645–1657. [Google Scholar] [CrossRef]
- Tongaonkar, P.; Punj, V.; Subramanian, A.; Tran, D.Q.; Trinh, K.K.; Schaal, J.B.; Laragione, T.; Ouellette, A.J.; Gulko, P.S.; Selsted, M.E. RTD-1 therapeutically normalizes synovial gene signatures in rat autoimmune arthritis and suppresses proinflammatory mediators in RA synovial fibroblasts. Physiol. Genom. 2019, 51, 657–667. [Google Scholar] [CrossRef]
- Kim, K.; Punj, V.; Kim, J.M.; Lee, S.; Ulmer, T.S.; Lu, W.; Rice, J.C.; An, W. MMP-9 facilitates selective proteolysis of the histone H3 tail at genes necessary for proficient osteoclastogenesis. Genes Dev. 2016, 30, 208–219. [Google Scholar] [CrossRef]
- Kramer, A.; Green, J.; Pollard, J., Jr.; Tugendreich, S. Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics 2014, 30, 523–530. [Google Scholar] [CrossRef]
- Punj, V.; Matta, H.; Chaudhary, P.M. A computational profiling of changes in gene expression and transcription factors induced by vFLIP K13 in primary effusion lymphoma. PLoS ONE 2012, 7, e37498. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.M.; Shin, Y.; Lee, S.; Kim, M.Y.; Punj, V.; Shin, H.I.; Kim, K.; Koh, J.M.; Jeong, D.; An, W. MacroH2A.2 inhibits prostate cancer-induced osteoclastogenesis through cooperation with HP1alpha and H1.2. Oncogene 2018, 37, 5749–5765. [Google Scholar] [CrossRef]
- Wohlschlegel, J.A. Identification of SUMO-conjugated proteins and their SUMO attachment sites using proteomic mass spectrometry. Methods Mol. Biol. 2009, 497, 33–49. [Google Scholar] [CrossRef]
- Florens, L.; Carozza, M.J.; Swanson, S.K.; Fournier, M.; Coleman, M.K.; Workman, J.L.; Washburn, M.P. Analyzing chromatin remodeling complexes using shotgun proteomics and normalized spectral abundance factors. Methods 2006, 40, 303–311. [Google Scholar] [CrossRef]
- McAlister, G.C.; Nusinow, D.P.; Jedrychowski, M.P.; Wuhr, M.; Huttlin, E.L.; Erickson, B.K.; Rad, R.; Haas, W.; Gygi, S.P. MultiNotch MS3 enables accurate, sensitive, and multiplexed detection of differential expression across cancer cell line proteomes. Anal. Chem. 2014, 86, 7150–7158. [Google Scholar] [CrossRef]
- Choi, M.; Chang, C.Y.; Clough, T.; Broudy, D.; Killeen, T.; MacLean, B.; Vitek, O. MSstats: An R package for statistical analysis of quantitative mass spectrometry-based proteomic experiments. Bioinformatics 2014, 30, 2524–2526. [Google Scholar] [CrossRef]
- Jonas, S.; Izaurralde, E. Towards a molecular understanding of microRNA-mediated gene silencing. Nat. Rev. Genet. 2015, 16, 421–433. [Google Scholar] [CrossRef]
- Gosline, S.J.; Gurtan, A.M.; JnBaptiste, C.K.; Bosson, A.; Milani, P.; Dalin, S.; Matthews, B.J.; Yap, Y.S.; Sharp, P.A.; Fraenkel, E. Elucidating MicroRNA Regulatory Networks Using Transcriptional, Post-transcriptional, and Histone Modification Measurements. Cell Rep. 2016, 14, 310–319. [Google Scholar] [CrossRef]
- Huang, Y.; Liu, Y.; Li, L.; Su, B.; Yang, L.; Fan, W.; Yin, Q.; Chen, L.; Cui, T.; Zhang, J.; et al. Involvement of inflammation-related miR-155 and miR-146a in diabetic nephropathy: Implications for glomerular endothelial injury. BMC Nephrol. 2014, 15, 142. [Google Scholar] [CrossRef]
- Wang, C.; Zhang, W.; Zhang, L.; Chen, X.; Liu, F.; Zhang, J.; Guan, S.; Sun, Y.; Chen, P.; Wang, D.; et al. miR-146a-5p mediates epithelial-mesenchymal transition of oesophageal squamous cell carcinoma via targeting Notch2. Br. J. Cancer 2016, 115, 1548–1554. [Google Scholar] [CrossRef]
- Wang, N.; Chen, F.E.; Long, Z.W. Mechanism of MicroRNA-146a/Notch2 Signaling Regulating IL-6 in Graves Ophthalmopathy. Cell Physiol. Biochem. 2017, 41, 1285–1297. [Google Scholar] [CrossRef] [PubMed]
- Long, J.P.; Dong, L.F.; Chen, F.F.; Fan, Y.F. miR-146a-5p targets interleukin-1 receptor-associated kinase 1 to inhibit the growth, migration, and invasion of breast cancer cells. Oncol. Lett. 2019, 17, 1573–1580. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Jiang, Q.; Jiang, X.Q.; Li, D.Q.; Jiang, X.C.; Wu, X.B.; Cao, Y.L. miR-146a promoted breast cancer proliferation and invasion by regulating NM23-H1. J. Biochem. 2020, 167, 41–48. [Google Scholar] [CrossRef]
- King, J.K.; Ung, N.M.; Paing, M.H.; Contreras, J.R.; Alberti, M.O.; Fernando, T.R.; Zhang, K.; Pellegrini, M.; Rao, D.S. Regulation of Marginal Zone B-Cell Differentiation by MicroRNA-146a. Front. Immunol. 2016, 7, 670. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Luly, F.R.; Leveque, M.; Licursi, V.; Cimino, G.; Martin-Chouly, C.; Theret, N.; Negri, R.; Cavinato, L.; Ascenzioni, F.; Del Porto, P. MiR-146a is over-expressed and controls IL-6 production in cystic fibrosis macrophages. Sci. Rep. 2019, 9, 16259. [Google Scholar] [CrossRef]
- Javidan, A.; Jiang, W.; Okuyama, M.; Thiagarajan, D.; Yang, L.; Moorleghen, J.J.; Muniappan, L.; Subramanian, V. miR-146a Deficiency Accelerates Hepatic Inflammation Without Influencing Diet-induced Obesity in Mice. Sci. Rep. 2019, 9, 12626. [Google Scholar] [CrossRef]
- Ye, E.A.; Steinle, J.J. miR-146a Attenuates Inflammatory Pathways Mediated by TLR4/NF-kappaB and TNFalpha to Protect Primary Human Retinal Microvascular Endothelial Cells Grown in High Glucose. Mediat. Inflamm. 2016, 2016, 3958453. [Google Scholar] [CrossRef]
- Saika, S.; Muragaki, Y.; Okada, Y.; Miyamoto, T.; Ohnishi, Y.; Ooshima, A.; Kao, W.W. Sonic hedgehog expression and role in healing corneal epithelium. Investig. Ophthalmol. Vis. Sci. 2004, 45, 2577–2585. [Google Scholar] [CrossRef]
- Kucerova, R.; Dora, N.; Mort, R.L.; Wallace, K.; Leiper, L.J.; Lowes, C.; Neves, C.; Walczysko, P.; Bruce, F.; Fowler, P.A.; et al. Interaction between hedgehog signalling and PAX6 dosage mediates maintenance and regeneration of the corneal epithelium. Mol. Vis. 2012, 18, 139–150. [Google Scholar]
- Fan, N.W.; Ho, T.C.; Wu, C.W.; Tsao, Y.P. Pigment epithelium-derived factor peptide promotes limbal stem cell proliferation through hedgehog pathway. J. Cell Mol. Med. 2019, 23, 4759–4769. [Google Scholar] [CrossRef] [PubMed]
- Tandon, A.; Tovey, J.C.; Sharma, A.; Gupta, R.; Mohan, R.R. Role of transforming growth factor Beta in corneal function, biology and pathology. Curr. Mol. Med. 2010, 10, 565–578. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, D.; Pinto, F.; Senoo, M. Inhibition of TGF-beta signaling supports high proliferative potential of diverse p63(+) mouse epithelial progenitor cells in vitro. Sci. Rep. 2017, 7, 6089. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Li, D.Q.; Tong, L.; Stewart, P.; Chu, C.; Pflugfelder, S.C. Targeted inhibition of p57 and p15 blocks transforming growth factor beta-inhibited proliferation of primary cultured human limbal epithelial cells. Mol. Vis. 2006, 12, 983–994. [Google Scholar]
- Liu, J.; Sato, C.; Cerletti, M.; Wagers, A. Notch signaling in the regulation of stem cell self-renewal and differentiation. Curr. Top. Dev. Biol. 2010, 92, 367–409. [Google Scholar] [CrossRef]
- Sato, C.; Zhao, G.; Ilagan, M.X. An overview of notch signaling in adult tissue renewal and maintenance. Curr. Alzheimer Res. 2012, 9, 227–240. [Google Scholar] [CrossRef]
- Aguirre, A.; Rubio, M.E.; Gallo, V. Notch and EGFR pathway interaction regulates neural stem cell number and self-renewal. Nature 2010, 467, 323–327. [Google Scholar] [CrossRef]
- Youngstrom, D.W.; Hankenson, K.D. Contextual Regulation of Skeletal Physiology by Notch Signaling. Curr. Osteoporos. Rep. 2019, 17, 217–225. [Google Scholar] [CrossRef]
- Blanco-Obregon, D.; Katz, M.J.; Durrieu, L.; Gandara, L.; Wappner, P. Context-specific functions of Notch in Drosophila blood cell progenitors. Dev. Biol. 2020, 462, 101–115. [Google Scholar] [CrossRef]
- Li, J.; Chen, S.Y.; Zhao, X.Y.; Zhang, M.C.; Xie, H.T. Rat Limbal Niche Cells Prevent Epithelial Stem/Progenitor Cells From Differentiation and Proliferation by Inhibiting Notch Signaling Pathway In Vitro. Investig. Ophthalmol. Vis. Sci. 2017, 58, 2968–2976. [Google Scholar] [CrossRef]
- Gonzalez, S.; Uhm, H.; Deng, S.X. Notch Inhibition Prevents Differentiation of Human Limbal Stem/Progenitor Cells in vitro. Sci. Rep. 2019, 9, 10373. [Google Scholar] [CrossRef] [PubMed]
- Guan, Y.J.; Li, J.; Yang, X.; Du, S.; Ding, J.; Gao, Y.; Zhang, Y.; Yang, K.; Chen, Q. Evidence that miR-146a attenuates aging- and trauma-induced osteoarthritis by inhibiting Notch1, IL-6, and IL-1 mediated catabolism. Aging Cell 2018, 17, e12752. [Google Scholar] [CrossRef] [PubMed]
- Hung, P.S.; Liu, C.J.; Chou, C.S.; Kao, S.Y.; Yang, C.C.; Chang, K.W.; Chiu, T.H.; Lin, S.C. miR-146a enhances the oncogenicity of oral carcinoma by concomitant targeting of the IRAK1, TRAF6 and NUMB genes. PLoS ONE 2013, 8, e79926. [Google Scholar] [CrossRef]
- Forloni, M.; Dogra, S.K.; Dong, Y.; Conte, D.; Jr Ou, J.; Zhu, L.J.; Deng, A.; Mahalingam, M.; Green, M.R.; Wajapeyee, N. miR-146a promotes the initiation and progression of melanoma by activating Notch signaling. Elife 2014, 3, e01460. [Google Scholar] [CrossRef] [PubMed]
- Raimo, M.; Orso, F.; Grassi, E.; Cimino, D.; Penna, E.; De Pitta, C.; Stadler, M.B.; Primo, L.; Calautti, E.; Quaglino, P.; et al. miR-146a Exerts Differential Effects on Melanoma Growth and Metastatization. Mol. Cancer Res. 2016, 14, 548–562. [Google Scholar] [CrossRef] [PubMed]
- Fischer, A.; Schumacher, N.; Maier, M.; Sendtner, M.; Gessler, M. The Notch target genes Hey1 and Hey2 are required for embryonic vascular development. Genes Dev. 2004, 18, 901–911. [Google Scholar] [CrossRef]
- Hayashi, Y.; Osanai, M.; Lee, G.H. NOTCH2 signaling confers immature morphology and aggressiveness in human hepatocellular carcinoma cells. Oncol. Rep. 2015, 34, 1650–1658. [Google Scholar] [CrossRef]
- Kulkarni, M.; Leszczynska, A.; Wei, G.; Winkler, M.A.; Tang, J.; Funari, V.A.; Deng, N.; Liu, Z.; Punj, V.; Deng, S.X.; et al. Genome-wide analysis suggests a differential microRNA signature associated with normal and diabetic human corneal limbus. Sci. Rep. 2017, 7, 3448. [Google Scholar] [CrossRef]
- Liu, K.; Lin, Q.; Wei, Y.; He, R.; Shao, X.; Ding, Z.; Zhang, J.; Zhu, M.; Weinstein, L.S.; Hong, Y.; et al. Galphas regulates asymmetric cell division of cortical progenitors by controlling Numb mediated Notch signaling suppression. Neurosci. Lett. 2015, 597, 97–103. [Google Scholar] [CrossRef]
- Shinin, V.; Gayraud-Morel, B.; Tajbakhsh, S. Template DNA-strand co-segregation and asymmetric cell division in skeletal muscle stem cells. Methods Mol. Biol. 2009, 482, 295–317. [Google Scholar] [CrossRef]
- Movahedan, A.; Afsharkhamseh, N.; Sagha, H.M.; Shah, J.R.; Milani, B.Y.; Milani, F.Y.; Logothetis, H.D.; Chan, C.C.; Djalilian, A.R. Loss of Notch1 disrupts the barrier repair in the corneal epithelium. PLoS ONE 2013, 8, e69113. [Google Scholar] [CrossRef] [PubMed]
- Fan, X.; Mikolaenko, I.; Elhassan, I.; Ni, X.; Wang, Y.; Ball, D.; Brat, D.J.; Perry, A.; Eberhart, C.G. Notch1 and notch2 have opposite effects on embryonal brain tumor growth. Cancer Res. 2004, 64, 7787–7793. [Google Scholar] [CrossRef] [PubMed]
- Muddashetty, R.S.; Nalavadi, V.C.; Gross, C.; Yao, X.; Xing, L.; Laur, O.; Warren, S.T.; Bassell, G.J. Reversible inhibition of PSD-95 mRNA translation by miR-125a, FMRP phosphorylation, and mGluR signaling. Mol. Cell 2011, 42, 673–688. [Google Scholar] [CrossRef]
- Mukherji, S.; Ebert, M.S.; Zheng, G.X.; Tsang, J.S.; Sharp, P.A.; Van Oudenaarden, A. MicroRNAs can generate thresholds in target gene expression. Nat. Genet. 2011, 43, 854–859. [Google Scholar] [CrossRef] [PubMed]
- Subtelny, A.O.; Eichhorn, S.W.; Chen, G.R.; Sive, H.; Bartel, D.P. Poly(A)-tail profiling reveals an embryonic switch in translational control. Nature 2014, 508, 66–71. [Google Scholar] [CrossRef]
- Ghazalpour, A.; Bennett, B.; Petyuk, V.A.; Orozco, L.; Hagopian, R.; Mungrue, I.N.; Farber, C.R.; Sinsheimer, J.; Kang, H.M.; Furlotte, N.; et al. Comparative analysis of proteome and transcriptome variation in mouse. PLoS Genet. 2011, 7, e1001393. [Google Scholar] [CrossRef]
- Pascal, L.E.; True, L.D.; Campbell, D.S.; Deutsch, E.W.; Risk, M.; Coleman, I.M.; Eichner, L.J.; Nelson, P.S.; Liu, A.Y. Correlation of mRNA and protein levels: Cell type-specific gene expression of cluster designation antigens in the prostate. BMC Genom. 2008, 9, 246. [Google Scholar] [CrossRef]
- Haider, S.; Pal, R. Integrated analysis of transcriptomic and proteomic data. Curr. Genom. 2013, 14, 91–110. [Google Scholar] [CrossRef]
- Hernandez-Valladares, M.; Vaudel, M.; Selheim, F.; Berven, F.; Bruserud, O. Proteogenomics approaches for studying cancer biology and their potential in the identification of acute myeloid leukemia biomarkers. Expert Rev. Proteom. 2017, 14, 649–663. [Google Scholar] [CrossRef]
- Park, J.W.; Ko, J.H.; Kim, B.H.; Ryu, J.S.; Kim, H.J.; Kim, M.K.; Oh, J.Y. Inhibition of mTOR by Rapamycin Aggravates Corneal Epithelial Stem Cell Deficiency by Upregulating Inflammatory Response. Stem Cells 2019, 37, 1212–1222. [Google Scholar] [CrossRef]

| Case Number | Age | Gender | Cause of Death | History of Eye Diseases |
|---|---|---|---|---|
| N16-13 | 87 | M | Ruptured abdominal aortic aneurysm | IOL |
| N16-14 | 76 | F | Cardiac pulmonary failure | -- |
| N17-04 | 75 | M | Cardiac Arrest | -- |
| N17-06 | 93 | F | Myocardial infarction | IOL |
| N17-07 | 73 | M | Cardiac arrest | IOL, Cataract |
| N17-10 | 80 | F | Cerebrovascular/Stroke | IOL, Cataract |
| N17-11 | 52 | M | Hypertension | IOL |
| N17-14 | 51 | F | Intracerebral hemorrhage | Cataract |
| N17-24 | 75 | M | Cardiac arrest | -- |
| N18-19 | 73 | M | Multi-system organ failure | -- |
| N18-23 | 57 | F | Cerebrovascular accident | -- |
| N18-36 | 74 | M | Cardiopulmonary arrest | -- |
| N19-04 | 71 | M | Cardiopulmonary arrest | IOL, Cataract |
| N19-13 | 66 | M | Cardiopulmonary arrest | Lasik |
| N19-35 | 35 | M | Potassium chloride overdose | – |
| N20-03 | 64 | F | Cardiac arrest | – |
| N20-05 | 73 | M | Shortness of breath | – |
| Antigen | Antibody | Source | MW (kDa) | Dilution | Application |
|---|---|---|---|---|---|
| Notch-1 | Rabbit mAb ab52627 | Abcam | 125 | 1:500, 1:20, 1:100 | WB, IHC, IF |
| Numb | Rabbit mAb 2761S | Cell Signaling | 72, 74 | 1:500 | WB |
Notch-2 | Goat pAb ab4147 Rabbit mAb 4530S | Abcam Cell Signaling | 110 | 1:20, 1:100 1:500, 1:50, 1:50 | IHC, IF WB, IHC, IF |
| K15 | Mouse mAb ab52816 | Abcam | 45 | 1:5000, 1:100 | WB, IHC |
| Hey1 Hes1 β-Actin | Rabbit pAb ab22614 Mouse mAb ab119776 Rabbit mAb 11988S Mouse mAb A5441 | Abcam Abcam Cell Signaling Sigma | 34 30 42 | 1:50 1:50 1:500 1:2000 | IHC IHC, IF WB WB |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Poe, A.J.; Kulkarni, M.; Leszczynska, A.; Tang, J.; Shah, R.; Jami-Alahmadi, Y.; Wang, J.; Kramerov, A.A.; Wohlschlegel, J.; Punj, V.; et al. Integrated Transcriptome and Proteome Analyses Reveal the Regulatory Role of miR-146a in Human Limbal Epithelium via Notch Signaling. Cells 2020, 9, 2175. https://doi.org/10.3390/cells9102175
Poe AJ, Kulkarni M, Leszczynska A, Tang J, Shah R, Jami-Alahmadi Y, Wang J, Kramerov AA, Wohlschlegel J, Punj V, et al. Integrated Transcriptome and Proteome Analyses Reveal the Regulatory Role of miR-146a in Human Limbal Epithelium via Notch Signaling. Cells. 2020; 9(10):2175. https://doi.org/10.3390/cells9102175
Chicago/Turabian StylePoe, Adam J., Mangesh Kulkarni, Aleksandra Leszczynska, Jie Tang, Ruchi Shah, Yasaman Jami-Alahmadi, Jason Wang, Andrei A. Kramerov, James Wohlschlegel, Vasu Punj, and et al. 2020. "Integrated Transcriptome and Proteome Analyses Reveal the Regulatory Role of miR-146a in Human Limbal Epithelium via Notch Signaling" Cells 9, no. 10: 2175. https://doi.org/10.3390/cells9102175
APA StylePoe, A. J., Kulkarni, M., Leszczynska, A., Tang, J., Shah, R., Jami-Alahmadi, Y., Wang, J., Kramerov, A. A., Wohlschlegel, J., Punj, V., Ljubimov, A. V., & Saghizadeh, M. (2020). Integrated Transcriptome and Proteome Analyses Reveal the Regulatory Role of miR-146a in Human Limbal Epithelium via Notch Signaling. Cells, 9(10), 2175. https://doi.org/10.3390/cells9102175






