The Role of Protein Disorder in Nuclear Transport and in Its Subversion by Viruses
Abstract
:1. Intrinsically Disordered Proteins Have Unique Features
2. Active Transport of Molecules across the Nuclear Envelope
2.1. Classical Nuclear Import
2.2. Classical Nuclear Export
3. Intrinsic Disorder Is an Inherent Feature of the NPC
Advantages of Intrinsic Disorder within the NPC
4. Flexibility and Dynamics Are Inherent Features of Nuclear Transport Receptors
4.1. Importin α/β
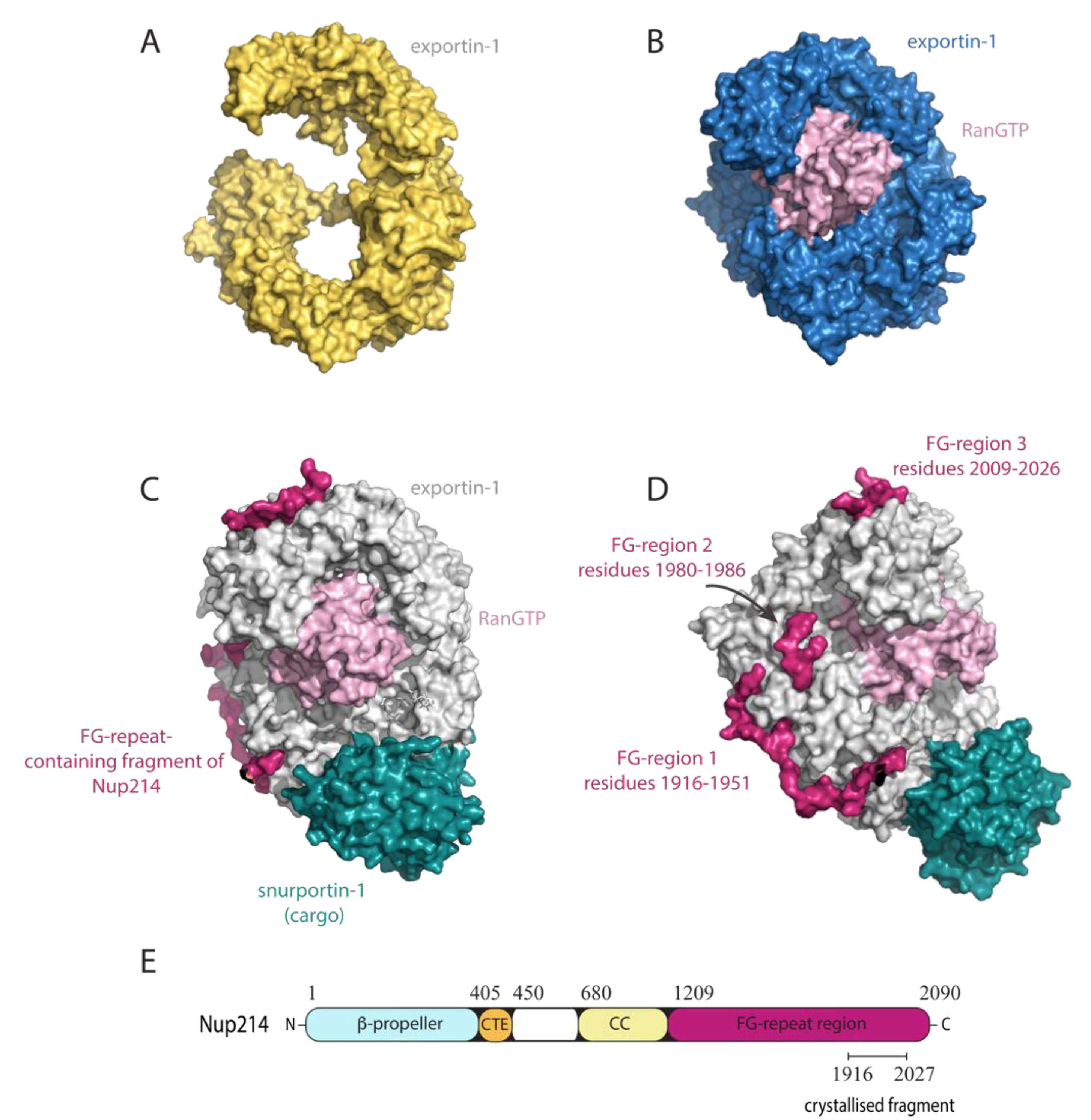
4.2. Exportin-1
5. Nuclear Transport Signals on Cargo Are Frequently Disordered
5.1. Nuclear Localisation Signals (NLSs)
5.2. Nuclear Export Signals (NESs)
5.3. Advantages of Disordered NLS/NES Motifs
6. Intrinsic Disorder Can Regulate the Nuclear Transport of Cargo
6.1. The Integrase Interactor 1 (INI1) Protein
6.2. Inhibitors of κBs
7. Intrinsic Disorder in Viral Proteins
8. Viral Subversion of Host Nuclear Transport Machinery
8.1. FG-Nups Are Targeted by Viruses
8.1.1. Adenovirus Types 2 and 5
8.1.2. Dengue and Zika Virus
8.1.3. Poliovirus and Rhinovirus
8.1.4. Encephalomyocarditis Virus (EMCV)
8.1.5. Hepatitis C Virus (HCV)
8.2. Intrinsic Disorder Can Modulate Viral Cargo NLS/NES Accessibility
8.2.1. Human Immunodeficiency Virus Type 1 (HIV-1) Rev
8.2.2. Hepatitis C Virus (HCV) Core
8.2.3. Hendra Virus P/V/W
8.2.4. Simian Virus 40 (SV-40) Large T-Antigen
9. Therapeutic Targeting of Nuclear Transport to Limit Viral Infection
9.1. Nuclear Import Inhibitors
9.2. Nuclear Export Inhibitors
9.3. Cargo-Specific Transport Inhibitors
10. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Uversky, V.N. Natively unfolded proteins: A point where biology waits for physics. Protein Sci. 2002, 11, 739–756. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Uversky, V.N. What does it mean to be natively unfolded? Eur. J. Biochem. 2002, 269, 2–12. [Google Scholar] [CrossRef] [PubMed]
- Croft, S.N.; Walker, E.J.; Ghildyal, R. Human Rhinovirus 3C protease cleaves RIPK1, concurrent with caspase 8 activation. Sci. Rep. 2018, 8, 1569. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dyson, H.J.; Wright, P.E. Intrinsically unstructured proteins and their functions. Nat. Rev. Mol. Cell Biol. 2005, 6, 197–208. [Google Scholar] [CrossRef]
- Wright, P.E.; Dyson, H.J. Linking folding and binding. Curr. Opin. Struct. Biol. 2009, 19, 31–38. [Google Scholar] [CrossRef] [Green Version]
- Shammas, S.L.; Crabtree, M.D.; Dahal, L.; Wicky, B.I.; Clarke, J. Insights into Coupled Folding and Binding Mechanisms from Kinetic Studies. J. Biol. Chem. 2016, 291, 6689–6695. [Google Scholar] [CrossRef] [Green Version]
- Ittisoponpisan, S.; Alhuzimi, E.; Sternberg, M.J.; David, A. Landscape of Pleiotropic Proteins Causing Human Disease: Structural and System Biology Insights. Hum. Mutat. 2017, 38, 289–296. [Google Scholar] [CrossRef] [Green Version]
- Iakoucheva, L.M.; Radivojac, P.; Brown, C.J.; O’Connor, T.R.; Sikes, J.G.; Obradovic, Z.; Dunker, A.K. The importance of intrinsic disorder for protein phosphorylation. Nucleic Acids Res. 2004, 32, 1037–1049. [Google Scholar] [CrossRef] [Green Version]
- Wright, P.E.; Dyson, H.J. Intrinsically disordered proteins in cellular signalling and regulation. Nat. Rev. Mol. Cell. Biol. 2015, 16, 18–29. [Google Scholar] [CrossRef]
- Timney, B.L.; Raveh, B.; Mironska, R.; Trivedi, J.M.; Kim, S.J.; Russel, D.; Wente, S.R.; Sali, A.; Rout, M.P. Simple rules for passive diffusion through the nuclear pore complex. J. Cell Biol. 2016, 215, 57–76. [Google Scholar] [CrossRef]
- Kimura, F.; Suzu, S.; Nakamura, Y.; Nakata, Y.; Yamada, M.; Kuwada, N.; Matsumura, T.; Yamashita, T.; Ikeda, T.; Sato, K.; et al. Cloning and characterization of a novel RING-B-box-coiled-coil protein with apoptotic function. J. Biol. Chem. 2003, 278, 25046–25054. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tessier, T.M.; MacNeil, K.M.; Mymryk, J.S. Piggybacking on Classical Import and Other Non-Classical Mechanisms of Nuclear Import Appear Highly Prevalent within the Human Proteome. Biology 2020, 9, 188. [Google Scholar] [CrossRef] [PubMed]
- Berneking, L.; Schnapp, M.; Rumm, A.; Trasak, C.; Ruckdeschel, K.; Alawi, M.; Grundhoff, A.; Kikhney, A.G.; Koch-Nolte, F.; Buck, F.; et al. Immunosuppressive Yersinia Effector YopM Binds DEAD Box Helicase DDX3 to Control Ribosomal S6 Kinase in the Nucleus of Host Cells. PLoS Pathog. 2016, 12, e1005660. [Google Scholar] [CrossRef]
- Kimura, M.; Imamoto, N. Biological significance of the importin-beta family-dependent nucleocytoplasmic transport pathways. Traffic 2014, 15, 727–748. [Google Scholar] [CrossRef] [PubMed]
- Goldfarb, D.S.; Corbett, A.H.; Mason, D.A.; Harreman, M.T.; Adam, S.A. Importin alpha: A multipurpose nuclear-transport receptor. Trends Cell Biol. 2004, 14, 505–514. [Google Scholar] [CrossRef]
- Kutay, U.; Bischoff, F.R.; Kostka, S.; Kraft, R.; Gorlich, D. Export of importin alpha from the nucleus is mediated by a specific nuclear transport factor. Cell 1997, 90, 1061–1071. [Google Scholar] [CrossRef] [Green Version]
- Lindsay, M.E.; Holaska, J.M.; Welch, K.; Paschal, B.M.; Macara, I.G. Ran-binding protein 3 is a cofactor for Crm1-mediated nuclear protein export. J. Cell Biol. 2001, 153, 1391–1402. [Google Scholar] [CrossRef] [Green Version]
- Fornerod, M.; Ohno, M.; Yoshida, M.; Mattaj, I.W. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell 1997, 90, 1051–1060. [Google Scholar] [CrossRef] [Green Version]
- Gorlich, D.; Pante, N.; Kutay, U.; Aebi, U.; Bischoff, F.R. Identification of different roles for RanGDP and RanGTP in nuclear protein import. EMBO J. 1996, 15, 5584–5594. [Google Scholar] [CrossRef] [Green Version]
- Smith, A.; Brownawell, A.; Macara, I.G. Nuclear import of Ran is mediated by the transport factor NTF2. Curr. Biol. 1998, 8, 1403–1406. [Google Scholar] [CrossRef] [Green Version]
- Ribbeck, K.; Gorlich, D. Kinetic analysis of translocation through nuclear pore complexes. EMBO J. 2001, 20, 1320–1330. [Google Scholar] [CrossRef] [PubMed]
- Moore, M.S.; Blobel, G. Purification of a Ran-interacting protein that is required for protein import into the nucleus. Proc. Natl. Acad. Sci. USA 1994, 91, 10212–10216. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bischoff, F.R.; Ponstingl, H. Catalysis of guanine nucleotide exchange on Ran by the mitotic regulator RCC1. Nature 1991, 354, 80–82. [Google Scholar] [CrossRef] [PubMed]
- Fried, H.; Kutay, U. Nucleocytoplasmic transport: Taking an inventory. Cell Mol. Life Sci. 2003, 60, 1659–1688. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rout, M.P.; Wente, S.R. Pores for thought: Nuclear pore complex proteins. Trends Cell Biol. 1994, 4, 357–365. [Google Scholar] [CrossRef]
- Denning, D.P.; Patel, S.S.; Uversky, V.; Fink, A.L.; Rexach, M. Disorder in the nuclear pore complex: The FG repeat regions of nucleoporins are natively unfolded. Proc. Natl. Acad. Sci. USA 2003, 100, 2450–2455. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Port, S.A.; Monecke, T.; Dickmanns, A.; Spillner, C.; Hofele, R.; Urlaub, H.; Ficner, R.; Kehlenbach, R.H. Structural and Functional Characterization of CRM1-Nup214 Interactions Reveals Multiple FG-Binding Sites Involved in Nuclear Export. Cell Rep. 2015, 13, 690–702. [Google Scholar] [CrossRef] [Green Version]
- Koyama, M.; Shirai, N.; Matsuura, Y. Structural insights into how Yrb2p accelerates the assembly of the Xpo1p nuclear export complex. Cell Rep. 2014, 9, 983–995. [Google Scholar] [CrossRef] [Green Version]
- Koyama, M.; Hirano, H.; Shirai, N.; Matsuura, Y. Crystal structure of the Xpo1p nuclear export complex bound to the SxFG/PxFG repeats of the nucleoporin Nup42p. Genes Cells 2017, 22, 861–875. [Google Scholar] [CrossRef] [Green Version]
- Bayliss, R.; Kent, H.M.; Corbett, A.H.; Stewart, M. Crystallization and initial X-ray diffraction characterization of complexes of FxFG nucleoporin repeats with nuclear transport factors. J. Struct. Biol. 2000, 131, 240–247. [Google Scholar] [CrossRef]
- Mohr, D.; Frey, S.; Fischer, T.; Guttler, T.; Gorlich, D. Characterisation of the passive permeability barrier of nuclear pore complexes. EMBO J. 2009, 28, 2541–2553. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Keminer, O.; Peters, R. Permeability of single nuclear pores. Biophys. J. 1999, 77, 217–228. [Google Scholar] [CrossRef] [Green Version]
- Frey, S.; Gorlich, D. FG/FxFG as well as GLFG repeats form a selective permeability barrier with self-healing properties. EMBO J. 2009, 28, 2554–2567. [Google Scholar] [CrossRef] [PubMed]
- Frey, S.; Gorlich, D. A saturated FG-repeat hydrogel can reproduce the permeability properties of nuclear pore complexes. Cell 2007, 130, 512–523. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rout, M.P.; Aitchison, J.D.; Magnasco, M.O.; Chait, B.T. Virtual gating and nuclear transport: The hole picture. Trends Cell Biol. 2003, 13, 622–628. [Google Scholar] [CrossRef] [PubMed]
- Lim, R.Y.; Fahrenkrog, B.; Koser, J.; Schwarz-Herion, K.; Deng, J.; Aebi, U. Nanomechanical basis of selective gating by the nuclear pore complex. Science 2007, 318, 640–643. [Google Scholar] [CrossRef]
- Dunker, A.K.; Lawson, J.D.; Brown, C.J.; Williams, R.M.; Romero, P.; Oh, J.S.; Oldfield, C.J.; Campen, A.M.; Ratliff, C.M.; Hipps, K.W.; et al. Intrinsically disordered protein. J. Mol. Graph. Model. 2001, 19, 26–59. [Google Scholar] [CrossRef] [Green Version]
- Dyson, H.J.; Wright, P.E. How Do Intrinsically Disordered Viral Proteins Hijack the Cell? Biochemistry 2018, 57, 4045–4046. [Google Scholar] [CrossRef] [Green Version]
- Dunker, A.K.; Obradovic, Z. The protein trinity--linking function and disorder. Nat. Biotechnol. 2001, 19, 805–806. [Google Scholar] [CrossRef]
- Yang, W.; Gelles, J.; Musser, S.M. Imaging of single-molecule translocation through nuclear pore complexes. Proc. Natl. Acad. Sci. USA 2004, 101, 12887–12892. [Google Scholar] [CrossRef] [Green Version]
- Kubitscheck, U.; Grunwald, D.; Hoekstra, A.; Rohleder, D.; Kues, T.; Siebrasse, J.P.; Peters, R. Nuclear transport of single molecules: Dwell times at the nuclear pore complex. J. Cell Biol. 2005, 168, 233–243. [Google Scholar] [CrossRef] [PubMed]
- Hough, L.E.; Dutta, K.; Sparks, S.; Temel, D.B.; Kamal, A.; Tetenbaum-Novatt, J.; Rout, M.P.; Cowburn, D. The molecular mechanism of nuclear transport revealed by atomic-scale measurements. Elife 2015, 4. [Google Scholar] [CrossRef] [PubMed]
- Raveh, B.; Karp, J.M.; Sparks, S.; Dutta, K.; Rout, M.P.; Sali, A.; Cowburn, D. Slide-and-exchange mechanism for rapid and selective transport through the nuclear pore complex. Proc. Natl. Acad. Sci USA 2016, 113, E2489–E2497. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schneider, R.; Maurin, D.; Communie, G.; Kragelj, J.; Hansen, D.F.; Ruigrok, R.W.; Jensen, M.R.; Blackledge, M. Visualizing the molecular recognition trajectory of an intrinsically disordered protein using multinuclear relaxation dispersion NMR. J. Am. Chem. Soc. 2015, 137, 1220–1229. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kobe, B. Autoinhibition by an internal nuclear localization signal revealed by the crystal structure of mammalian importin alpha. Nat. Struct. Biol. 1999, 6, 388–397. [Google Scholar] [CrossRef]
- Cingolani, G.; Petosa, C.; Weis, K.; Muller, C.W. Structure of importin-beta bound to the IBB domain of importin-alpha. Nature 1999, 399, 221–229. [Google Scholar] [CrossRef]
- Zachariae, U.; Grubmuller, H. Importin-beta: Structural and dynamic determinants of a molecular spring. Structure 2008, 16, 906–915. [Google Scholar] [CrossRef] [Green Version]
- Vetter, I.R.; Arndt, A.; Kutay, U.; Gorlich, D.; Wittinghofer, A. Structural view of the Ran-Importin beta interaction at 2.3 A resolution. Cell 1999, 97, 635–646. [Google Scholar] [CrossRef] [Green Version]
- Liu, S.M.; Stewart, M. Structural basis for the high-affinity binding of nucleoporin Nup1p to the Saccharomyces cerevisiae importin-beta homologue, Kap95p. J. Mol. Biol. 2005, 349, 515–525. [Google Scholar] [CrossRef]
- Lee, S.J.; Imamoto, N.; Sakai, H.; Nakagawa, A.; Kose, S.; Koike, M.; Yamamoto, M.; Kumasaka, T.; Yoneda, Y.; Tsukihara, T. The adoption of a twisted structure of importin-beta is essential for the protein-protein interaction required for nuclear transport. J. Mol. Biol. 2000, 302, 251–264. [Google Scholar] [CrossRef]
- Bhardwaj, A.; Cingolani, G. Conformational selection in the recognition of the snurportin importin beta binding domain by importin beta. Biochemistry 2010, 49, 5042–5047. [Google Scholar] [CrossRef] [PubMed]
- Forwood, J.K.; Lange, A.; Zachariae, U.; Marfori, M.; Preast, C.; Grubmuller, H.; Stewart, M.; Corbett, A.H.; Kobe, B. Quantitative structural analysis of importin-beta flexibility: Paradigm for solenoid protein structures. Structure 2010, 18, 1171–1183. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yoshimura, S.H.; Kumeta, M.; Takeyasu, K. Structural mechanism of nuclear transport mediated by importin beta and flexible amphiphilic proteins. Structure 2014, 22, 1699–1710. [Google Scholar] [CrossRef] [Green Version]
- Monecke, T.; Haselbach, D.; Voss, B.; Russek, A.; Neumann, P.; Thomson, E.; Hurt, E.; Zachariae, U.; Stark, H.; Grubmuller, H.; et al. Structural basis for cooperativity of CRM1 export complex formation. Proc. Natl. Acad. Sci. USA 2013, 110, 960–965. [Google Scholar] [CrossRef] [Green Version]
- Dolker, N.; Blanchet, C.E.; Voss, B.; Haselbach, D.; Kappel, C.; Monecke, T.; Svergun, D.I.; Stark, H.; Ficner, R.; Zachariae, U.; et al. Structural determinants and mechanism of mammalian CRM1 allostery. Structure 2013, 21, 1350–1360. [Google Scholar] [CrossRef] [Green Version]
- Koyama, M.; Matsuura, Y. An allosteric mechanism to displace nuclear export cargo from CRM1 and RanGTP by RanBP1. EMBO J. 2010, 29, 2002–2013. [Google Scholar] [CrossRef] [Green Version]
- Fox, A.M.; Ciziene, D.; McLaughlin, S.H.; Stewart, M. Electrostatic interactions involving the extreme C terminus of nuclear export factor CRM1 modulate its affinity for cargo. J. Biol. Chem. 2011, 286, 29325–29335. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guttler, T.; Madl, T.; Neumann, P.; Deichsel, D.; Corsini, L.; Monecke, T.; Ficner, R.; Sattler, M.; Gorlich, D. NES consensus redefined by structures of PKI-type and Rev-type nuclear export signals bound to CRM1. Nat. Struct. Mol. Biol. 2010, 17, 1367–1376. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lange, A.; Mills, R.E.; Lange, C.J.; Stewart, M.; Devine, S.E.; Corbett, A.H. Classical nuclear localization signals: Definition, function, and interaction with importin alpha. J. Biol. Chem. 2007, 282, 5101–5105. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smith, K.M.; Di Antonio, V.; Bellucci, L.; Thomas, D.R.; Caporuscio, F.; Ciccarese, F.; Ghassabian, H.; Wagstaff, K.M.; Forwood, J.K.; Jans, D.A.; et al. Contribution of the residue at position 4 within classical nuclear localization signals to modulating interaction with importins and nuclear targeting. Biochim. Biophys. Acta Mol. Cell Res. 2018, 1865, 1114–1129. [Google Scholar] [CrossRef]
- Marfori, M.; Lonhienne, T.G.; Forwood, J.K.; Kobe, B. Structural basis of high-affinity nuclear localization signal interactions with importin-alpha. Traffic 2012, 13, 532–548. [Google Scholar] [CrossRef] [PubMed]
- Fontes, M.R.; Teh, T.; Kobe, B. Structural basis of recognition of monopartite and bipartite nuclear localization sequences by mammalian importin-alpha. J. Mol. Biol. 2000, 297, 1183–1194. [Google Scholar] [CrossRef] [PubMed]
- Chelsky, D.; Ralph, R.; Jonak, G. Sequence requirements for synthetic peptide-mediated translocation to the nucleus. Mol. Cell Biol. 1989, 9, 2487–2492. [Google Scholar] [CrossRef] [PubMed]
- Fontes, M.R.; Teh, T.; Toth, G.; John, A.; Pavo, I.; Jans, D.A.; Kobe, B. Role of flanking sequences and phosphorylation in the recognition of the simian-virus-40 large T-antigen nuclear localization sequences by importin-alpha. Biochem. J. 2003, 375, 339–349. [Google Scholar] [CrossRef] [PubMed]
- Conti, E.; Uy, M.; Leighton, L.; Blobel, G.; Kuriyan, J. Crystallographic analysis of the recognition of a nuclear localization signal by the nuclear import factor karyopherin alpha. Cell 1998, 94, 193–204. [Google Scholar] [CrossRef] [Green Version]
- Morita, E.H.; Shirakawa, M.; Hayashi, F.; Imagawa, M.; Kyogoku, Y. Structure of the Oct-3 POU-homeodomain in solution, as determined by triple resonance heteronuclear multidimensional NMR spectroscopy. Protein Sci. 1995, 4, 729–739. [Google Scholar] [CrossRef] [Green Version]
- Galea, C.A.; Wang, Y.; Sivakolundu, S.G.; Kriwacki, R.W. Regulation of cell division by intrinsically unstructured proteins: Intrinsic flexibility, modularity, and signaling conduits. Biochemistry 2008, 47, 7598–7609. [Google Scholar] [CrossRef] [Green Version]
- Cervantes, C.F.; Bergqvist, S.; Kjaergaard, M.; Kroon, G.; Sue, S.C.; Dyson, H.J.; Komives, E.A. The RelA nuclear localization signal folds upon binding to IkappaBalpha. J. Mol. Biol. 2011, 405, 754–764. [Google Scholar] [CrossRef] [Green Version]
- Yamagishi, R.; Okuyama, T.; Oba, S.; Shimada, J.; Chaen, S.; Kaneko, H. Comprehensive analysis of the dynamic structure of nuclear localization signals. Biochem. Biophys. Rep. 2015, 4, 392–396. [Google Scholar] [CrossRef] [Green Version]
- la Cour, T.; Kiemer, L.; Molgaard, A.; Gupta, R.; Skriver, K.; Brunak, S. Analysis and prediction of leucine-rich nuclear export signals. Protein Eng. Des. Sel. 2004, 17, 527–536. [Google Scholar] [CrossRef] [Green Version]
- Dong, X.; Biswas, A.; Suel, K.E.; Jackson, L.K.; Martinez, R.; Gu, H.; Chook, Y.M. Structural basis for leucine-rich nuclear export signal recognition by CRM1. Nature 2009, 458, 1136–1141. [Google Scholar] [CrossRef] [PubMed]
- Fung, H.Y.; Fu, S.C.; Chook, Y.M. Nuclear export receptor CRM1 recognizes diverse conformations in nuclear export signals. Elife 2017, 6. [Google Scholar] [CrossRef] [PubMed]
- Xu, D.; Farmer, A.; Collett, G.; Grishin, N.V.; Chook, Y.M. Sequence and structural analyses of nuclear export signals in the NESdb database. Mol. Biol. Cell 2012, 23, 3677–3693. [Google Scholar] [CrossRef] [PubMed]
- Ward, J.J.; Sodhi, J.S.; McGuffin, L.J.; Buxton, B.F.; Jones, D.T. Prediction and functional analysis of native disorder in proteins from the three kingdoms of life. J. Mol. Biol. 2004, 337, 635–645. [Google Scholar] [CrossRef] [PubMed]
- Xu, D.; Grishin, N.V.; Chook, Y.M. NESdb: A database of NES-containing CRM1 cargoes. Mol. Biol. Cell 2012, 23, 3673–3676. [Google Scholar] [CrossRef] [PubMed]
- Fu, S.C.; Imai, K.; Horton, P. Prediction of leucine-rich nuclear export signal containing proteins with NESsential. Nucleic Acids. Res. 2011, 39, e111. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dellaire, G.; Farrall, R.; Bickmore, W.A. The Nuclear Protein Database (NPD): Sub-nuclear localisation and functional annotation of the nuclear proteome. Nucleic Acids Res. 2003, 31, 328–330. [Google Scholar] [CrossRef] [Green Version]
- Frege, T.; Uversky, V.N. Intrinsically disordered proteins in the nucleus of human cells. Biochem. Biophys. Rep. 2015, 1, 33–51. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.; Perumal, N.B.; Oldfield, C.J.; Su, E.W.; Uversky, V.N.; Dunker, A.K. Intrinsic disorder in transcription factors. Biochemistry 2006, 45, 6873–6888. [Google Scholar] [CrossRef] [Green Version]
- Craig, E.; Zhang, Z.K.; Davies, K.P.; Kalpana, G.V. A masked NES in INI1/hSNF5 mediates hCRM1-dependent nuclear export: Implications for tumorigenesis. EMBO J. 2002, 21, 31–42. [Google Scholar] [CrossRef]
- Romero, P.; Obradovic, Z.; Li, X.; Garner, E.C.; Brown, C.J.; Dunker, A.K. Sequence complexity of disordered protein. Proteins 2001, 42, 38–48. [Google Scholar] [CrossRef]
- He, B.; Wang, K.; Liu, Y.; Xue, B.; Uversky, V.N.; Dunker, A.K. Predicting intrinsic disorder in proteins: An overview. Cell Res. 2009, 19, 929–949. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Das, S.; Cano, J.; Kalpana, G.V. Multimerization and DNA binding properties of INI1/hSNF5 and its functional significance. J. Biol. Chem. 2009, 284, 19903–19914. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Borg, N.A. Ubiquitin Signaling to NF-Kb; Academic Press: Oxford, UK, 2016. [Google Scholar]
- Latimer, M.; Ernst, M.K.; Dunn, L.L.; Drutskaya, M.; Rice, N.R. The N-terminal domain of IkappaB alpha masks the nuclear localization signal(s) of p50 and c-Rel homodimers. Mol. Cell Biol. 1998, 18, 2640–2649. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jacobs, M.D.; Harrison, S.C. Structure of an IkappaBalpha/NF-kappaB complex. Cell 1998, 95, 749–758. [Google Scholar] [CrossRef] [Green Version]
- Muller, C.W.; Rey, F.A.; Sodeoka, M.; Verdine, G.L.; Harrison, S.C. Structure of the NF-kappa B p50 homodimer bound to DNA. Nature 1995, 373, 311–317. [Google Scholar] [CrossRef]
- Huang, D.B.; Huxford, T.; Chen, Y.Q.; Ghosh, G. The role of DNA in the mechanism of NFkappaB dimer formation: Crystal structures of the dimerization domains of the p50 and p65 subunits. Structure 1997, 5, 1427–1436. [Google Scholar] [CrossRef] [Green Version]
- Ghosh, G.; van Duyne, G.; Ghosh, S.; Sigler, P.B. Structure of NF-kappa B p50 homodimer bound to a kappa B site. Nature 1995, 373, 303–310. [Google Scholar] [CrossRef]
- Westerheide, S.D.; Raynes, R.; Powell, C.; Xue, B.; Uversky, V.N. HSF transcription factor family, heat shock response, and protein intrinsic disorder. Curr. Protein Pept. Sci. 2012, 13, 86–103. [Google Scholar] [CrossRef]
- Hatos, A.; Hajdu-Soltesz, B.; Monzon, A.M.; Palopoli, N.; Alvarez, L.; Aykac-Fas, B.; Bassot, C.; Benitez, G.I.; Bevilacqua, M.; Chasapi, A.; et al. DisProt: Intrinsic protein disorder annotation in 2020. Nucleic Acids Res. 2020, 48, D269–D276. [Google Scholar] [CrossRef] [Green Version]
- Goh, G.K.; Dunker, A.K.; Uversky, V.N. Correlating Flavivirus virulence and levels of intrinsic disorder in shell proteins: Protective roles vs. immune evasion. Mol. Biosyst. 2016, 12, 1881–1891. [Google Scholar] [CrossRef] [PubMed]
- Goh, G.K.; Dunker, A.K.; Uversky, V.N. Protein intrinsic disorder and influenza virulence: The 1918 H1N1 and H5N1 viruses. Virol. J. 2009, 6, 69. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goh, G.K.; Dunker, A.K.; Foster, J.A.; Uversky, V.N. Shell Disorder Analysis Suggests That Pangolins Offered a Window for a Silent Spread of an Attenuated SARS-CoV-2 Precursor among Humans. J. Proteome Res. 2020, 19, 4543–4552. [Google Scholar] [CrossRef] [PubMed]
- Goh, G.K.; Dunker, A.K.; Foster, J.A.; Uversky, V.N. Nipah shell disorder, modes of infection, and virulence. Microb. Pathog. 2020, 141, 103976. [Google Scholar] [CrossRef] [PubMed]
- Goh, G.K.; Dunker, A.K.; Foster, J.A.; Uversky, V.N. Zika and Flavivirus Shell Disorder: Virulence and Fetal Morbidity. Biomolecules 2019, 9, 710. [Google Scholar] [CrossRef] [Green Version]
- Cohen, S.; Au, S.; Pante, N. How viruses access the nucleus. Biochim. Biophys. Acta 2011, 1813, 1634–1645. [Google Scholar] [CrossRef] [Green Version]
- Yarbrough, M.L.; Mata, M.A.; Sakthivel, R.; Fontoura, B.M. Viral subversion of nucleocytoplasmic trafficking. Traffic 2014, 15, 127–140. [Google Scholar] [CrossRef]
- Le Sage, V.; Mouland, A.J. Viral subversion of the nuclear pore complex. Viruses 2013, 5, 2019–2142. [Google Scholar] [CrossRef]
- Trotman, L.C.; Mosberger, N.; Fornerod, M.; Stidwill, R.P.; Greber, U.F. Import of adenovirus Dann involves the nuclear pore complex receptor CAN/Nup214 and histone H1. Nat. Cell Biol. 2001, 3, 1092–1100. [Google Scholar] [CrossRef]
- Cassany, A.; Ragues, J.; Guan, T.; Begu, D.; Wodrich, H.; Kann, M.; Nemerow, G.R.; Gerace, L. Nuclear import of adenovirus DNA involves direct interaction of hexon with an N-terminal domain of the nucleoporin Nup214. J. Virol. 2015, 89, 1719–1730. [Google Scholar] [CrossRef] [Green Version]
- Strunze, S.; Engelke, M.F.; Wang, I.H.; Puntener, D.; Boucke, K.; Schleich, S.; Way, M.; Schoenenberger, P.; Burckhardt, C.J.; Greber, U.F. Kinesin-1-mediated capsid disassembly and disruption of the nuclear pore complex promote virus infection. Cell Host Microbe 2011, 10, 210–223. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wodrich, H.; Cassany, A.; D’Angelo, M.A.; Guan, T.; Nemerow, G.; Gerace, L. Adenovirus core protein pVII is translocated into the nucleus by multiple import receptor pathways. J. Virol. 2006, 80, 9608–9618. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Jesus-Gonzalez, L.A.; Cervantes-Salazar, M.; Reyes-Ruiz, J.M.; Osuna-Ramos, J.F.; Farfan-Morales, C.N.; Palacios-Rapalo, S.N.; Perez-Olais, J.H.; Cordero-Rivera, C.D.; Hurtado-Monzon, A.M.; Ruiz-Jimenez, F.; et al. The Nuclear Pore Complex: A Target for NS3 Protease of Dengue and Zika Viruses. Viruses 2020, 12, 583. [Google Scholar] [CrossRef] [PubMed]
- Watters, K.; Palmenberg, A.C. Differential processing of nuclear pore complex proteins by rhinovirus 2A proteases from different species and serotypes. J. Virol. 2011, 85, 10874–10883. [Google Scholar] [CrossRef] [Green Version]
- Park, Y.C.; Burkitt, V.; Villa, A.R.; Tong, L.; Wu, H. Structural basis for self-association and receptor recognition of human TRAF2. Nature 1999, 398, 533–538. [Google Scholar] [CrossRef]
- Park, N.; Skern, T.; Gustin, K.E. Specific cleavage of the nuclear pore complex protein Nup62 by a viral protease. J. Biol. Chem. 2010, 285, 28796–28805. [Google Scholar] [CrossRef] [Green Version]
- Park, N.; Katikaneni, P.; Skern, T.; Gustin, K.E. Differential targeting of nuclear pore complex proteins in poliovirus-infected cells. J. Virol. 2008, 82, 1647–1655. [Google Scholar] [CrossRef] [Green Version]
- Ghildyal, R.; Jordan, B.; Li, D.; Dagher, H.; Bardin, P.G.; Gern, J.E.; Jans, D.A. Rhinovirus 3C protease can localize in the nucleus and alter active and passive nucleocytoplasmic transport. J. Virol. 2009, 83, 7349–7352. [Google Scholar] [CrossRef] [Green Version]
- Gustin, K.E.; Sarnow, P. Inhibition of nuclear import and alteration of nuclear pore complex composition by rhinovirus. J. Virol. 2002, 76, 8787–8796. [Google Scholar] [CrossRef] [Green Version]
- Gustin, K.E.; Sarnow, P. Effects of poliovirus infection on nucleo-cytoplasmic trafficking and nuclear pore complex composition. EMBO J. 2001, 20, 240–249. [Google Scholar] [CrossRef] [Green Version]
- Castello, A.; Izquierdo, J.M.; Welnowska, E.; Carrasco, L. RNA nuclear export is blocked by poliovirus 2A protease and is concomitant with nucleoporin cleavage. J. Cell Sci. 2009, 122, 3799–3809. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, N.; Schweers, N.J.; Gustin, K.E. Selective Removal of FG Repeat Domains from the Nuclear Pore Complex by Enterovirus 2A(pro). J. Virol. 2015, 89, 11069–11079. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Belov, G.A.; Lidsky, P.V.; Mikitas, O.V.; Egger, D.; Lukyanov, K.A.; Bienz, K.; Agol, V.I. Bidirectional increase in permeability of nuclear envelope upon poliovirus infection and accompanying alterations of nuclear pores. J. Virol. 2004, 78, 10166–10177. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oberste, M.S.; Gotuzzo, E.; Blair, P.; Nix, W.A.; Ksiazek, T.G.; Comer, J.A.; Rollin, P.; Goldsmith, C.S.; Olson, J.; Kochel, T.J. Human febrile illness caused by encephalomyocarditis virus infection, Peru. Emerg. Infect. Dis. 2009, 15, 640–646. [Google Scholar] [CrossRef] [PubMed]
- Czechowicz, J.; Huaman, J.L.; Forshey, B.M.; Morrison, A.C.; Castillo, R.; Huaman, A.; Caceda, R.; Eza, D.; Rocha, C.; Blair, P.J.; et al. Prevalence and risk factors for encephalomyocarditis virus infection in Peru. Vector Borne Zoonotic Dis. 2011, 11, 367–374. [Google Scholar] [CrossRef]
- Porter, F.W.; Bochkov, Y.A.; Albee, A.J.; Wiese, C.; Palmenberg, A.C. A picornavirus protein interacts with Ran-GTPase and disrupts nucleocytoplasmic transport. Proc. Natl. Acad. Sci. USA 2006, 103, 12417–12422. [Google Scholar] [CrossRef] [Green Version]
- Porter, F.W.; Palmenberg, A.C. Leader-induced phosphorylation of nucleoporins correlates with nuclear trafficking inhibition by cardioviruses. J. Virol. 2009, 83, 1941–1951. [Google Scholar] [CrossRef] [Green Version]
- Bardina, M.V.; Lidsky, P.V.; Sheval, E.V.; Fominykh, K.V.; van Kuppeveld, F.J.; Polyakov, V.Y.; Agol, V.I. Mengovirus-induced rearrangement of the nuclear pore complex: Hijacking cellular phosphorylation machinery. J. Virol. 2009, 83, 3150–3161. [Google Scholar] [CrossRef] [Green Version]
- Ciomperlik, J.J.; Basta, H.A.; Palmenberg, A.C. Three cardiovirus Leader proteins equivalently inhibit four different nucleocytoplasmic trafficking pathways. Virology 2015, 484, 194–202. [Google Scholar] [CrossRef] [Green Version]
- Lidsky, P.V.; Hato, S.; Bardina, M.V.; Aminev, A.G.; Palmenberg, A.C.; Sheval, E.V.; Polyakov, V.Y.; van Kuppeveld, F.J.; Agol, V.I. Nucleocytoplasmic traffic disorder induced by cardioviruses. J. Virol. 2006, 80, 2705–2717. [Google Scholar] [CrossRef] [Green Version]
- Shindo, Y.; Iwamoto, K.; Mouri, K.; Hibino, K.; Tomita, M.; Kosako, H.; Sako, Y.; Takahashi, K. Conversion of graded phosphorylation into switch-like nuclear translocation via autoregulatory mechanisms in ERK signalling. Nat. Commun. 2016, 7, 10485. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kehlenbach, R.H.; Gerace, L. Phosphorylation of the nuclear transport machinery down-regulates nuclear protein import in vitro. J. Biol. Chem. 2000, 275, 17848–17856. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mishra, A.; Sipma, W.; Veenhoff, L.M.; Van der Giessen, E.; Onck, P.R. The Effect of FG-Nup Phosphorylation on NPC Selectivity: A One-Bead-Per-Amino-Acid Molecular Dynamics Study. Int. J. Mol. Sci. 2019, 20, 596. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Neufeldt, C.J.; Joyce, M.A.; Levin, A.; Steenbergen, R.H.; Pang, D.; Shields, J.; Tyrrell, D.L.; Wozniak, R.W. Hepatitis C virus-induced cytoplasmic organelles use the nuclear transport machinery to establish an environment conducive to virus replication. PLoS Pathog. 2013, 9, e1003744. [Google Scholar] [CrossRef] [Green Version]
- Levin, A.; Neufeldt, C.J.; Pang, D.; Wilson, K.; Loewen-Dobler, D.; Joyce, M.A.; Wozniak, R.W.; Tyrrell, D.L. Functional characterization of nuclear localization and export signals in hepatitis C virus proteins and their role in the membranous web. PLoS ONE 2014, 9, e114629. [Google Scholar] [CrossRef] [PubMed]
- Henderson, B.R.; Percipalle, P. Interactions between HIV Rev and nuclear import and export factors: The Rev nuclear localisation signal mediates specific binding to human importin-beta. J. Mol. Biol. 1997, 274, 693–707. [Google Scholar] [CrossRef]
- Wang, P.; Palese, P.; O’Neill, R.E. The NPI-1/NPI-3 (karyopherin alpha) binding site on the influenza a virus nucleoprotein NP is a nonconventional nuclear localization signal. J. Virol. 1997, 71, 1850–1856. [Google Scholar] [CrossRef] [Green Version]
- Weber, F.; Kochs, G.; Gruber, S.; Haller, O. A classical bipartite nuclear localization signal on Thogoto and influenza A virus nucleoproteins. Virology 1998, 250, 9–18. [Google Scholar] [CrossRef] [Green Version]
- Atkinson, S.C.; Audsley, M.D.; Lieu, K.G.; Marsh, G.A.; Thomas, D.R.; Heaton, S.M.; Paxman, J.J.; Wagstaff, K.M.; Buckle, A.M.; Moseley, G.W.; et al. Recognition by host nuclear transport proteins drives disorder-to-order transition in Hendra virus V. Sci. Rep. 2018, 8, 358. [Google Scholar] [CrossRef] [Green Version]
- Ghildyal, R.; Ho, A.; Dias, M.; Soegiyono, L.; Bardin, P.G.; Tran, K.C.; Teng, M.N.; Jans, D.A. The respiratory syncytial virus matrix protein possesses a Crm1-mediated nuclear export mechanism. J. Virol. 2009, 83, 5353–5362. [Google Scholar] [CrossRef] [Green Version]
- Huang, S.; Chen, J.; Chen, Q.; Wang, H.; Yao, Y.; Chen, J.; Chen, Z. A second CRM1-dependent nuclear export signal in the influenza A virus NS2 protein contributes to the nuclear export of viral ribonucleoproteins. J. Virol. 2013, 87, 767–778. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anderson, G.; Wang, R.; Bandyopadhyay, A.; Goodin, M. The Nucleocapsid Protein of Potato Yellow dwarf Virus: Protein Interactions and Nuclear Import Mediated by a Non-Canonical Nuclear Localization Signal. Front. Plant. Sci. 2012, 3, 14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fernandez, J.; Machado, A.K.; Lyonnais, S.; Chamontin, C.; Gartner, K.; Leger, T.; Henriquet, C.; Garcia, C.; Portilho, D.M.; Pugniere, M.; et al. Transportin-1 binds to the HIV-1 capsid via a nuclear localization signal and triggers uncoating. Nat. Microbiol. 2019, 4, 1840–1850. [Google Scholar] [CrossRef] [PubMed]
- Sparrer, K.M.; Pfaller, C.K.; Conzelmann, K.K. Measles virus C protein interferes with Beta interferon transcription in the nucleus. J. Virol. 2012, 86, 796–805. [Google Scholar] [CrossRef] [Green Version]
- Kobayashi, T.; Kamitani, W.; Zhang, G.; Watanabe, M.; Tomonaga, K.; Ikuta, K. Borna disease virus nucleoprotein requires both nuclear localization and export activities for viral nucleocytoplasmic shuttling. J. Virol. 2001, 75, 3404–3412. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, F.J.; Beeche, A.A.; Hunter, J.J.; Chin, D.J.; Hope, T.J. Characterization of the nuclear export signal of human T-cell lymphotropic virus type 1 Rex reveals that nuclear export is mediated by position-variable hydrophobic interactions. Mol. Cell Biol. 1996, 16, 5147–5155. [Google Scholar] [CrossRef] [Green Version]
- Siomi, H.; Shida, H.; Nam, S.H.; Nosaka, T.; Maki, M.; Hatanaka, M. Sequence requirements for nucleolar localization of human T cell leukemia virus type I pX protein, which regulates viral RNA processing. Cell 1988, 55, 197–209. [Google Scholar] [CrossRef]
- DiMattia, M.A.; Watts, N.R.; Stahl, S.J.; Rader, C.; Wingfield, P.T.; Stuart, D.I.; Steven, A.C.; Grimes, J.M. Implications of the HIV-1 Rev dimer structure at 3.2 A resolution for multimeric binding to the Rev response element. Proc. Natl. Acad. Sci. USA 2010, 107, 5810–5814. [Google Scholar] [CrossRef] [Green Version]
- Daugherty, M.D.; Liu, B.; Frankel, A.D. Structural basis for cooperative RNA binding and export complex assembly by HIV Rev. Nat. Struct. Mol. Biol. 2010, 17, 1337–1342. [Google Scholar] [CrossRef] [Green Version]
- Hutten, S.; Walde, S.; Spillner, C.; Hauber, J.; Kehlenbach, R.H. The nuclear pore component Nup358 promotes transportin-dependent nuclear import. J. Cell Sci. 2009, 122, 1100–1110. [Google Scholar] [CrossRef] [Green Version]
- Gu, L.; Tsuji, T.; Jarboui, M.A.; Yeo, G.P.; Sheehy, N.; Hall, W.W.; Gautier, V.W. Intermolecular masking of the HIV-1 Rev NLS by the cellular protein HIC: Novel insights into the regulation of Rev nuclear import. Retrovirology 2011, 8, 17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arnold, M.; Nath, A.; Hauber, J.; Kehlenbach, R.H. Multiple importins function as nuclear transport receptors for the Rev protein of human immunodeficiency virus type 1. J. Biol. Chem. 2006, 281, 20883–20890. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ye, X.; Gorin, A.; Frederick, R.; Hu, W.; Majumdar, A.; Xu, W.; McLendon, G.; Ellington, A.; Patel, D.J. RNA architecture dictates the conformations of a bound peptide. Chem. Biol. 1999, 6, 657–669. [Google Scholar] [CrossRef] [Green Version]
- Casu, F.; Duggan, B.M.; Hennig, M. The arginine-rich RNA-binding motif of HIV-1 Rev is intrinsically disordered and folds upon RRE binding. Biophys. J. 2013, 105, 1004–1017. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Daugherty, M.D.; Booth, D.S.; Jayaraman, B.; Cheng, Y.; Frankel, A.D. HIV Rev response element (RRE) directs assembly of the Rev homooligomer into discrete asymmetric complexes. Proc. Natl. Acad. Sci. USA 2010, 107, 12481–12486. [Google Scholar] [CrossRef] [PubMed]
- Auer, M.; Gremlich, H.U.; Seifert, J.M.; Daly, T.J.; Parslow, T.G.; Casari, G.; Gstach, H. Helix-loop-helix motif in HIV-1 Rev. Biochemistry 1994, 33, 2988–2996. [Google Scholar] [CrossRef] [PubMed]
- Behrens, R.T.; Aligeti, M.; Pocock, G.M.; Higgins, C.A.; Sherer, N.M. Nuclear Export Signal Masking Regulates HIV-1 Rev Trafficking and Viral RNA Nuclear Export. J. Virol. 2017, 91. [Google Scholar] [CrossRef] [Green Version]
- Fineberg, K.; Fineberg, T.; Graessmann, A.; Luedtke, N.W.; Tor, Y.; Lixin, R.; Jans, D.A.; Loyter, A. Inhibition of nuclear import mediated by the Rev-arginine rich motif by RNA molecules. Biochemistry 2003, 42, 2625–2633. [Google Scholar] [CrossRef]
- Malim, M.H.; Cullen, B.R. HIV-1 structural gene expression requires the binding of multiple Rev monomers to the viral RRE: Implications for HIV-1 latency. Cell 1991, 65, 241–248. [Google Scholar] [CrossRef]
- Faust, O.; Grunhaus, D.; Shimshon, O.; Yavin, E.; Friedler, A. Protein Regulation by Intrinsically Disordered Regions: A Role for Subdomains in the IDR of the HIV-1 Rev Protein. Chembiochem 2018, 19, 1618–1624. [Google Scholar] [CrossRef]
- Jayaraman, B.; Fernandes, J.D.; Yang, S.; Smith, C.; Frankel, A.D. Highly Mutable Linker Regions Regulate HIV-1 Rev Function and Stability. Sci. Rep. 2019, 9, 5139. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, R.; Matsuura, Y.; Suzuki, T.; Ando, A.; Chiba, J.; Harada, S.; Saito, I.; Miyamura, T. Nuclear localization of the truncated hepatitis C virus core protein with its hydrophobic C terminus deleted. J. Gen. Virol. 1995, 76 Pt 1, 53–61. [Google Scholar] [CrossRef]
- Song, J.; Nagano-Fujii, M.; Wang, F.; Florese, R.; Fujita, T.; Ishido, S.; Hotta, H. Nuclear localization and intramolecular cleavage of N-terminally deleted NS5A protein of hepatitis C virus. Virus Res. 2000, 69, 109–117. [Google Scholar] [CrossRef]
- Kunkel, M.; Watowich, S.J. Biophysical characterization of hepatitis C virus core protein: Implications for interactions within the virus and host. FEBS Lett. 2004, 557, 174–180. [Google Scholar] [CrossRef] [Green Version]
- Boulant, S.; Vanbelle, C.; Ebel, C.; Penin, F.; Lavergne, J.P. Hepatitis C virus core protein is a dimeric alpha-helical protein exhibiting membrane protein features. J. Virol. 2005, 79, 11353–11365. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Duvignaud, J.B.; Savard, C.; Fromentin, R.; Majeau, N.; Leclerc, D.; Gagne, S.M. Structure and dynamics of the N-terminal half of hepatitis C virus core protein: An intrinsically unstructured protein. Biochem. Biophys. Res. Commun. 2009, 378, 27–31. [Google Scholar] [CrossRef] [PubMed]
- Shaw, M.L.; Cardenas, W.B.; Zamarin, D.; Palese, P.; Basler, C.F. Nuclear localization of the Nipah virus W protein allows for inhibition of both virus- and toll-like receptor 3-triggered signaling pathways. J. Virol. 2005, 79, 6078–6088. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Audsley, M.D.; Jans, D.A.; Moseley, G.W. Nucleocytoplasmic trafficking of Nipah virus W protein involves multiple discrete interactions with the nuclear import and export machinery. Biochem. Biophys. Res. Commun. 2016, 479, 429–433. [Google Scholar] [CrossRef]
- Xiao, C.Y.; Hubner, S.; Elliot, R.M.; Caon, A.; Jans, D.A. A consensus cAMP-dependent protein kinase (PK-A) site in place of the CcN motif casein kinase II site simian virus 40 large T-antigen confers PK-A-mediated regulation of nuclear import. J. Biol. Chem. 1996, 271, 6451–6457. [Google Scholar] [CrossRef] [Green Version]
- Hubner, S.; Xiao, C.Y.; Jans, D.A. The protein kinase CK2 site (Ser111/112) enhances recognition of the simian virus 40 large T-antigen nuclear localization sequence by importin. J. Biol. Chem. 1997, 272, 17191–17195. [Google Scholar] [CrossRef] [Green Version]
- Yang, S.N.Y.; Atkinson, S.C.; Wang, C.; Lee, A.; Bogoyevitch, M.A.; Borg, N.A.; Jans, D.A. The broad spectrum antiviral ivermectin targets the host nuclear transport importin alpha/beta1 heterodimer. Antiviral Res. 2020, 177, 104760. [Google Scholar] [CrossRef] [PubMed]
- King, C.R.; Tessier, T.M.; Dodge, M.J.; Weinberg, J.B.; Mymryk, J.S. Inhibition of Human Adenovirus Replication by the Importin alpha/beta1 Nuclear Import Inhibitor Ivermectin. J. Virol. 2020, 94. [Google Scholar] [CrossRef] [PubMed]
- Kosyna, F.K.; Nagel, M.; Kluxen, L.; Kraushaar, K.; Depping, R. The importin alpha/beta-specific inhibitor Ivermectin affects HIF-dependent hypoxia response pathways. Biol. Chem. 2015, 396, 1357–1367. [Google Scholar] [CrossRef] [PubMed]
- Fraser, J.E.; Rawlinson, S.M.; Wang, C.; Jans, D.A.; Wagstaff, K.M. Investigating dengue virus nonstructural protein 5 (NS5) nuclear import. Methods Mol. Biol. 2014, 1138, 301–328. [Google Scholar] [CrossRef] [PubMed]
- Lv, C.; Liu, W.; Wang, B.; Dang, R.; Qiu, L.; Ren, J.; Yan, C.; Yang, Z.; Wang, X. Ivermectin inhibits DNA polymerase UL42 of pseudorabies virus entrance into the nucleus and proliferation of the virus in vitro and vivo. Antiviral Res. 2018, 159, 55–62. [Google Scholar] [CrossRef] [PubMed]
- Raza, S.; Shahin, F.; Zhai, W.; Li, H.; Alvisi, G.; Yang, K.; Chen, X.; Chen, Y.; Chen, J.; Hu, C.; et al. Ivermectin Inhibits Bovine Herpesvirus 1 DNA Polymerase Nuclear Import and Interferes with Viral Replication. Microorganisms 2020, 8, 409. [Google Scholar] [CrossRef] [Green Version]
- Mastrangelo, E.; Pezzullo, M.; De Burghgraeve, T.; Kaptein, S.; Pastorino, B.; Dallmeier, K.; de Lamballerie, X.; Neyts, J.; Hanson, A.M.; Frick, D.N.; et al. Ivermectin is a potent inhibitor of flavivirus replication specifically targeting NS3 helicase activity: New prospects for an old drug. J. Antimicrob. Chemother. 2012, 67, 1884–1894. [Google Scholar] [CrossRef] [Green Version]
- Lundberg, L.; Pinkham, C.; Baer, A.; Amaya, M.; Narayanan, A.; Wagstaff, K.M.; Jans, D.A.; Kehn-Hall, K. Nuclear import and export inhibitors alter capsid protein distribution in mammalian cells and reduce Venezuelan Equine Encephalitis Virus replication. Antiviral Res. 2013, 100, 662–672. [Google Scholar] [CrossRef]
- Barrows, N.J.; Campos, R.K.; Powell, S.T.; Prasanth, K.R.; Schott-Lerner, G.; Soto-Acosta, R.; Galarza-Munoz, G.; McGrath, E.L.; Urrabaz-Garza, R.; Gao, J.; et al. A Screen of FDA-Approved Drugs for Inhibitors of Zika Virus Infection. Cell Host Microbe 2016, 20, 259–270. [Google Scholar] [CrossRef] [Green Version]
- Wagstaff, K.M.; Sivakumaran, H.; Heaton, S.M.; Harrich, D.; Jans, D.A. Ivermectin is a specific inhibitor of importin alpha/beta-mediated nuclear import able to inhibit replication of HIV-1 and dengue virus. Biochem. J. 2012, 443, 851–856. [Google Scholar] [CrossRef] [Green Version]
- Fraser, J.E.; Watanabe, S.; Wang, C.; Chan, W.K.; Maher, B.; Lopez-Denman, A.; Hick, C.; Wagstaff, K.M.; Mackenzie, J.M.; Sexton, P.M.; et al. A nuclear transport inhibitor that modulates the unfolded protein response and provides in vivo protection against lethal dengue virus infection. J. Infect. Dis. 2014, 210, 1780–1791. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tay, M.Y.; Fraser, J.E.; Chan, W.K.; Moreland, N.J.; Rathore, A.P.; Wang, C.; Vasudevan, S.G.; Jans, D.A. Nuclear localization of dengue virus (DENV) 1-4 non-structural protein 5; protection against all 4 DENV serotypes by the inhibitor Ivermectin. Antiviral Res. 2013, 99, 301–306. [Google Scholar] [CrossRef] [PubMed]
- Gotz, V.; Magar, L.; Dornfeld, D.; Giese, S.; Pohlmann, A.; Hoper, D.; Kong, B.W.; Jans, D.A.; Beer, M.; Haller, O.; et al. Influenza A viruses escape from MxA restriction at the expense of efficient nuclear vRNP import. Sci. Rep. 2016, 6, 23138. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Caly, L.; Druce, J.D.; Catton, M.G.; Jans, D.A.; Wagstaff, K.M. The FDA-approved drug ivermectin inhibits the replication of SARS-CoV-2 in vitro. Antiviral Res. 2020, 178, 104787. [Google Scholar] [CrossRef] [PubMed]
- Ketkar, H.; Yang, L.; Wormser, G.P.; Wang, P. Lack of efficacy of ivermectin for prevention of a lethal Zika virus infection in a murine system. Diagn. Microbiol. Infect. Dis. 2019, 95, 38–40. [Google Scholar] [CrossRef]
- Hintersteiner, M.; Ambrus, G.; Bednenko, J.; Schmied, M.; Knox, A.J.; Meisner, N.C.; Gstach, H.; Seifert, J.M.; Singer, E.L.; Gerace, L.; et al. Identification of a small molecule inhibitor of importin beta mediated nuclear import by confocal on-bead screening of tagged one-bead one-compound libraries. ACS Chem. Biol. 2010, 5, 967–979. [Google Scholar] [CrossRef] [Green Version]
- Soderholm, J.F.; Bird, S.L.; Kalab, P.; Sampathkumar, Y.; Hasegawa, K.; Uehara-Bingen, M.; Weis, K.; Heald, R. Importazole, a small molecule inhibitor of the transport receptor importin-beta. ACS Chem. Biol. 2011, 6, 700–708. [Google Scholar] [CrossRef] [Green Version]
- Sun, Q.; Carrasco, Y.P.; Hu, Y.; Guo, X.; Mirzaei, H.; Macmillan, J.; Chook, Y.M. Nuclear export inhibition through covalent conjugation and hydrolysis of Leptomycin B by CRM1. Proc. Natl. Acad. Sci. USA 2013, 110, 1303–1308. [Google Scholar] [CrossRef] [Green Version]
- Wolff, B.; Sanglier, J.J.; Wang, Y. Leptomycin B is an inhibitor of nuclear export: Inhibition of nucleo-cytoplasmic translocation of the human immunodeficiency virus type 1 (HIV-1) Rev protein and Rev-dependent mRNA. Chem. Biol. 1997, 4, 139–147. [Google Scholar] [CrossRef] [Green Version]
- Elton, D.; Simpson-Holley, M.; Archer, K.; Medcalf, L.; Hallam, R.; McCauley, J.; Digard, P. Interaction of the influenza virus nucleoprotein with the cellular CRM1-mediated nuclear export pathway. J. Virol. 2001, 75, 408–419. [Google Scholar] [CrossRef] [Green Version]
- Newlands, E.S.; Rustin, G.J.; Brampton, M.H. Phase I trial of elactocin. Br. J. Cancer 1996, 74, 648–649. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hing, Z.A.; Fung, H.Y.; Ranganathan, P.; Mitchell, S.; El-Gamal, D.; Woyach, J.A.; Williams, K.; Goettl, V.M.; Smith, J.; Yu, X.; et al. Next-generation XPO1 inhibitor shows improved efficacy and in vivo tolerability in hematological malignancies. Leukemia 2016, 30, 2364–2372. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Etchin, J.; Berezovskaya, A.; Conway, A.S.; Galinsky, I.A.; Stone, R.M.; Baloglu, E.; Senapedis, W.; Landesman, Y.; Kauffman, M.; Shacham, S.; et al. KPT-8602, a second-generation inhibitor of XPO1-mediated nuclear export, is well tolerated and highly active against AML blasts and leukemia-initiating cells. Leukemia 2017, 31, 143–150. [Google Scholar] [CrossRef] [PubMed]
- Neira, J.L.; Bintz, J.; Arruebo, M.; Rizzuti, B.; Bonacci, T.; Vega, S.; Lanas, A.; Velazquez-Campoy, A.; Iovanna, J.L.; Abian, O. Identification of a Drug Targeting an Intrinsically Disordered Protein Involved in Pancreatic Adenocarcinoma. Sci. Rep. 2017, 7, 39732. [Google Scholar] [CrossRef] [PubMed]
- Iconaru, L.I.; Ban, D.; Bharatham, K.; Ramanathan, A.; Zhang, W.; Shelat, A.A.; Zuo, J.; Kriwacki, R.W. Discovery of Small Molecules that Inhibit the Disordered Protein, p27(Kip1). Sci. Rep. 2015, 5, 15686. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hammoudeh, D.I.; Follis, A.V.; Prochownik, E.V.; Metallo, S.J. Multiple independent binding sites for small-molecule inhibitors on the oncoprotein c-Myc. J. Am. Chem. Soc. 2009, 131, 7390–7401. [Google Scholar] [CrossRef] [PubMed]
- Follis, A.V.; Hammoudeh, D.I.; Wang, H.; Prochownik, E.V.; Metallo, S.J. Structural rationale for the coupled binding and unfolding of the c-Myc oncoprotein by small molecules. Chem. Biol. 2008, 15, 1149–1155. [Google Scholar] [CrossRef] [Green Version]


| Virus | Protein | NLS/NES | Sequence | Ref. |
|---|---|---|---|---|
| PYDV 1 | M | NLS | 213-KKTVSDPLKNLLKR-226 | [133] |
| HIV-1 2 | Capsid | NLS | 84-HPVHAGPIAPGQMREPRGSDIA-105 | [134] |
| Measles | C | NLS | 41-PPARKRRQ-48 | [135] |
| HCV 3 | Core | NLS1 | 6-KPQRKTKRN-14 | [126] |
| HCV | Core | NLS2 | 38-PRRGPR-43 | [126] |
| HCV | Core | NLS3 | 58-PRGRRQPIPKARRS-71 | [126] |
| BDV 4 | N | NLS | 3-PKRRLVDDT-11 | [136] |
| HTLV-1 5 | Rex | NES | 81-ALSAQLYSSLSLDSP-95 | [137] |
| HTLV-1 | Rex | NLS | 1-MPKTRRRPRRSQRKRPPTP-19 | [138] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wubben, J.M.; Atkinson, S.C.; Borg, N.A. The Role of Protein Disorder in Nuclear Transport and in Its Subversion by Viruses. Cells 2020, 9, 2654. https://doi.org/10.3390/cells9122654
Wubben JM, Atkinson SC, Borg NA. The Role of Protein Disorder in Nuclear Transport and in Its Subversion by Viruses. Cells. 2020; 9(12):2654. https://doi.org/10.3390/cells9122654
Chicago/Turabian StyleWubben, Jacinta M., Sarah C. Atkinson, and Natalie A. Borg. 2020. "The Role of Protein Disorder in Nuclear Transport and in Its Subversion by Viruses" Cells 9, no. 12: 2654. https://doi.org/10.3390/cells9122654
APA StyleWubben, J. M., Atkinson, S. C., & Borg, N. A. (2020). The Role of Protein Disorder in Nuclear Transport and in Its Subversion by Viruses. Cells, 9(12), 2654. https://doi.org/10.3390/cells9122654






